Abstract
The IFN-γ-inducible chemokines CXCL9 and CXCL10 are implicated in the pathogenesis of T-cell-mediated immunity in the CNS. However, in various CNS immune pathologies the cellular localization of these chemokines differs with CXCL9 produced by macrophage/microglia while CXCL10 is produced by both macrophage/microglia and astrocytes. Here we determined the mechanism for the microglial cell-restricted expression of the Cxcl9 gene induced by IFN-γ. In cultured glial cells the induction of the CXCL9 (in microglia) and CXCL10 (in microglia and astrocytes) mRNAs by IFN-γ was not inhibited by cycloheximide. Of various transcription factors involved with IFN-γ-mediated gene regulation, PU.1 was identified as a constitutively expressed nuclear factor in microglia but not in astrocytes. STAT1 and PU.1 bound constitutively to the Cxcl9 gene promoter in microglia and this increased significantly following IFN-γ-treatment with IRF-8 identified as an additional late binding factor. However in astrocytes, STAT1 alone bound to the Cxcl9 gene promoter. STAT-1 was critical for IFN-γ-induction of both the Cxcl9 and Cxcl10 genes in microglia and in microglia and astrocytes, respectively. The siRNA-mediated knockdown of PU.1 in microglia markedly impaired IFN-γ-induced CXCL9 but not STAT1 or IRF-8. Cells of the D1A astrocyte line showed partial reprogramming to a myeloid-like phenotype after transduction with PU.1 and, in addition to the expression of CD11b, acquired the ability to produce CXCL9 in response to IFN-γ. Thus, PU.1 not only is crucial for the induction of CXCL9 by IFN-γ in microglia but also is a key determinant factor for the cell-specific expression of this chemokine by these myeloid cells.
Keywords: astrocyte, microglia, IFN-γ, CXCL9, PU.1
Introduction
Three structurally related chemokines known as CXCL9 (also known as monokine induced by gamma or MIG) (1), CXCL10 (also known as interferon inducible protein 10 or IP-10) (2, 3) and CXCL11 (also known as IFN-gamma-inducible T cell alpha chemoattractant or ITAC) (4, 5) bind the common receptor CXCR3 (6) and comprise the IFN-γ-inducible non-ELR CXC chemokine subgroup. These three chemokines can stimulate the chemotaxis of activated CXCR3-positive T-cells and NK cells in vitro (4, 6–9). Additionally, CXCL9 and CXCL10 are reported to be pivotal for T-cell migration in various experimental disease models including transplant rejection (10–12) and infectious and chronic inflammatory diseases (13–15). Thus, the CXCR3 ligands appear to be fundamental for T-and NK-cell trafficking in cell-mediated immunity.
As the original functional names suggest, CXCL9, CXCL10 and CXCL11 were all identified based on the common property that their genes were induced or upregulated in cells exposed to IFN-γ (1, 2, 4). Following the binding of IFN-γ to its receptor the receptor-associated tyrosine kinases JAK1 and JAK2 are activated by tyrosine phosphorylation. The latent cytoplasmic transcription factor STAT1 is subsequently recruited to the receptor and activated via tyrosine phosphorylation mediated by the JAKS. After disengaging from the receptor, activated STAT1 molecules form homodimers, that translocate to the nucleus and bind to the gamma activated sequence (GAS) element to modulate the transcription of IFN-γ-responsive genes (reviewed in (16)). In addition to STAT1, the transcription factors STAT2 (17), IRF-1 (18), IRF-4, IRF-8 and PU.1 (19) and CIITA (20) have been shown to be positive regulators of IFN-γ-modulated gene transcription, while the transcription factors STAT3 (21), IRF-2 (18), IRF-8 (22), PML (23) and TEL (22) have been shown to act as negative regulators of gene expression in response to IFN-γ. The induction of IFN-γ-dependent genes can be mediated by the interaction of certain IRFs (e.g. IRF-8 (19)) with members of the Ets transcription factor family. Two specific members of this family, PU.1 and TEL, have been reported to play a role in immune cell specific signaling by IFN-γ and are capable of interacting with IRF-8 and IRF-4 (19).
We have proposed an important role for CXCL9 and CXCL10 in the positioning of mononuclear leukocytes in the CNS during experimental autoimmune encephalomyelitis (EAE), corralling these cells to the perivascular space (24, 25). This function is supported by the unique spatial production of CXCL9 and CXCL10, with CXCL9 being predominantly localized to the lesion-associated microglia/macrophage populations, while CXCL10 is found largely in astrocytes surrounding the perivascular mononuclear cell infiltrates. These studies identified IFN-γ as the principal factor responsible for the induction of the gene for CXCL9 in microglia and CXCL10 in microglia and astrocytes, in vitro and in vivo (24). Similarly, in other models of CNS inflammation including Toxoplasma encephalitis (26), lymphocytic choriomeningitis (LCM) virus-induced neurological disease (27) and cerebral malaria (28) the expression of the gene for CXCL9 is found in microglia but not astrocytes and this is critically dependent on IFN-γ. These observations raised the question as to how the common stimulating factor IFN-γ could differentially regulate the expression of these chemokine genes in the glial cells of the CNS. We theorized that a variety of different transcription factors might be involved in the differential activation of the genes for CXCL9 and CXCL10 mediated by IFN-γ. Therefore, the major objective of this study was to determine the mechanisms responsible for the IFN-γ-mediated, cell-specific induction of the gene for CXCL9 in microglia.
Materials and Methods
Animals
Wild type mice (C57BL/6 strain) were obtained from the Animal Resources Centre, Canning Vale, WA, Australia. STAT1−/− mice (C57BL/6 strain) (29) were kindly provided by Dr. Joan Durbin (Center for Vaccines and Immunity, The Research Institute at Nationwide Children’s Hospital, Ohio State University, Columbus OH) and a local breeding colony maintained at the University of Sydney. Animals were kept under pathogen free conditions in the animal facility at the University of Sydney. Ethical approval for the use of all mice in this study was obtained from the University of Sydney Animal Care and Ethics Committee.
Induction of EAE
EAE was induced in C57BL/6 mice as described previously (24) and the brain and spinal cord removed for histological processing and analysis as described below.
Glial Cell Culture
Primary mixed glial cell cultures, primary microglial cultures and primary astrocyte cultures were prepared as described previously (24). The degree of microglial contamination of primary astrocyte cultures was determined by flow cytometry using phycoerythrin-conjugated antibody against CD11b (BD Biosciences, North Ryde, NSW) as described below and was routinely <0.1%. The microglial cell line EOC-13 (30) was obtained from the American Type Culture Collection (ATCC number CRL-2468). The EOC-13 cells were maintained in DMEM supplemented with 10% FBS and 20% conditioned media from LADMAC cells (ATCC number CRL-2430) as a source of CSF-1. Prior to treatment, EOC-13 cells were cultured for 72 h without CSF-1 to induce a resting state. Prior to treatment of cultures, growth medium was removed and cells washed once with phosphate-buffered saline (PBS). Murine recombinant IFN-γ (1000 U/ml; Sigma-Aldrich, Castle Hill, NSW) diluted in DMEM/FBS media was added to the different cell cultures for the specified times. In some experiments, cycloheximide (20 μg/ml; Sigma-Aldrich) was added 2 h prior to IFN-γ to inhibit protein synthesis. Cells were washed with PBS before total RNA was extracted with TriReagent (Sigma-Aldrich) according to the Manufacturer’s protocol.
RNase Protection Assay (RPA) Plasmid Constructs
The construction and characterization of the RPA plasmids used as probes for CXCL9, CXCL10, STAT1 and RPL32 were reported previously (31, 32). New probes were synthesized for IRF-8, IRF-4, IRF-1, CIITA and PU.1. For all probes the sub-cloning of the specific cDNA sequences was performed by PCR-assisted directional cloning as described previously (33). Briefly, cDNA was reverse transcribed from 10 μg total RNA (isolated from spleen of mice) by reverse transcriptase-polymerase chain reaction (RT-PCR) using oligo(dT) primers12-18 (Invitrogen, Rowville, VIC) and 1 μl of Superscript II RT (0.5 μg/μl) (Invitrogen) as recommended by the Manufacturer. The synthesized cDNA was then used as a template and subjected to amplification by PCR from the specific oligonucleotide sequences shown in Table I. Each cDNA product was then directionally cloned into pGEM-4Z plasmid (Promega, Madison, WI) that incorporate T7 and SP6 RNA polymerase promoters flanking the cloning region defined by the EcoRI or HindIII restriction enzyme sites. The presence of the correct insert sequence was confirmed by sequencing analysis provided by SUPAMAC (Sydney University Prince Alfred Macromolecular Analysis Centre, Sydney, Australia). Digestion of the transformed plasmids with either EcoRI or HindIII, transcription with T7 or SP6 RNA polymerase yielded anti-sense (T7) or sense (SP6) RNA probes, respectively. The genomic clone RPL32-4A served as a probe for the ribosomal protein L32. This was included as an internal control for RNA loading during RPA analysis.
Table I.
Primer sequences used to generate RPA probes
| Target | Upstream Primer | Downstream Primer |
|---|---|---|
| CIITA | CCAGAGGCTGAGAAACCCCTCAGA | GTTCGAACGTCCGTGGGTGTAGTCACTACA |
| IRF-1 | AATGAATTCCTTCAGCTGCAAAGAGGAACCAGA | ATAAAGCTTAGGCTGTCCATCCACATGATGGAG |
| IRF-4 | AATGAATTCCTGGATGGCTCCAGATGGGCTTTA | AATAAGCTTTGATTTGTTGAGCAAAGTAATACA |
| IRF-8 | AATGAATTCGGCTGTGCCAGGGCCGCGTGTTCT | AATAAGCTTCCGGAAACATGCGGAAAGCCTGGT |
| PU.1 | AATGAATTCACAACAACGAGTTTGAGAACTTCC | AATAAGCTTCAAGGTTTGATAAGGGAAGCACAT |
RNase Protection Assay
RPA was performed as described previously (33). The RNA samples (3 μg of total RNA) were hybridized with [32P]UTP-labeled probe sets (containing cRNA probes for CXCL9, CXCL10, IRF-1, IRF-4, IRF-8, CIITA, STAT1, PU.1 and L32). For quantification, autoradiographs were scanned and analyzed by densitometry using NIH Image J software v1.38 (Wayne Rasband, National Institutes of Health, USA). The densitometric value for each transcript was expressed as a ratio to the L32 RNA, which served as a control for RNA loading.
Immunoblot Analysis
Protein lysates were prepared by lysing cells in RIPA buffer containing 100 mM Tris PH 7.6, 150 mM NaCl, 1 mM EDTA, 1% deoxcyholic acid, 1% Triton X-100, 0.1% SDS, 20 mM PMSF, 50 mM NaF, protease and phosphatase inhibitors (Invitrogen). Protein concentration of the samples was estimated by BCA Assay (Pierce, Paddington, QLD). Between 25 and 50 μg of protein was separated on a precast NuPAGE gradient gel (4 – 12%) (Invitrogen) by electrophoresis at 40 mA. Following electrotransfer to a PVDF membrane, membranes were blocked with 5% milk in Tris-buffered saline plus 1% Tween 20 (TBS-T) with 50mM NaF. Membranes were then incubated with primary antibody (Table II) overnight at 4°C, followed by washing in TBS-T, and then 1 h incubation with peroxidase-coupled secondary antibody (Sigma-Aldrich). Protein signals were detected by ECL reagent (Pierce, Scoresby, VIC) and visualized on Hyperfilm (Amersham, Sydney, Australia).
