Abstract
We evaluated the roles of five single-nucleotide polymorphisms (SNPs) within PDCD1, and haplotypes defined by these SNPs, for the development of systemic lupus erythematosus (SLE) and specific sub-phenotypes (nephritis, antiphospholipid antibody positive, arthritis and double-stranded DNA positive) within a multiethnic US cohort of 1036 patients. Family based analyses were performed using 844 simplex families from four ethnic groups (Caucasian, Asian, Hispanic and African American). Subjects were genotyped for five ‘tag’ SNPs (selected from 15) to provide complete genetic information in all main ethnic groups. We employed transmission disequilibrium testing to assess risk for SLE by allele or haplotype, and multiple logistic regression analysis of SLE cases to examine associations with specific sub-phenotypes. In family based analyses, a haplotype containing the PD1.3A allele was significantly associated with SLE susceptibility among Caucasian families (P = 0.01). Among Hispanic families, two novel SNPs were associated with SLE risk (P = 0.005 and 0.01). In multivariate logistic regression analyses, five haplotypes were associated with specific sub-phenotypes among the different ethnic groups. These results suggest that PDCD1 genetic variation influences the risk and expression of SLE and that these associations vary according to ethnic background.
Keywords: systemic lupus erythematosus, PDCD1, family-based methods, haplotypes
Introduction
Systemic lupus erythematosus (SLE) is one of the most clinically and serologically diverse of the chronic autoimmune diseases. Previous studies of patients with SLE have suggested a significant genetic component in the pathogenesis. Several genome-wide screens support the genetic hypothesis in human SLE, and have reported many susceptibility loci as reviewed by Tsao.1 One of the regions demonstrating suggestive linkage is within the long arm of chromosome 2 at 2q37.2 Further mapping of this region has identified a locus, SLEB2, involved in susceptibility to SLE.3
The SLEB2 locus contains a candidate gene, PDCD1, which might provide the biological explanation for linkage at this locus. Nishimura et al.,4,5 have demonstrated that disruption of the PDCD1 gene in mice leads to the development of a lupus-like syndrome, with the development of nephritis and arthritis as well as the production of IgG3 autoantibodies.4,5 The first human study identified an allelic variant in the PDCD1 gene that was associated with susceptibility to SLE in both European and Mexican family cohorts.6 This variation involves a G to A change (PD1.3A/G) in intron 4 of the PDCD1 gene.6 Further investigation of this gene variant has shown positive association in case–control analyses with lupus nephritis (LN) in two separate Swedish cohorts7,8 but not in a US cohort of European American descent.7 Furthermore, a bi-ethnic (African American and European American) case–control cohort revealed a protective association of the PD1.3A allele with presence of antiphospholipid antibodies (APLAs).9 This same cohort failed to show an association between PD1.3A and SLE susceptibility.
We have now analyzed 15 single-nucleotide polymorphisms (SNPs) within PDCD1 in order to: (i) determine allele and haplotype frequencies and haplotype structure of the gene and (ii) select a number of ‘tag’ SNPs that can be used for association studies with SLE and its sub-phenotypes in different ethnic populations. With the determination that five tag SNPs could informatively define the genetic structure of our multiethnic US cohort, we typed these five SNPs among our SLE simplex family cohort and performed transmission disequilibrium testing to evaluate association with SLE susceptibility. Finally, as genetic linkages among SLE cohorts often have been more striking when assessed according to specific clinical phenotype,10 we further characterized the role of PDCD1 gene polymorphism by SLE clinical phenotypes. For these analyses, we conducted a nested case–control study within our multiethnic US cohort to evaluate the role of PDCD1 in the development of LN, APLA positivity, arthritis and double-stranded DNA (dsDNA) antibody positivity. We chose to focus on these specific phenotypes based on features of the mouse model in which PDCD1 is disrupted.4,5
Results
Identification of tag SNPs and haplotype construction
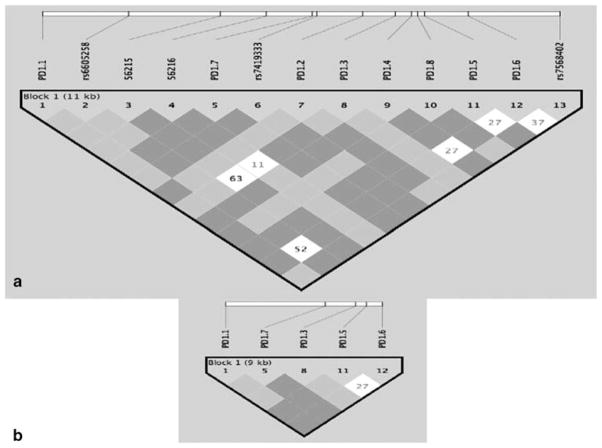
Within 10 kb containing the entire PDCD1 gene, we studied 15 SNPs for which we have determined allele frequencies in Swedish and Mexican population controls (Table 1 and Figure 1a). Using the ‘Tagger’ routine of Haploview 3.2, we selected seven groups of SNPs with linkage disequilibrium (LD) of R2>0.7 within each group and R2<0.7 between SNPs in different groups (Table 2). SNP PD1.9 (Tag group 5, Table 2) was excluded because it defined the same haplotype as SNPs PD1.5T and PD1.6A together and was difficult to genotype (by sequencing). Assays C_56215 and C_56216 (Tag group 7 in Table 2) were excluded because they defined a very rare haplotype with a frequency of <2% in all populations. Therefore, by using only five ‘tag’ SNPs, we could obtain similar information about all haplotypes as we could with 15 SNPs (Figure 1b). From each of the groups having similar ‘tag’ SNPs, we selected one SNP that was the most convenient for genotyping. The five ‘tag’ SNPs and the six corresponding haplotypes with frequencies > 2% are presented in Table 3.