Table II.
Details of antibodies used in this study
| Antigen | Species | Antibody | Cat. | Manufacturer | Dilution* |
|---|---|---|---|---|---|
| p-STAT1 Y701 |
Mouse | Rb Polyclonal | 9171 | Cell Signaling | IB: 1:1000 |
| STAT1 | Mouse | Rb Polyclonal | 9172 | Cell Signaling | IB: 1:1000 EMSA: 1 μg |
| STAT1 CT | Mouse | Rb Polyclonal | 06-501 | Upstate | ChIP: 4μg |
| PU.1 | Mouse | Rb Monoclonal | 2258 | Cell Signaling | IB: 1:1000 IC-IF: 1:200 P-IHC: Citrate 1:100 |
| PU.1 | Mouse | Gt Polyclonal | Sc-5949 | Santa Cruz | ChIP: 4 μg EMSA: 1 μg |
| IRF-1 | Mouse | Rb Polyclonal | 4966 | Cell Signaling | IB: 1:1000 |
| IRF-8 | Mouse | Gt Polyclonal | Sc-6058 | Santa Cruz Biotechnology |
IB: 1:500 ChIP: 4μg |
| GFAP | Cow | Rb Polyclonal | Z0334 | Dakocytomation | P-IHC: 1:1000 |
| GFAP | Mouse | Ms Monoclonal | G3893 | Sigma-Aldrich | P-IHC: 1:1000 |
| CD11b-APC | Mouse | Rt Monoclonal | 553312 | BD Biosciences | FACS: 1:250 |
| F4/80 | Mouse | Rt Monoclonal | MCA-497 | Serotec | IF: 1:200 |
| GAPDH | Mouse | Ms Monoclonal | MAB374 | Chemicon | IB: 1:20 000 |
IB: Immunoblot; IC-IF: Immunoctyochemistry-Immunofluorescence; P-IHC: Paraffin Immunohistochemistry ChIP: Chromatin Immunoprecipitation; FACS: Fluorescent Activated Cell Sorting; EMSA: Electrophoretic Mobility Shift Assay
Tissue Processing and Dual Label Paraffin Immunohistochemistry
Brains removed from mice for immunohistochemical examination were placed immediately in ice-cold PBS-buffered 4% paraformaldehyde (pH 7.4; Sigma-Aldrich) and stored for 24 h at 4°C before processing and embedding in paraffin. Sections (5 μm) were deparaffinized in xylene and rehydrated through a series of graded ethanol. For antigen unmasking, slides were boiled in 10 mM sodium citrate (pH 6.0) and then maintained at 95°C for 15 min. Slides were then cooled at RT for 30 min and washed in dH2O. Endogenous peroxidase activity was inhibited by incubating sections in 3% H2O. Sections were then washed in TBS-T and sections were subsequently blocked for 30 min (TBS-T + 5% normal horse serum (Vector Laboratories, Brisbane, QLD). Sections were incubated overnight at 4°C with diluted primary antibody (Table II). After washing in TBS-T, biotinylated secondary antibody (Vector Laboratories) in TBS-T was added for 1 hour, and following washing, HRP-coupled streptavidin (Vector Laboratories) was applied for 30 min. DAB color reagent (Vector Laboratories) with nickel was applied as the immunoperoxidase substrate according to the Manufacturer’s instructions to visualize specific nuclear signals. Sections were then washed with dH2O, and prior to the second antigen staining, sections were incubated for 15 min with avidin and then biotin (Vector Laboratories) to prevent interaction of the second set of labeling reagents with the first. Slides were subsequently stained for the second antigen following the same procedure as detailed above. Specific signals for the second antigen were visualized with NovaRed color reagent (Vector Laboratories).
Immunocytochemistry
Mixed glial cells or EOC-13 cells were plated on autoclaved coverslips in 24-well plates and grown for 24 hours. Cells were treated with IFN-γ (1000 U/ml) for the times specified, or left untreated as controls. Cells were washed with PBS, fixed for 15 min with 4% PBS-buffered paraformaldehyde (pH 7.2) and washed again and subsequently fixed in ice-cold (−20°C) methanol for 10 minutes. After rinsing in PBS, cells were then blocked in 10% BSA + 0.01% Triton-X for 30 min, incubated with primary antibody (Table II) for 1 hour, followed by washing with PBS and then incubated with fluorescent conjugated secondary antibody (Alexa 488 or Alexa 594, Invitrogen). Coverslips were then placed on glass slides with mounting medium containing DAPI to visualize nuclei (Vector). Dual-label staining on cells was performed as described above for single staining except that a mixture of two primary primary antibodies was applied. Subsequently, after washing, a cocktail of fluorescent conjugated secondary antibodies was applied as appropriate and the remaining steps performed as described above.
RNA Interference
DharmaFECT® 1 siRNA transfection reagent, SMARTpool small interfering RNA (siRNAs) specific for murine PU.1 (Spi-1) and siGENOME RISC-free control siRNA were purchased from Dharmacon (Lafayette, CO). EOC-13 (5 × 105) cells in six-well plates were transfected with 100 nM of PU.1 siRNAs or control siRNAs using DharmaFECT® 1 (3 μl/well) according to the manufacturer’s instructions. After 36 h, EOC-13 cells were treated with IFN-γ (1000 U/ml) for 24 h or left untreated. Supernatants were collected and CXCL9 protein production was analyzed by ELISA. Protein lysates were prepared and immunoblotting was performed as described above to detect PU.1, STAT1 or IRF-8 protein. Total RNA was extracted and RNA levels were assessed by RPA as described above.
ELISA
Cell supernatants were assayed for CXCL9 and CXCL10 production by ELISA. Quantikine ELISA kits for CXCL9 and CXCL10 were obtained from R&D Systems (Kirrawee, NSW) and used according to the Manufacturer’s instructions. A standard curve was generated with each assay with the limit of detection for CXCL9 = 3 pg/ml and for CXCL10 = 2 pg/ml. All samples were measured in duplicate.
Chromatin Immunoprecipitation
Cells were cultured in T75 flasks and stimulated with IFN-γ (1000U/ml) for the specified times in 10 ml DMEM with 1% FBS. Cells were fixed by the addition of 37% formaldehyde to the medium to a final concentration of 1% and incubated at RT for 10 min. Cells were rinsed with ice cold PBS and scraped off the plates and centrifuged (200 × g; 5 min). The cell pellet was re-suspended in hypotonic buffer (10 mM KCl, 20 mM HEPES, 1 mM MgCl2, 1 mM DTT) and incubated for 5 min on ice before the addition of 10% NP-40 (Sigma-Aldrich) to a final concentration of 0.5% and incubated for a further 5 min. After centrifugation (200 × g; 5 min) the disrupted nuclei were resuspended in TE (pH 8.0). Sonication was performed using a Bioruptor (Cosmo Bio Co Ltd, Tokyo, JP). Samples were then centrifuged (1800 × g; 15 min) and chromatin-containing supernatant was snap frozen and stored at −80C for subsequent analysis.
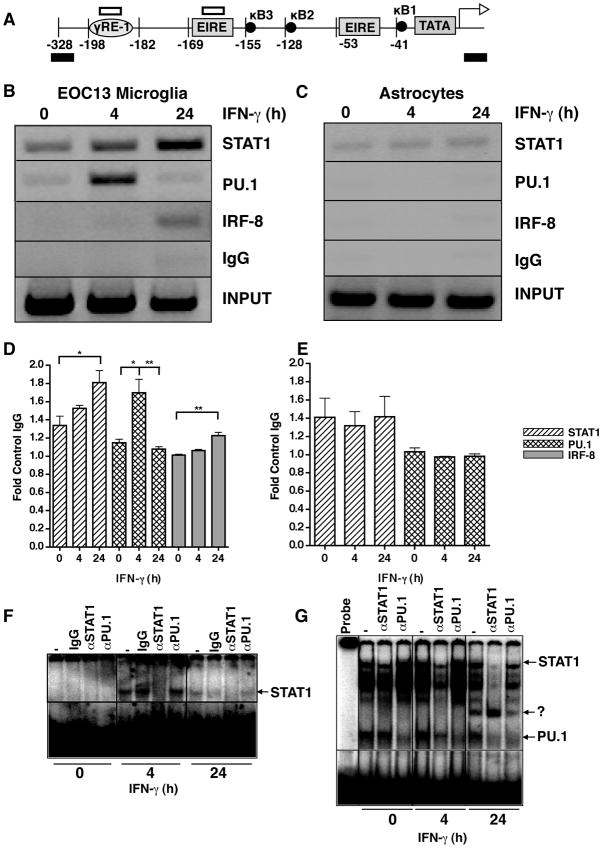
Equal amounts of DNA from supernatants were re-suspended to 500 μl with 2x RIPA buffer (2.0% NP-40, 1.0% sodium deoxycholate, 0.2% SDS, PBS). Samples were precleared with 40 μl salmon sperm DNA/protein A agarose beads (50% slurry; Upstate Biotechnology, Murarrie, QLD). Beads were pelleted and precleared supernatant was immunoprecipitated overnight at 4°C with 4 μg of either anti-STAT1 (Upstate), anti-PU.1 or anti-IRF-8 (both from Santa Cruz Biotechnology, Santa Cruz, CA) antibody. In each experiment, one sample with an irrelevant antibody (anti-IgG) was included. Immune complexes were recovered by adding 60 μl of salmon sperm DNA/protein A agarose beads (50% slurry; Upstate) and rotating at 4°C for a minimum of 2h. Extensive washing with low salt buffer (0.1% SDS, 1.0% Triton-X-100, 2 mM EDTA, 20 mM Tris HCl pH 8.0, 150 mM NaCl), high salt buffer (0.1% SDS, 1.0% Triton-X-100, 2 mM EDTA, 20 mM Tris HCl pH 8.0, 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1.0% NP-40, 1.0% Na-DC, 1 mM EDTA, 10 mM Tris HCl pH 8.0), and TE buffer was performed. Immunoprecipitated DNA was eluted from the beads by rotating with elution buffer (1% SDS, 0.1 M NaHCO3) for 30 min. One sample that did not undergo immunoprecipitation was saved as input control. DNA-protein crosslinks were reversed in immunoprecipitated samples and input controls by addition of 0.2 M NaCl and incubation at 65°C overnight. The remaining protein was digested by shaking in PK buffer (20 μg PK, 0.63 M Tris HCl, pH 6.5, 156 mM EDTA) for 2h at 37°C. The DNA was then purified using spin columns (Qiagen, Doncaster, VIC) and re-suspended in 50 μl H2O. An aliquot (5 μl) of IP DNA or input control (1 μl) were then subjected to PCR analysis using primers directed against the Cxcl9 gene promoter (Figure 4A; upstream - 5′ AGCTTTGACTTGTGAGGAAAGG 3′; downstream - 5′ TATTGAGTCACTGTGTTGGAGTTGA 3′).
Figure 4.
Interaction of STAT1, PU.1 and IRF-8 with the Cxcl9 gene promoter in microglia and astrocytes. A schematic illustration of the murine Cxcl9 gene promoter (A) showing the location of the primer sites for ChIP analysis (black bars) and the sites corresponding to the oligonucleotide probe used for EMSA (open bars). ChIP analysis was performed on EOC microglia (B) or astrocytes (C). Cells were incubated in the absence or presence of IFN-γ (1000U/ml) for 0, 4 or 24 hours, and cross-linked with formaldehyde and soluble chromatin was subjected to immunoprecipitation with antibodies against STAT1, PU.1, IRF-8 or normal IgG as described in the Materials and Methods. Representative PCR images taken from 1 of 3 independent experiments is shown for EOC-13 cells (A) and astrocytes (B). Fold-change as compared with IgG was quantified for each transcription factor binding for EOC13 cells (D) and astrocytes (E). For statistical significance: *p<0.05, **p<0.01, ***p<0.001. Binding activity to γ-RE (F) or EIRE1 (G) radiolabeled oligonucleotides with nuclear extracts (5 μg) from EOC-13 cells treated with IFN-γ for the times shown and incubated with the indicated antibodies was performed as described in Materials and Methods.
Electrophoretic Mobility Shift Assay (EMSA)
For the preparation of nuclear extracts, cells were washed in 10ml cold PBS, re-suspended in 500μl Buffer A (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF, protease inhibitors (Invitrogen)) and incubated on ice for 15 min. Following this, NP-40 was added to give a final concentration of 0.5% v/v and the cells vortexed for 10 sec. Nuclear material was obtained by centrifugation at 3,800 × g for 20 sec, followed by addition of 150μl Buffer C (20 mM HEPES, 1.5 mM MgCl2, 420 mM NaCl, 02 mM EDTA, 25% (v/v) Glycerol, 0.5 mM PMSF, protease inhibitors) to re-suspend the nuclear pellet. This solution was subjected to vigorous agitation 4°C for 30 min, followed by centrifugation at 4°C and 8,200 × g for 10 min. Nuclear extracts (supernatants) were recovered and protein concentration was determined by Bradford assay according to the manufacturer’s instructions (Biorad).
Radiolabelled double-stranded DNA probes were prepared using oligonucleotides (Sigma-Aldrich) with sequences corresponding to the γ-RE1 (5′ CCTTACTATAAACTCC 3′) and EIRE1 (5′ ATGGAAGTAGAACATGCAGAAATTC 3′) sites of the Cxcl9 gene promoter (Figure 4A). Radiolabelled sense oligonucleotides were prepared by adding 3 μl [32P]-ATP, 1 μL 10 × PNK buffer, 1 μl T4 Polynucleotide kinase and 4μl H20 to 1 μl of each oligonucleotide (100 ng/μl) and then incubating the reaction mixture at 37°C for 30 min. TNE buffer (40 μl;10 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA pH 8.0) was added and the labelled probe denatured by boiling for 1 min at 98°C. Following this, the corresponding complimentary oligonucleotide (4 μl) was added for annealing and the reaction mixture allowed to cool slowly to RT. The labelled probe was purified on a G25 Column Matrix (GE Healthcare, Rydalmere, NSW) according to the Manufacturer’s protocol and stored at −20°C prior to use. EMSA was performed with 5 μg of nuclear extract in a total volume of 30μl of binding buffer (50 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 0.5 mM dithiothreitol, 4 mMTris-Cl (pH 7.5), 1μg polydeoxyinosinic-deoxycytidyl acid, and 0.5 μl 32P-labelled probe), and incubated at RT for 15 min. Bound and free DNA were then resolved by electrophoresis through a 6% polyacrylamide gel (40% Acrylamide/Bis-Acrylamide (19:1)) (Sigma-Aldrich) in 0.5x TBE Buffer at 250 V for 1.75 hr at 4°C. For supershift analysis, 1 μg of indicated antibody was added to the nuclear extracts and incubated at RT for 30 min prior to the addition and incubation with the radiolabelled probe. A sample containing probe but no nuclear extract was included as a negative control. After drying, the gel was placed on a phosphorimaging screen for 12 – 24 h before scanning using a Typhoon 8600 scanner (GE Healthcare).
Lentiviral Vectors and Cell Transduction
Murine PU.1 cDNA spanning the entire coding region was synthesised from EOC-13 cells by PCR using the primers: 5′AATGCGGCCGCACCAACCTGGAGCTCAGCTGG3′ (upstream) and 5′AATCTCGAGGGAGCCTGGCGGTCTCTGCGG3′ (downstream). After sequence verification the amplified cDNA was cloned into the vector pHAGE-pgk-IRES-Zs green using Not I and Xho I to produce pHAGE-PU.1. pHAGE-pgk-IRES-Zs green is a third generation, self-inactivating lentiviral vector (34) obtained from PlasmID, Harvard, MA. Replication incompetent lentiviral particles were produced in 293FT cells (Invitrogen), cultured in high glucose DMEM (Gibco) with 10% FBS. Culture plates (10 cm) containing 75% confluent 293FT cells were transfected with the packaging constructs pMD2.G, which expresses the envelope protein VSVG, (5μg, Addgene plasmid 12259), pMDLg/pRRE (10 μg, Addgene plasmid 12251) and pRSV-Rev (5μg, Addgene plasmid 12253) and 20 μg of the vector pHAGE-PU.1 using calcium phosphate-mediated DNA precipitation. Briefly, the plasmids were suspended in 500 μL CaCl2 (0.25M), added drop wise to 500 μL 2x HEBS (0.28 M NaCl, 0.05M HEPES, 1.5 mM Na2HPO4, pH 7.0), incubated at RT for 25 min and then added to the 293FT cells which were incubated for a further 12 hr. After 48 hr the culture medium was collected and filtered through a 0.45 μm membrane (Millipore, Billerica, MA). Virus produced using the ‘empty’ vector pHAGE-pgk-IRES-Zs-green (here termed pHAGE-GFP) was used as the negative control. C8-D1A astrocytes (ATCC; CRL-2541), were cultured in high glucose DMEM with 10% FBS. For viral transduction, undiluted viral supernatant (500 μl) with 8 μg/mL polybrene (hexadimethrine bromide, Sigma-Aldrich) was added to 75% confluent D1A cells in 6 well plates and incubated for 12 h. The transduced cells were washed and maintained for 3 passages in high glucose DMEM with 10% FBS. After trypsinsization, cell suspensions were analyzed by flow cytometry (FACSVantage DiVa; BD Biosciences) and >100000 GFP positive cells were collected. This enriched GFP-positive population was then maintained by tissue culture in high glucose DMEM with 10% FBS. For experimental studies the GFP-enriched cell populations as well as non-transduced D1A cells were treated with or without murine recombinant IFN-γ and analyzed as described above.
Flow Cytometry
Primary astrocytes, EOC-13 cells and non-transduced and virally-transduced D1A cells were stained to detect CD11b. For each cell population, 5 × 105 cells were incubated for 10 min with anti-CD16/32 antibody (BD-Biosciences) to block the Fc receptor. After washing (PBS containing 1% FBS/2mM EDTA), cells were then incubated for 30 min with phycoerythrin-conjugated anti-CD11b or an isotype control antibody (both BD-Biosciences) and washed. Stained cells were detected by flow cytometry using a FACSCalibur instrument (BD Biosciences) and the data analyzed using Flo-Jo Software (Tree Star, California, USA).
Statistical Analysis
Data was processed using Prism 4 software (GraphPad, La Jolla, CA). One-way ANOVA with Tukey’s post-test or one-tailed student’s t test were used as appropriate to compare groups with p< 0.05 being considered as statistically significant.
Results
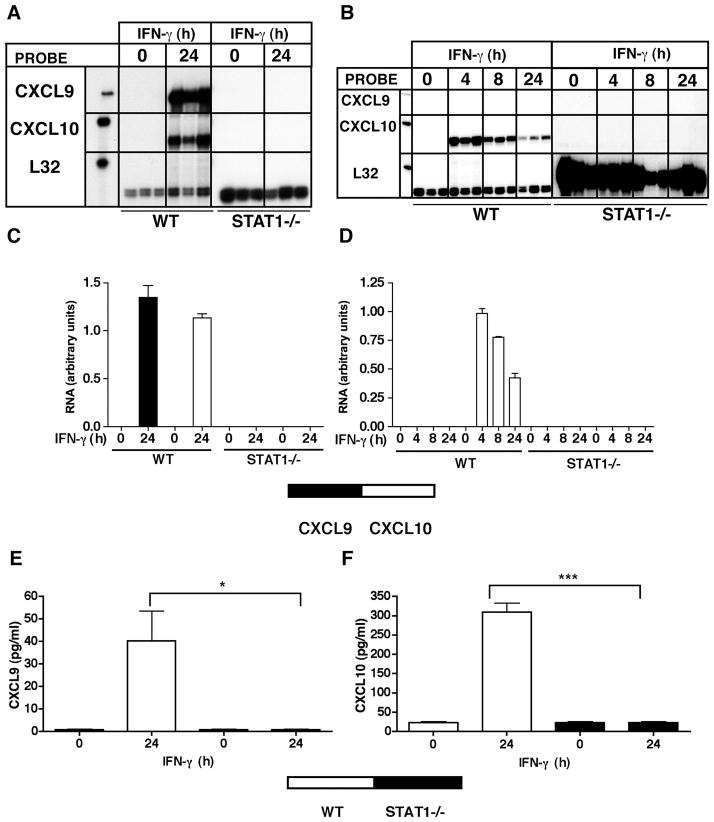
IFN-γ was shown to be a crucial stimulus for CXCR3 ligand gene expression by cultured glial cells, with CXCL9 mRNA induced only in microglia, whereas CXCL10 mRNA was induced in both astrocytes and microglia (24). Here these observations were confirmed and further extended to the protein level. In control, non-IFN-γ-treated cultures, little or no detectable CXCL9 or CXCL10 was produced (Figure 1A,B). However, following IFN-γ treatment, significant CXCL9 protein was released only from primary microglia and EOC-13 microglial cultures (Figure 1A), while significant CXCL10 protein was released from both astrocyte and microglial cultures (Figure 1B). Thus, these findings indicated that while both astrocytes and microglia responded to IFN-γ with the production of CXCL10, the production of CXCL9 was restricted to the microglial cells.
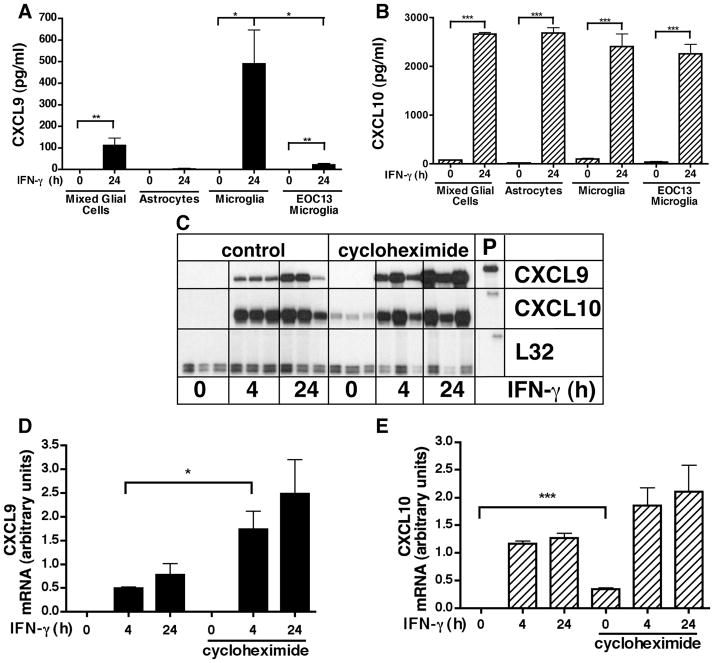
Figure 1. Regulation of CXCL9 and CXCL10 gene expression by IFN-γ in CNS glial cells.
CXCL9 (A) and CXCL10 (B) levels in supernatants from mixed glial cells, astrocytes, primary microglia and EOC-13 microglial cells. Supernatants were collected from IFN-γ-treated (24 hours) or untreated (0 hours) (n=3 per timepoint) cells were analyzed by ELISA. Autoradiographic images showing CXCL9 and CXCL10 mRNA levels in primary mixed glial cells (C). Cells were cultured as described in the Materials and Methods and treated either with medium alone (0 hour) or with medium containing recombinant IFN-γ (1000U/ml) for 4 hours or 24 hours with or without cycloheximide (CHX) 20μg/ml). In CHX treated groups, samples were pretreated for 30 min before treatment with IFN-γ. Chemokine mRNAs were detected by RPA analysis of total RNA (3 μg per sample) using a multiprobe set as described in the Materials and Methods. Quantification of CXCL9 (D) and CXCL10 (E) mRNA levels was performed by densitometry as described in the Materials and Methods. For statistical significance: *p<0.05, **p<0.01, ***p<0.001.
We next investigated whether de novo protein synthesis was required for the induction of the CXCL9 and CXCL10 mRNAs in microglia and/or astrocytes in response to IFN-γ. To address this question the IFN-γ stimulation of the glial cells was performed in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX; Fig. 1C). Compared with untreated controls, CHX treatment alone did not influence the level of CXCL9 mRNA (Figure 1C&D) but increased significantly the level of CXCL10 mRNA in cultured mixed glial cells (Figure 1C&E). Following exposure to IFN-γ, significant induction of both the CXCL9 and CXCL10 mRNA transcripts was observed by 4 hr. The presence of cycloheximide did not prevent the induction of CXCL9 mRNA mediated by IFN-γ and actually further potentiated significantly the response at both 4 and 24 hr post-stimulation (Figure 1C&D). Similarly, the presence of CHX did not prevent stimulation of CXCL10 RNA mediated by IFN-γ (Figure 1C&E). However, although there was a trend toward increased CXCL10 mRNA stimulation by the combination of IFN-γ and CHX, this response did not reach statistical significance. In all, these findings indicated that the IFN-γ-mediated induction of the genes for both CXL9 and CXCL10 in microglial cells was not dependent on new protein synthesis and in the case of CXCL9, new protein synthesis was associated with down regulation of this response.
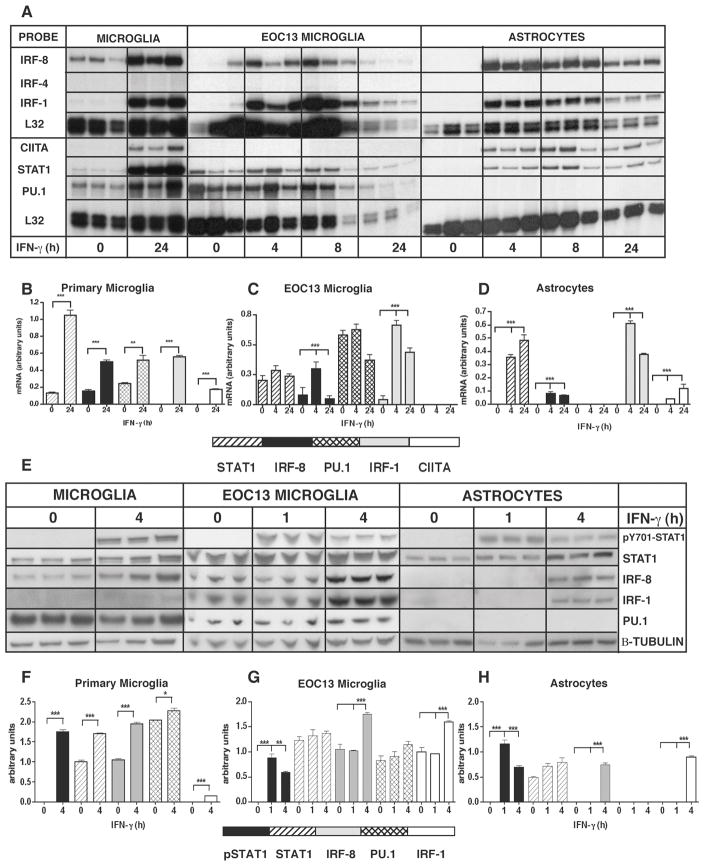
The above findings suggested that the differential expression of the genes encoding CXCL9 and CXCL10 is likely regulated at the transcriptional level. In light of this, it was hypothesized that the induction by IFN-γ of CXCL9 and CXCL10 mRNA in glial cells is mediated by cell-specific transcription factors. Therefore, the transcription factor profiles were investigated in microglia versus astrocytes. RNA transcripts for the canonical IFN-γ-signaling factor STAT1 (Figure 2A–D), as well as two other STAT factors, STAT2 and STAT3 were upregulated in both astrocytes and microglia following IFN-γ treatment (data not shown), and reached maximal levels after 24 h (Figure 2A–2D). In addition, PML mRNA transcripts were significantly induced in both astrocytes and microglia following IFN-γ treatment reaching maximal levels by 4 h (data not shown). CIITA mRNA was not only detectable in primary microglia, but also in astrocytes following IFN-γ treatment reaching maximal levels after 24 h of IFN-γ treatment (Figure 2A–D). Notably, and in contrast to primary microglia, EOC-13 microglia did not express detectable levels of CIITA mRNA. IRF-1 mRNA was induced following IFN-γ treatment of astrocytes, microglia and EOC-13 cells (Figure 2A–D). IRF-2 mRNA was detectable at low levels in untreated primary microglia, astrocytes and EOC-13 cells. Furthermore, IRF-2 mRNA levels were upregulated following IFN-γ treatment in microglial cells, but not in astrocytes. However, IRF-4 mRNA was not detectable in either untreated or IFN-γ-treated microglia, EOC-13 cells or astrocytes (Figure 2A–D). IRF-8 mRNA was detectable in untreated primary microglial cells (Figure 2A–D). IFN-γ treatment resulted in a significant increase in IRF-8 mRNA in these cells after 24 h of treatment, while EOC-13 cells showed maximal levels of IRF-8 mRNA after 4 h that were maintained at 8 h. Although not detectable in untreated astrocytes, IRF-8 mRNA was induced in astrocytes following IFN-γ treatment, reaching maximal levels after 4 h but remaining present until 24 h (Figure 2A–D). TEL mRNA transcripts were detectable in untreated astrocytes, microglia and EOC-13 microglia. In cell populations containing microglia, a time-dependent increase of TEL RNA levels in response to IFN-γ was observed, while IFN-γ treatment did not alter TEL mRNA level in astrocytes (data not shown). PU.1 mRNA was detectable in untreated primary microglial cells and EOC-13 microglia. The levels of PU.1 mRNA in primary microglia were significantly upregulated following IFN-γ treatment (Figure 2A–D). In contrast to the microglial cells, PU.1 was not detectable in untreated or IFN-γ-treated astrocytes. In summary, the results of this survey indicated that many of the transcription factor genes involved in IFN-γ-regulated gene expression were found to be expressed constitutively (e.g. STAT1, STAT3, IRF-2 and TEL) in both astrocytes and microglia or were induced (e.g. PML, CIITA and IRF-1) by IFN-γ in these cells. Differences were observed in the constitutive and/or IFN-γ-regulated expression of various transcription factor genes (e.g. STAT1, STAT3 and CIITA) between primary microglia and the EOC-13 microglial cells. Interestingly, IRF-8 RNA, which, as expected was found in microglia, was identified by this study as a novel IFN-γ-inducible transcription factor in cultured astrocytes. Finally, the expression of the gene for PU.1 was found to be cell-specific being detectable in microglia but not in astrocytes.
Figure 2. Expression of selected transcription factors in astrocytes and microglia.
All cells were cultured as described in the Materials and Methods and treated either with medium alone (0 hour) or medium containing recombinant IFN-γ (1000U/ml) at the time points shown. Total RNA was isolated from purified primary astrocytes, primary microglia and EOC-13 microglial cells and 3 μg was used for analysis by RPA (A) as described in the Materials and Methods section. The relative mRNA levels were quantified by densitometry and normalized to the L-32 loading control (B, C, D). PU.1 gene expression was detected only in microglial cells and not in astrocytes. Selected transcription factors were also examined at the protein level by immunoblot analysis (E) as described in Materials and Methods. Transcription factor levels in cultured glial cells were quantified by densitometry and normalized to the β-tubulin loading control (F, G, H). For statistical significance: *p<0.05, **p<0.01, ***p<0.001.
The products of a selected number of these transcription factor genes were next examined by immunoblot analysis to determine whether protein production correlated with the cellular RNA expression pattern (Figure 2E–H). Consistent with a response to IFN-γ, STAT1 phosphorylation was induced in all three cell types and STAT1 was present constitutively. In primary microglia and EOC-13 cells, PU.1 and IRF-8 were produced constitutively. IRF-1 was constitutively produced in EOC-13 microglial cells but not in primary microglia. Following IFN-γ treatment, the levels of all these transcription factors, as well as IRF-1, increased significantly in primary microglia and also in EOC-13 cells. However, while not detectable in untreated astrocytes, IRF-1 and IRF-8 were induced by 4 h following IFN-γ treatment. Finally, and in contrast to the microglial cells, PU.1 was not detectable in untreated nor in IFN-γ-treated astrocytes. In conclusion, these findings confirmed there is good concordance between the transcription factor mRNA and protein levels. They also highlighted that the constitutive and IFN-γ-regulated production of various transcription factors involved in the actions of IFN-γ show similarities as well as differences for microglia versus astrocytes.
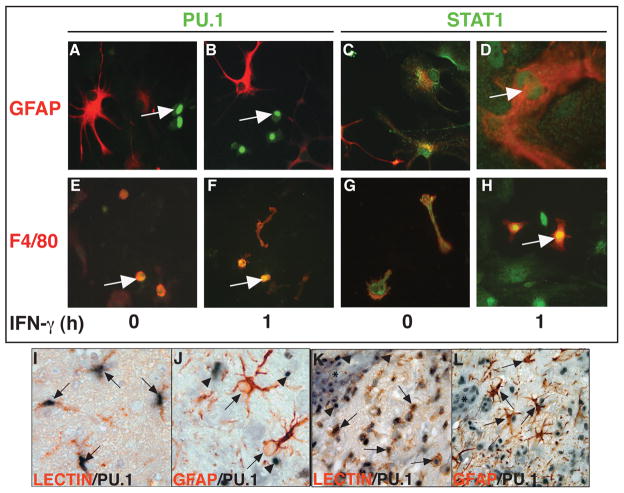
The function of many transcription factors is associated with changes in their intracellular localization between the cytoplasm and the nucleus. Here, the subcellular localization of PU.1 and STAT-1 in mixed glial cells in response to IFN-γ was examined by dual-label immunofluorescence microscopy (Figure 3A–H). In untreated mixed glial cells STAT1 was present in the cytoplasm of astrocytes (Figure 3C) and microglia (Figure 3G), whereas following IFN-γ treatment, STAT1 was localized to the nucleus of these cells (Figure 3D and H). In contrast to STAT1, PU.1 was restricted to F4/80+ microglia (Figure 3E,F) and absent from GFAP+ astrocytes (Figure 3A,B). Whether in the absence or presence of IFN-γ treatment, PU.1 was localized to the nucleus only (Figure 3E,F; arrows). These findings confirmed the differential production of PU.1 in microglia as compared with astrocytes and further extended this to demonstrate that PU.1 was localized to the nucleus but not apparently the cytoplasm and this localization pattern was not altered by IFN-γ. In contrast to PU.1, STAT1 was localized predominantly in the cytoplasm and occasionally found in the nucleus prior to IFN-γ treatment, however, STAT1 accumulated in the nucleus of both astrocytes and microglia following IFN-γ treatment.
Figure 3.
Localization of PU.1 and STAT1 in glial cells. The PU.1 and STAT1 proteins were analyzed by immunofluorescence staining of mixed glial cells (A–H) treated either with medium alone (0 hour) or medium containing recombinant IFN-γ (1000U/ml) for 1 hour as described in Materials and Methods. PU.1 (green; A,B,E,F, arrows) colocalized with F4/80+ (red; E,F) cells and not GFAP+ (red; A,B) cells. PU.1 was localized in the nucleus independent of IFN-γ treatment (E, F). In contrast, STAT1 (green; C,D,G,H) was found in the cytoplasm of GFAP+ (red; C) and F4/80+ (red; G) cells cultured in medium alone but was found in the nucleus of GFAP+ (D; arrow) and F4/80+ (H; arrow) cells following IFN-γ treatment. Dual-label immunohistochemical staining on brain sections from control mice (I,J) or mice at peak EAE (K,L) was performed as described in the Materials and Methods. PU.1 (purple) co-localised with lectin+ (red) microglia/macrophages (I,K; arrows). However, PU.1 (purple; arrowheads) did not co-localize with GFAP+ astrocytes (red; arrows) (J, L). Asterisks denote blood vessels (K,L). Original magnification of panels I and J: 1000x and K and L: 400x.
Although STAT1 has been shown previously to be expressed constitutively in the CNS of WT mice at both the RNA and protein level and can be upregulated further in EAE (32), little is known concerning the regulation and localization of PU.1 in the CNS. To address this question, the cellular localization of PU.1 was examined by dual-label immunohistochemistry in the brain. This revealed that in the normal brain, PU.1 protein was present predominantly in the nucleus of lectin-positive macrophage/microglia (Figure 3I; arrows) while GFAP-positive astrocytes (Figure 3J; arrows) were negative for PU.1 (Figure 3J; arrowheads). A similar pattern of cellular localization of PU.1 was observed in the CNS of mice with EAE (Figure 3K & 3L). However, the overall number of PU.1+ cells in the CNS was increased markedly in EAE and in addition to macrophage/microglia (Figure 3K; arrows) the majority of blood vessel (asterisk) -associated mononuclear cells were also positive for PU.1 (Figure 3K; arrowheads). Similar to normal brain, in the CNS affected by EAE, astrocytes remained negative for PU.1 (Figure 3L; arrows). In summary, the cell-specific localization of PU.1 seen in vitro was recapitulated in vivo in the inflammatory model of EAE. Furthermore, PU.1 was constitutively expressed and localized to the nucleus of the lectin+ microglia/macrophage population in the normal brain as well as in EAE.
The transcription factors STAT1, PU.1, IRF-1 and IRF-8 are known to be involved in the regulation of IFN-γ-dependent gene transcription (reviewed in (19)). To examine further the role of these transcription factors in the induction of the gene for CXCL9 mediated by IFN-γ, ChIP assays were performed to delineate transcription factor binding to the Cxcl9 gene promoter (Figure 4A) in EOC-13 microglial cells and in astrocytes. When compared with the IgG control, it was found that STAT1 and PU.1 were bound constitutively at low levels to the Cxcl9 gene promoter in EOC-13 microglia (Figure 4B,D). However, IRF-8 binding was not detectable to the Cxcl9 gene promoter in these untreated cells. Following treatment with IFN-γ for 4 h higher levels of STAT1 and PU.1 and low levels of IRF-8 were bound to the Cxcl9 gene promoter (Figure 4B). At 24 h after IFN-γ treatment the amount of STAT1 and IRF-8 bound to the Cxcl9 gene promoter increased further, while PU.1 binding decreased back to basal levels (Figure 4B,D). In contrast to EOC-13 cells, in untreated or IFN-γ treated astrocytes, PU.1 or IRF-8 binding to the Cxcl9 gene promoter was not increased when compared with the IgG control levels (Figure 4C,E). However, similar to microglia, in astrocytes STAT1 was bound constitutively to the Cxcl9 gene promoter but in contrast to microglia did not increase following IFN-γ treatment (Figure 4C,E). The preceding results provided support for the concept that STAT1 and PU.1 are involved in the transcriptional activation of the gene for CXCL9 by microglial cells in response to IFN-γ.
The specific binding of PU.1 to the Cxcl9 gene promoter has not been documented previously. In addition to the γRE-1 binding sites for STAT1, the Cxcl9 gene promoter (Figure 4A) contains the putative Ets factor binding sites EIRE1 and EIRE2 (35). Therefore, further analysis was performed using EMSA to determine whether oligonucleotide probes corresponding to the potential DNA response elements present in the Cxcl9 gene promoter were capable of binding STAT1 and PU.1 in nuclear extracts of IFN-γ-stimulated EOC-13 microglial cells. In extracts from unstimulated cells no bands were observed with the γRE-1 probe (Figure 4F). However, in extracts from cells stimulated with IFN-γ for 4 or 24 h a single band appeared (Figure 4F). Addition of an antibody to STAT1 (αSTAT1) to the nuclear extract displaced the band with the γRE-1 probe indicating that this binding complex induced by IFN-γ contained STAT1. However, the binding of this complex to the γRE-1 probe was not altered in the presence of PU.1 antibody (αPU.1). In contrast to the γRE-1 probe, with the EIRE-1 probe a number of bands were present with extracts from unstimulated cells (Figure 4G). Antibody shift analysis revealed these elements to contain either STAT1 (arrow, αSTAT1) or PU.1 (arrow, αPU.1). Following 4 and 24 h IFN-γ-stimulation, these complexes remained bound to the EIRE1 element. However, after 24 h of IFN-γ-stimulation the formation of an additional, undetermined complex containing neither STAT1, nor PU.1 was observed. No binding was detectable with the EIRE2 probe (data not shown).
The preceding studies established that both STAT1 and PU.1 were bound to the Cxcl9 gene promoter. We next asked whether these transcription factors were involved in the IFN-γ-induced expression of the CXCL9 and CXCL10 genes in the glial cells. Astrocytes or microglia derived from WT or Stat1−/− mice were used to examine the role of STAT1 in the induction of the genes for CXCL9 and CXCL10. Following treatment of primary microglia from WT mice with IFN-γ there was a significant induction of CXCL9 and CXCL10 mRNA transcripts (Figure 5A&C) and protein (Figure 5E&F). However, in microglia that were deficient for STAT1 the IFN-γ-mediated induction of CXCL9 and CXCL10 mRNA (Figure 5A&C) and protein (Figure 5E&F) did not occur. As expected, there was no detectable expression of CXCL9 in astrocytes with or without IFN-γ treatment, while CXCL10 mRNA levels were induced at 4, 8 and 24 hours after IFN-γ treatment (Figure 5B&D). In similarly treated astrocytes that were deficient for STAT1, the level of CXCL10 mRNA remained at basal levels and was not altered by IFN-γ treatment (Figure 5B&D). In summary, these findings demonstrated that STAT1 was necessary for the induction of both CXCL9 and CXCL10 in microglia and CXCL10 in astrocytes in response to IFN-γ.
Figure 5.
Essential role for STAT1 in the induction of CXCL9 and CXCL10 by microglia and astrocytes in response to IFN-γ. Autoradiographic images showing CXCL9 and CXCL10 RNA levels in primary microglia (A) or primary astrocytes (B) from Wt or Stat1−/− mice. Cells were cultured as described in the Materials and Methods and treated with or without IFN-γ (1000 U/ml) for the times shown and total RNA (3 μg per sample) analyzed by RPA using a multiprobe set as described in the Materials and Methods. The relative mRNA levels were quantified by densitometry and normalized to the L32 loading control for microglia (C) and astrocytes (D). Cultured WT or Stat1−/− mixed glial cells were treated with or without IFN-γ (1000 U/ml) and the supernatants collected and analyzed by ELISA for CXCL9 (E) and CXCL10 (F) as described in the Material and Methods. For statistical significance: *p < 0.05, ***p<0.001.
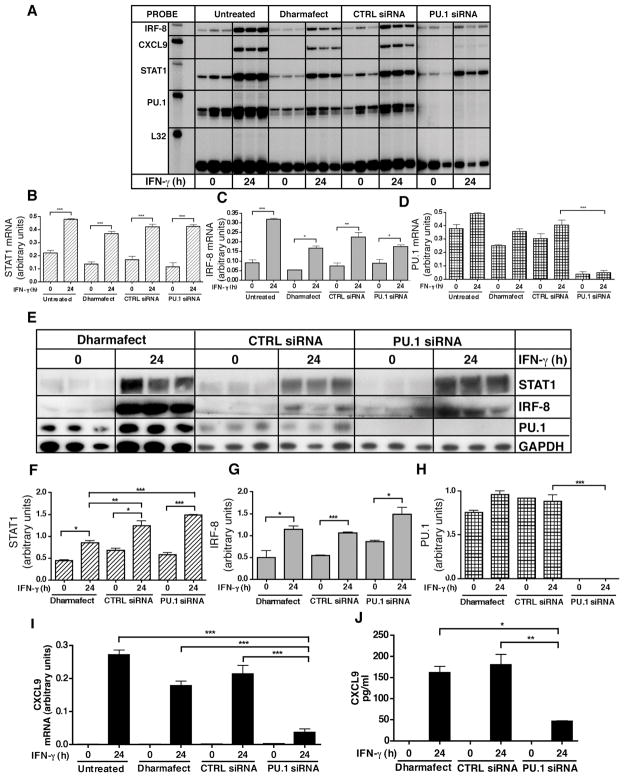
The role of PU.1 in mediating IFN-γ-induced expression of the gene for CXCL9 was determined following siRNA knockdown of PU.1 mRNA in EOC-13 microglial cells (Figure 6). In control cells the basal level of PU.1 mRNA was decreased significantly by PU.1 siRNA treatment when compared with untreated, Dharmafect® or control siRNA transfected cells (Figure 6A,D). In contrast, the level of IRF-8 and STAT1 mRNAs showed no significant differences between PU.1 siRNA, control siRNA or Dharmafect® transfected cells (Figure 6A–C). Following IFN-γ treatment, the level of PU.1 mRNA remained significantly lower in PU.1 siRNA treated cells compared with the Dharmafect and siRNA controls (Figure 6A,D). However, these cells were able to respond to IFN-γ with significant increases in both STAT1 and IRF-8 mRNAs irrespective of the siRNA transfection condition (Figure 6A–C). Similar observations were made at the protein level, with PU.1 not detectable in untreated or IFN-γ-treated cells transfected with PU.1 siRNA when compared with Dharmafect® or control siRNA transfected cells (Figure 6E,H). The level of STAT1 and IRF-8 showed no significant differences in PU.1 siRNA, control siRNA or DharmafectR treated cells (Figure 6E–G). Moreover, STAT1 and IRF-8 levels were upregulated significantly following IFN-γ treatment irrespective of the siRNA treatment condition (Figure 6E–G). Ultimately, these findings indicated that while transfection of EOC-13 cells with PU.1 siRNA effectively knocked down PU.1, this did not compromise the ability of IFN-γ to stimulate either STAT1 or IRF-8.
Figure 6.
siRNA mediated knockdown of PU.1 in EOC-13 cells and its impact on IFN-induced-CXCL9 production. siRNA knockdown was performed as described in the Materials and Methods. Non-transfected, transfection medium (Dharmafect®) alone, control (CTRL) siRNA transfected and PU.1 siRNA transfected EOC-13 cells were treated with medium alone or IFN-γ (1000 U/ml) for 24 hours. Total RNA was extracted and 3 ug RNA was subjected to RPA analysis as described in the Materials and Methods (A). Quantification of STAT1, IRF-8 and PU.1 mRNA levels was performed by densitometry and normalized to L32 loading control (B, C, D). Protein lysates were prepared and subjected to immunoblotting with anti-PU.1, anti-STAT1, anti-IRF-8 and anti-GAPDH antibodies (E). Quantification of STAT1, IRF-8 and PU.1 protein levels was performed by densitometry and normalized to GAPDH (F, G, H). Quantification of CXCL9 mRNA levels was performed by densitometry and normalized to the L32 loading control (I). Supernatants from PU.1 siRNA transfected, CTRL siRNA transfected or Dharmafect® transfected controls treated with or without IFN-γ treated were analyzed by ELISA for CXCL9 protein levels (J). For statistical significance: *p < 0.05, **p<0.01, ***p<0.001.
We next examined the production of CXCL9 mRNA and protein in response to IFN-γ by the EOC-13 microglial cells following PU.1 siRNA transfection. Both CXCL9 mRNA (Figure 6I) and protein (Figure 6J) levels were increased significantly in cells transfected with Dharmafect® or control siRNAs. However, the magnitude of this response was significantly decreased in cells transfected with PU.1 siRNA (Figure 6I,J). These findings indicated that in these microglial cells the induction of the gene for CXCL9 in response to IFN-γ is PU.1-dependent.
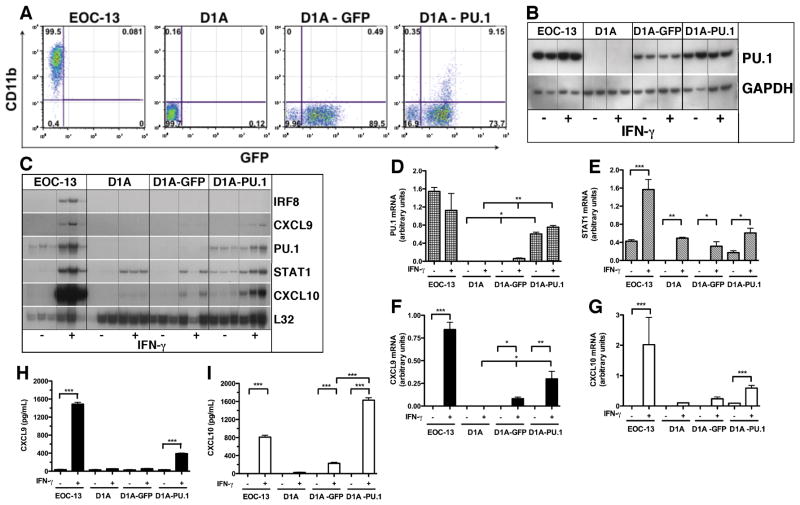
It was reported recently that retrovirally-transduced expression of the gene for PU.1 by fibroblasts can convert these cells into macrophage-like cells (36). Therefore, we asked whether acquisition of PU.1 by astrocytes could similarly convert these cells to macrophage-like cells and confer the ability to produce CXCL9 in response to IFN-γ. Unfortunately, attempts to express the gene for PU.1 in primary astrocytes by liposome-mediated transfection or virally-mediated transduction proved unsuccessful due to very low efficiency of transgene uptake and expression. As an alternative to primary astrocytes we used the astrocyte-like cell line, C8-D1A (37). These cells were transduced with a control lentiviral vector pHAGE-GFP or with pHAGE-PU.1 and the respective GFP-positive D1A cells collected by flow cytometry and maintained in culture for further study. Employing this strategy allowed for enrichment of 90% and 83% of GFP expressing D1A-GFP and D1A-PU.1 cells, respectively (Figure 7A). Analysis of PU.1 protein levels by western blot revealed that while there were high levels of PU.1 detectable in the EOC-13 positive control cells, PU.1 was not detectable in untreated or IFN-γ-treated D1A cells (Figure 7B). However and surprisingly, PU.1 was present at low levels in D1A-GFP cells while increased levels of PU.1 were found in D1A-PU.1 cells (Figure 7B). When analyzed at the RNA level, a very low level of PU.1 mRNA was detectable in D1A-GFP cells (Figure 7C&7D) while significantly higher levels of PU.1 mRNA were present in the D1A-PU.1 transduced cells (Figure 7C&7D). These findings confirmed the non-myeloid nature of the C8-D1A cell line but indicated that transduction of these cells with an empty pHAGE lentiviral vector alone was sufficient to induce PU.1, the levels of which, were increased significantly by transduction with pHAGE containing PU.1.
Figure 7.
Properties of PU.1 transduced C8-D1A astrocytic cells. Cells of the C8-D1A astrocyte cell line were transduced with the pHAGE lentiviral vector containing GFP alone (empty vector control) or GFP plus PU.1 as described in the Materials and Methods. Flow cytometry was used to collect and enrich for GFP-positive cells which were then analyzed for CD11b expression compared with EOC-13 microglial cells as a positive control (A). Following treatment with or without IFN-γ, cells were either lysed and the PU.1 protein was analyzed by immunoblotting (B) or total RNA was prepared and analyzed by RPA (C) as described in the Materials and Methods. The relative level of PU.1 (D), STAT1 (E), CXCL9 (F) or CXCL10 (G) mRNA was quantified by densitometry and normalized to the L32 loading control. The production of CXCL9 (H) and CXCL10 (I) by the different cell types in response to IFN-γ was determined by ELISA. For statistical significance: *p < 0.05, **p<0.01, ***p<0.001.
Since ectopically produced PU.1 is known to regulate the expression of a number of myeloid-lineage specific genes in fibroblasts including CD11b (36), we next examined the expression of CD11b by flow cytometry (Figure 7A). The results showed that in comparison with EOC-13 microglia for which over 99% of the cells were positive for CD11b, D1A cells did not express detectable levels of this myeloid marker. In the case of the D1A cells transduced with pHAGE, 0.5% D1A-GFP and 9% of D1A-PU.1 cells were positive for CD11b, respectively. These findings indicated that the presence of PU.1 is associated with a dose-dependent induction of CD11b expression in a subset of D1A cells.
We next examined whether the presence of PU.1 in D1A cells altered the response of these cells to IFN-γ (Figure 7C). In EOC-13 microglia, the IRF-8, STAT1 (Figure 7E), CXCL9 (Figure 7F) and CXCL10 (Figure 7G) mRNA transcripts were increased significantly after 4 hours exposure to IFN-γ. By contrast, in D1A cells STAT1 mRNA levels were increased significantly by IFN-γ treatment (Figure 7C&7E) while CXCL10 mRNA was induced at low levels (Figure 7C&7G). A similar response to IFN-γ was observed in D1A-GFP cells with the exception that there was a larger induction of CXCL10 mRNA (Figure 7C&7G). However, in D1A-PU.1 cells, STAT1 (Figure 7C&7E), CXCL9 (Figure 7C&7F) and CXCL10 (Figure 7C&7G) mRNA transcripts were all increased significantly after IFN-γ treatment. When compared with untreated controls CXCL9 protein secretion was increased significantly from IFN-γ treated EOC-13 microglia and D1A-PU.1 but not D1A or D1A-GFP cells (Figure 7H). The secretion of CXCL10 was increased significantly from IFN-γ treated EOC-13 and D1A-PU.1 cells (Figure 7I). Very low levels of CXCL10 were also produced by IFN-γ treated D1A and D1A-GFP cells (Figure 7I). In summary, the acquisition of PU.1 by D1A cells markedly altered the response of these cells to IFN-γ, notably, resulting in the induction of the gene for CXCL9.
Discussion
In the CNS, CXCL9 and CXCL10 are induced in an IFN-γ-dependent manner in a number of experimental neuroimmune pathologies including EAE (24, 38, 39), Toxoplasma encephalitis (26), lymphocytic choriomeningitis (27) and cerebral malaria (28). The findings described here confirmed not only that IFN-γ is a dominant regulator of CXCR3 ligand gene expression in glial cells in vitro, but there are cell-specific differences in the induction of the gene for CXCL9 which was expressed by microglia but not astrocytes. Differences in the spatial production of these chemokines during inflammation may have an important function in positioning of leukocytes in the brain during immune responses (24, 25). The mechanism by which IFN-γ can stimulate both astrocytes and microglia to induce CXCL10 and yet elicit the selective induction of CXCL9 in microglia was unknown. Our studies revealed that the IFN-γ-mediated induction of both CXCL10 and CXCL9 in glial cells did not require new protein synthesis and therefore was dependent on the presence of pre-existing factors in the cell. We reasoned that the mechanisms underlying the differential glial cell response to IFN-γ likely involved cell-specific transcription factor regulation of the promoters of these genes in microglia versus astrocytes. Although STAT1 is the canonical transcription factor responsible for most cellular responses to IFN-γ (17), there is much evidence to indicate that this process is more complex and involves additional positive and negative acting transcription factors (16, 19). Our findings revealed that many of the transcription factors that are known to be involved in IFN-γ-regulated gene transcription were found to be present constitutively and/or induced by IFN-γ in both astrocytes and microglia. However, the levels and or the kinetics of expression of some of these transcription factors in response to IFN-γ differed between astrocytes and microglia and between primary microglia and EOC-13 microglial cells. Significantly, of the transcription factors examined, PU.1 alone was found to be produced constitutively by microglia but not detectable in astrocytes, which is in accordance with a previously reported study in the rat (40). In all, these findings pointed to PU.1 as a good candidate for a transcription factor involved in the cell-specific response of microglia to IFN-γ.
PU.1 is known to have a crucial role in the differentiation and function of myeloid cells and can regulate the expression of a number of genes in these cells (41). Microglia are myeloid lineage cells that colonize the CNS during development and constitute the tissue resident macrophages of the CNS (42). Consistent with their myeloid lineage, we observed that microglia produced PU.1 and this factor was localized to the nucleus in vitro and in vivo, irrespective of whether these cells were activated or not. It is likely that PU.1 has an important role in the function of resting as well as activated microglial cells by regulating expression of myeloid specific genes such as CD40 (43, 44). Our findings here provide further evidence that PU.1 is involved in modulating the transcriptional response of microglial cells to IFN-γ. Moreover, the constitutive presence of PU.1 in microglia is consistent with the cycloheximide resistance we observed for IFN-γ-induced expression of the gene for CXCL9.
Evidence that PU.1 might be involved in activation of the gene for CXCL9 in response to IFN-γ came from the results of promoter binding studies. We found that PU.1 was bound constitutively at low levels to the Cxcl9 gene promoter in non-stimulated EOC-13 microglial cells. Importantly, the recruitment of PU.1 to the Cxcl9 gene promoter was increased significantly by 4 hours following IFN-γ treatment. IFN-γ similarly increased STAT1 binding to the Cxcl9 gene promoter by 4 hours, however, and in contrast to PU.1, STAT1 binding increased further at 24 hours after cytokine treatment. The binding of STAT1 to the Cxcl9 gene promoter has been described previously (45). However, our findings indicate that in addition to STAT1, PU.1 also bound to the Cxcl9 gene promoter and this is increased by IFN-γ and therefore is consistent with PU.1 being a transcriptional regulator of the CXCL9 gene. Interestingly, we also observed that IRF-8 was recruited to the Cxcl9 gene promoter as a late binding factor induced by IFN-γ. The kinetics of this process rules out a role for IRF-8 in the transcriptional activation of the gene for CXCL9. We can only speculate as to the function of IRF-8 at this time but it is intriguing that IRF-8 binding occurs concomitant with decreased PU.1 binding suggesting perhaps, that IRF-8 might be involved in downregulation of CXCL9 gene transcription. Consistent with this possibility, IRF-8 has been shown previously to be an important negative transcriptional regulator of “second-wave” gene expression induced by IFN-γ (19).
The Ets family of transcription factors of which PU.1 is a member, trigger the transcription of many genes by binding to cis-regulatory elements with the core DNA sequence GGAA (46). In the promoter of the murine gene for CXCL9, two putative regions that contain a composite Ets/ISRE (EIRE) binding site with this core sequence motif were identified (35). The forced expression of IRF-4 in the murine interleukin-3 (IL-3)-dependent pro-B cell line, Ba/F3, activates constitutive expression of the gene for CXCL9 due to the binding of IRF-4 and PU.1 to the EIRE1 site in the Cxcl9 gene promoter (35). Our data revealed that PU.1 and STAT1 could bind to the EIRE1 but not the EIRE2 DNA recognition motif. However, the binding of either PU.1 or STAT1 to the EIRE1 DNA was not altered in nuclear extracts from IFN-γ-treated cells suggesting that other regions of the Cxcl9 gene promoter may also be engaged in binding these factors in response to IFN-γ stimulation. An additional factor that bound to the EIRE1 site motif was also detected but was present only in extracts from cells treated for 24 hours with IFN-γ. Although the identity of this novel EIRE1 binding factor requires confirmation, preliminary studies by us suggest that it may be IRF-8 (Ellis and Campbell, unpublished data).
The γRE-1 site in the murine Cxcl9 gene promoter is a unique binding element with partial homology to the more classical GAS element found in the promoters of many IFN-γ-responsiveness genes that bind STAT1 homodimers (45, 47). The γRE-1 site is indispensible for IFN-γ mediated transcriptional activation of the gene for CXCL9 (47). Here we confirmed that a γ-RE-1 DNA recognition motif binds STAT1 in nuclear extracts from IFN-γ-treated but not untreated microglial cells. This is consistent with the known restriction of the γRE-1 for binding phosphotyrosine-STAT1 -the formation of which is induced by IFN-γ. Moreover, our finding that STAT1-deficient microglia were refractory to IFN-γ-induced Cxcl9 gene expression highlighted the crucial requirement for STAT1 in the transcriptional activation of the gene for CXCL9 in microglial cells.
Currently the significance of the constitutive binding of PU.1 and STAT1 to the promoter of the gene for CXCL9 remains unknown. Non-phosphorylated STAT1 was assumed to be a latent transcription factor. However, as shown here and as previously reported by others (48, 49) this factor can be detected in the nucleus and is bound to the promoters of certain genes in unstimulated cells. Although the function of such constitutive, nuclear localized STAT1 is unknown, recent studies provide strong evidence that it is transcriptionally active maintaining the expression of a subset of IFN-regulated genes independently of phosphorylated STAT1 (50). However, whether unphosphorylated STAT1 bound to the promoters of the Cxcl9 and Cxcl10 genes has a regulatory role remains to be determined.
It is now clear that IFN-γ-regulated gene transcription is determined by a multi-step process that involves the assembly of macromolecular transcription factor complexes (also known as enhanceosomes) that bind to specific promoter elements within target genes. In the case of PU.1, this transcription factor regulates many genes including CD40 (43), CCL5 (51), TLR9 (52) and gp91phox (53). In each instance this requires the assembly of PU.1 with one or more transcription factors that include IRF-1, IRF-2, STAT-1 or IRF-8 in a specific sequence to activate gene transcription in response to IFN-γ. The findings of the present study demonstrated that in microglia, STAT1, PU.1 and IRF-8 bound to the Cxcl9 gene promoter with specific dynamics in response to IFN-γ. However our data do not allow us to make any conclusions as to possible interactions between PU.1 and other transcription factors. Identifying the nature and kinetics of such interactions will need to be addressed by further studies in order to gain a more complete understanding of the transcriptional mechanisms that control the expression of the Cxcl9 gene.
Manipulation of the intracellular levels of PU.1 allowed us to assess the functional significance of PU.1 in IFN-γ-induced expression of the gene for CXCL9. While the siRNA-mediated depletion of PU.1 in EOC microglia did not compromise the IFN-γ response of these cells in terms of STAT1 activation or upregulation of IRF-8, the stimulation of Cxcl9 gene expression was markedly impaired. Thus, the transcriptional activation of the gene for CXCL9 in microglia in response to IFN-γ is dependent not only on STAT1 but also PU.1. Could the absence of PU.1 from astrocytes account for the inability of IFN-γ to induce the gene for CXCL9 in these cells? As has been described for fibroblasts (36) we found that PU.1 cDNA expressed ectopically in an astrocyte cell line via lentiviral transduction could partially reprogramme a sub-population of these cells to express the myeloid lineage marker CD11b. Surprisingly, our findings also revealed that transduction of this astrocyte cell line with an empty lentiviral vector was sufficient to induce low levels of endogenous Pu.1 gene expression although this was not associated with significant induction of CD11b. It is of interest that the Pu.1 gene was originally discovered as a consequence of insertional activation by the murine retrovirus Spleen focus-forming virus (54). However, whether such a mechanism explains the activation of the endogenous Pu.1 gene observed in the present study is unclear. Nevertheless, our studies make it clear that ectopic expression of PU.1 in the astrocytic cells can increase the IFN-γ responsiveness of a number of genes such as CXCL10 indicating that PU.1 likely has a more general role in regulating the cellular response to IFN-γ. Significantly, the acquisition of PU.1 also conferred on these cells the ability to respond to IFN-γ with the induction of the gene for CXCL9. Therefore our findings show not only is PU.1 a key transcriptional activator of the gene for CXCL9 in the microglial response to IFN-γ, but also that its transgenic production in non-myeloid cells can make these cells permissive for IFN-γ-induced CXCL9 production.
In conclusion, it is apparent that the transcriptional regulation of the Cxcl9 gene is a multi-step process that involves cell-specific transcription factors, as well as the likely combinatorial effects of different transcription factors within the cell. In microglia, IFN-γ-induced Cxcl9 gene transcription is dependent on not only STAT1 but also the myeloid lineage factor PU.1. The inability of IFN-γ to induce Cxcl9 gene expression in astrocytes correlates with the absence of PU.1 but can be counteracted by the ectopic expression of this transcription factor in these cells. Thus, these studies have identified a novel molecular mechanism by which the cytokine IFN-γ can induce cell-specific expression of the Cxcl9 gene in microglia versus astrocytes in the CNS.
Acknowledgments
We thank Jane Radford and Barbara Hernandez (Department of Pathology, University of Sydney) for expert technical assistance with tissue processing and routine histology and Laura Parker for breeding and screening of the mice used in this study.
Footnotes
This work was supported by NIH Grant R01 NS044905 and a start up grant from the University of Sydney (to ILC). SLC was supported by an Endeavour International Postgraduate Award and International Postgraduate Award from the University of Sydney. DG was supported by an Australian Postgraduate Award. MJH and MM were the recipients of Deutsche Forschungsgemeinschaft postdoctoral awards (HO3298/1-1) and (Mu17-07/3-1), respectively. MM was also supported by the fund “Innovative Medical Research” of the University of Münster Medical School, Germany.
Publisher's Disclaimer: “This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.”
References
- 1.Farber JM. A macrophage mRNA selectively induced by γ-interferon encodes a member of the platelet factor 4 family of cytokines. Proc Natl Acad Sci USA. 1990;87:5238–5242. doi: 10.1073/pnas.87.14.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luster AD, Unkeless JC, Ravetch JV. γ-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 3.Vanguri P, Farber JM. Identification of CRG-2. An interferon-inducible mRNA predicted to encode a murine monokine. J Biol Chem. 1990;265:15049–15057. [PubMed] [Google Scholar]
- 4.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, Gladue RP, Lin W, Boyd JG, Moser B, Wood DE, Sahagan BG, Neote K. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rani MRS, Foster GR, Leung S, Leaman D, Stark GR, Ransohoff RM. Characterization of β-R1, a gene that is selectively induced by interferon β (IFN-β) compared with IFN-α. J Biol Chem. 1996;271:22878–22884. doi: 10.1074/jbc.271.37.22878. [DOI] [PubMed] [Google Scholar]
- 6.Loetscher M, Gerber B, Loetscher P, Jones SA, Piali L, Clark Lewis I, Baggiolini M, Moser B. Chemokine receptor specific for IP10 and Mig: structure, function, and expression in activated T-lymphocytes. J Exp Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao F, Rabin RL, Yannelli JR, Koniaris LG, Vanguri P, Farber JM. Human Mig chemokine: biochemical and functional characterization. J Exp Med. 1995;182:1301–1314. doi: 10.1084/jem.182.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taub DD, Lloyd AR, Conlon K, Wang JM, Ortaldo JR, Harada A, Matsushima K, Kelvin DJ, Oppenheim JJ. Recombinant human-interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med. 1993;177:1809–1814. doi: 10.1084/jem.177.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taub DD, Longo DL, Murphy WJ. Human interferon-inducible protein-10 induces mononuclear cell infiltration in mice and promotes the migration of human T lymphocytes into the peripheral tissues and human peripheral blood lymphocytes-SCID mice. Blood. 1996;87:1423–1431. [PubMed] [Google Scholar]
- 10.Belperio JA, Keane MP, Burdick MD, Lynch JP, 3rd, Zisman DA, Xue YY, Li K, Ardehali A, Ross DJ, Strieter RM. Role of CXCL9/CXCR3 chemokine biology during pathogenesis of acute lung allograft rejection. J Immunol. 2003;171:4844–4852. doi: 10.4049/jimmunol.171.9.4844. [DOI] [PubMed] [Google Scholar]
- 11.Hancock WW, Gao W, Csizmadia V, Faia KL, Shemmeri N, Luster AD. Donor-derived IP-10 initiates deveopment of acute allograft rejection. J Exp Med. 2001;193:975–980. doi: 10.1084/jem.193.8.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hancock WW, Lu B, Gao W, Csizmadia V, Faia KL, King JA, Smiley ST, Ling M, Gerard NP, Gerard C. Requirement of the chemokine receptor CXCR3 for acute allograft rejection. J Exp Med. 2000;192:1515–1519. doi: 10.1084/jem.192.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang D, Liang J, Hodge J, Lu B, Zhu Z, Yu S, Fan J, Gao Y, Yin Z, Homer R, Gerard C, Noble PW. Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J Clin Invest. 2004;114:291–299. doi: 10.1172/JCI16861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan IA, Maclean JA, Lee FS, Casciotti L, DeHaan E, Schwartzman JD, Luster AD. IP-10 is critical for effector T cell trafficking and host survival in Toxoplasma gondii infection. Immunity. 2000;12:483–494. doi: 10.1016/s1074-7613(00)80200-9. [DOI] [PubMed] [Google Scholar]
- 15.Zeng X, Moore TA, Newstead MW, Deng JC, Kunkel SL, Luster AD, Standiford TJ. Interferon-inducible protein 10, but not monokine induced by gamma interferon, promotes protective type 1 immunity in murine Klebsiella pneumoniae pneumonia. Infect Immun. 2005;73:8226–8236. doi: 10.1128/IAI.73.12.8226-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 17.Darnell J, JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Ann Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 19.Kanno Y, Levi BZ, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25:770–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- 20.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 21.Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem. 2006;281:14111–14118. doi: 10.1074/jbc.M511797200. [DOI] [PubMed] [Google Scholar]
- 22.Kuwata T, Gongora C, Kanno Y, Sakaguchi K, Tamura T, Kanno T, Basrur V, Martinez R, Appella E, Golub T, Ozato K. Gamma interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Mol Cell Biol. 2002;22:7439–7448. doi: 10.1128/MCB.22.21.7439-7448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi YH, Bernardi R, Pandolfi PP, Benveniste EN. The promyelocytic leukemia protein functions as a negative regulator of IFN-{gamma} signaling. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0604800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter SL, Muller M, Manders PM, Campbell IL. Induction of the genes for Cxcl9 and Cxcl10 is dependent on IFN-gamma but shows differential cellular expression in experimental autoimmune encephalomyelitis and by astrocytes and microglia in vitro. Glia. 2007;55:1728–1739. doi: 10.1002/glia.20587. [DOI] [PubMed] [Google Scholar]
- 25.Muller M, Carter SL, Hofer MJ, Manders P, Getts DR, Getts MT, Dreykluft A, Lu B, Gerard C, King NJ, Campbell IL. CXCR3 Signaling Reduces the Severity of Experimental Autoimmune Encephalomyelitis by Controlling the Parenchymal Distribution of Effector and Regulatory T Cells in the Central Nervous System. J Immunol. 2007;179:2774–2786. doi: 10.4049/jimmunol.179.5.2774. [DOI] [PubMed] [Google Scholar]
- 26.Strack A, Asensio VC, CampbelI IL, Schluter D, Deckert M. Chemokines are differentially expressed by astrocytes, microglia and inflammatory leukocytes in Toxoplasma encephalitis and critically regulated by interferon-gamma. Acta Neuropathol. 2002;103:458–468. doi: 10.1007/s00401-001-0491-7. [DOI] [PubMed] [Google Scholar]
- 27.Hofer MJ, Carter SL, Muller M, Campbell IL. Unaltered neurological disease and mortality in CXCR3-deficient mice infected intracranially with lymphocytic choriomeningitis virus-Armstrong. Viral Immunol. 2008;21:425–433. doi: 10.1089/vim.2008.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miu J, Mitchell AJ, Muller M, Carter SL, Manders PM, McQuillan JA, Saunders BM, Ball HJ, Lu B, Campbell IL, Hunt NH. Chemokine gene expression during fatal murine cerebral malaria and protection due to CXCR3 deficiency. J Immunol. 2008;180:1217–1230. doi: 10.4049/jimmunol.180.2.1217. [DOI] [PubMed] [Google Scholar]
- 29.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 30.Walker WS, Gatewood J, Olivas E, Askew D, Havenith CEG. Mouse microglial cell lines differing in constitutive and interferon-γ-inducible antigen-presenting activities for naive and memory CD4+ and CD8+ T cells. J Immunol. 1995;63:163–174. doi: 10.1016/0165-5728(95)00146-8. [DOI] [PubMed] [Google Scholar]
- 31.Asensio VC, I, Campbell L. Chemokine gene expression in the brain of mice with lymphocytic choriomeningitis. J Virol. 1997;71:7832–7840. doi: 10.1128/jvi.71.10.7832-7840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maier J, Kincaid C, Pagenstecher A, Campbell IL. Regulation of signal transducer and activator of transcription and suppressor of cytokine signaling gene expression in the brain of mice with astrocyte-targeted production of IL-12 and experimental autoimmune encephalomyelitis. Am J Pathol. 2002;160:271–288. doi: 10.1016/S0002-9440(10)64371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ousman SS, I, Campbell L. Regulation of murine interferon regulatory factor (IRF) gene expression in the central nervous system determined by multiprobe RNase protection assay. Methods Mol Med. 2005;116:115–134. doi: 10.1385/1-59259-939-7:115. [DOI] [PubMed] [Google Scholar]
- 34.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci U S A. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uranishi M, Iida S, Sanda T, Ishida T, Tajima E, Ito M, Komatsu H, Inagaki H, Ueda R. Multiple myeloma oncogene 1 (MUM1)/interferon regulatory factor 4 (IRF4) upregulates monokine induced by interferon-gamma (MIG) gene expression in B-cell malignancy. Leukemia. 2005;19:1471–1478. doi: 10.1038/sj.leu.2403833. [DOI] [PubMed] [Google Scholar]
- 36.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBP{alpha}/{beta} convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alliot F, Pessac B. Astrocytic cell clones derived from established cultures of 8-day postnatal mouse cerebella. Brain Res. 1984;306:283–291. doi: 10.1016/0006-8993(84)90377-9. [DOI] [PubMed] [Google Scholar]
- 38.Glabinski AR, Krakowski M, Han Y, Owens T, Ransohoff RM. Chemokine expression in GKO mice (lacking interferon-gamma) with experimental autoimmune encephalomyelitis. J Neurovirol. 1999;5:95–101. doi: 10.3109/13550289909029750. [DOI] [PubMed] [Google Scholar]
- 39.Tran EH, Prince EN, Owens T. IFN-gamma shapes immune invasion of the central nervous system via regulation of chemokines. J Immunol. 2000;164:2759–2768. doi: 10.4049/jimmunol.164.5.2759. [DOI] [PubMed] [Google Scholar]
- 40.Walton MR, Gibbons H, MacGibbon GA, Sirimanne E, Saura J, Gluckman PD, Dragunow M. PU.1 expression in microglia. J Neuroimmunol. 2000;104:109–115. doi: 10.1016/s0165-5728(99)00262-3. [DOI] [PubMed] [Google Scholar]
- 41.Lloberas J, Soler C, Celada A. The key role of PU.1/SPI-1 in B cells, myeloid cells and macrophages. Immunol Today. 1999;20:184–189. doi: 10.1016/s0167-5699(99)01442-5. [DOI] [PubMed] [Google Scholar]
- 42.Ransohoff RM, V, Perry H. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen VT, Benveniste EN. Involvement of STAT-1 and Ets family members in IFN-(gamma) induction of CD40 transcription in microglia/macrophages. J Biol Chem. 2000;275:23674–23684. doi: 10.1074/jbc.M002482200. [DOI] [PubMed] [Google Scholar]
- 44.Weigelt K, Ernst W, Walczak Y, Ebert S, Loenhardt T, Klug M, Rehli M, Weber BH, Langmann T. Dap12 expression in activated microglia from retinoschisin-deficient retina and its PU.1-dependent promoter regulation. J Leukoc Biol. 2007 doi: 10.1189/jlb.0707447. [DOI] [PubMed] [Google Scholar]
- 45.Guyer NB, Severns CW, Wong P, Feghali CA, Wright TM. IFN-gamma induces a p91/Stat1 alpha-related transcription factor with distinct activation and binding properties. J Immunol. 1995;155:3472–3480. [PubMed] [Google Scholar]
- 46.Blair DG, Athanasiou M. Ets and retroviruses - transduction and activation of members of the Ets oncogene family in viral oncogenesis. Oncogene. 2000;19:6472–6481. doi: 10.1038/sj.onc.1204046. [DOI] [PubMed] [Google Scholar]
- 47.Wong P, Sevens CW, Guyer NB, Wright TM. A unique palindromic element mediates gamma interferon induction of mig gene expression. Mol Cell Biol. 1994;14:914–922. doi: 10.1128/mcb.14.2.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF-1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer T, Begitt A, Lodige I, van Rossum M, Vinkemeier U. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 2002;21:344–354. doi: 10.1093/emboj/21.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. 2009;106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Ma X. Interferon regulatory factor 8 regulates RANTES gene transcription in cooperation with interferon regulatory factor-1, NF-kappaB, and PU.1. J Biol Chem. 2006;281:19188–19195. doi: 10.1074/jbc.M602059200. [DOI] [PubMed] [Google Scholar]
- 52.Schroder K, Lichtinger M, Irvine KM, Brion K, Trieu A, Ross IL, Ravasi T, Stacey KJ, Rehli M, Hume DA, Sweet MJ. PU.1 and ICSBP control constitutive and IFN-gamma-regulated Tlr9 gene expression in mouse macrophages. J Leukoc Biol. 2007;81:1577–1590. doi: 10.1189/jlb.0107036. [DOI] [PubMed] [Google Scholar]
- 53.Eklund EA, Jalava A, Kakar R. PU.1, interferon regulatory factor 1, and interferon consensus sequence-binding protein cooperate to increase gp91(phox) expression. J Biol Chem. 1998;273:13957–13965. doi: 10.1074/jbc.273.22.13957. [DOI] [PubMed] [Google Scholar]
- 54.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]