Table 1.
PDCD1 SNPs studied
| Number | Marker name and alleles | Applied biosystems assay | rs number | Position on chromosome 2, bp (build 35) | Sequence | Location | Allele | Frequency in Swedish controls, % | Frequency in Mexican controls, % |
|---|---|---|---|---|---|---|---|---|---|
| 1 | PD1.1A/G | — | 112787 | GGATGGGCCG/AGGAAGGCAG | Promoter | A | 1.0 | 30.4 | |
| 2 | C/T | C_26891642 | rs6605258 | 117750 | CTGGCTGCCC/TGCTGGCCCTG | Intron 1 | T | 37 | 32 |
| 3 | G/A | C_56215 | — | 118468 | AATCACCCCG/ATTGCATGGCA | Intron 1 | G | 33 | 87 |
| 4 | C/G | C_56216 | — | 118563 | GCGGGTATCC/GCAAGCCCTG | Intron 1 | C | 33 | 87 |
| 5 | PD1.7T/C | C_26891639 | rs7421861 | 119060 | AGAGCCAGGT/CGCTGTGCAGA | Intron 1 | T | 80.1 | 59.4 |
| 6 | C/G | C_26891638 | rs7419333 | 119098 | AAGGAAGAGC/GCTCTGCAGTG | Intron 1 | G | 80.1 | 59.4 |
| 7 | PD1.2A/G | — | 119804 | ATGGTGACCG/AGCATCTCTGT | Intron 2 | A | 1.0 | 30.4 | |
| 8 | PD1.10C/G | — | 120232 | TAAGACCCCC/GCACCTAGGAG | Intron 3 | G | 6.9 | NA | |
| 9 | PD1.3A/G | rs11568821 | 120512 | CCCACCTGCG/AGTCTCCGGGG | Intron 4 | A | 7.5 | 2.5 | |
| 10 | PD1.9C/T | — | 120575 | CCTGGCCCAC/TGCGTGTGCCC | Intron 4 | T | 7.5 | 22.1 | |
| 11 | PD1.4A/G | rs6705653 | 120865 | AAGCAGCTCG/ATAGTGGGGGG | Intron 4 | G | 55.7 | 64.3 | |
| 12 | PD1.8C/T | — | 120991 | ACCCCTCAGC/TCGTGCCTGTG | Exon 5. Ala-Val | T | 1.0 | 30.0 | |
| 13 | PD1.5C/T | rs2227981 | 121151 | GGGCTCAGCC/TGACGGCCCTC | Exon 5. Ala-Ala | C | 55.7 | 64.3 | |
| 14 | PD1.6G/A | C_172862 | rs10204525 | 122103 | GCCCCCCATG/ATGCCCACCCT | 3′UTR | A | 6.5 | 52.5 |
| 15 | T/C | C_26891643 | rs7568402 | 122866 | GTCCCTCCCT/CAAGTACACAG | 3′UTR | T | 59.4 | 31.3 |
Figure 1.
(a) LD structure of PDCD1 gene in 16 Mexican trios; (b) LD structure using six tag SNPs. Figure generated using the Haploview program.
Table 2.
PDCD1 ‘tag’ groups
| ‘Tag’ group | Markers in the groupa |
|---|---|
| 1 | PD1.1, PD1.2, PD1.8 |
| 2 | PD-1.7, C_26891638 |
| 3 | PD1.3 |
| 4 | PD1.6 |
| 5 | PD1.9 |
| 6 | PD1.5, PD1.4, C_26891643, C_26891642 |
| 7 | C_56215, C_56216 |
Abbreviations: LD, linkage disequilibrium; SNPs, single-nucleotide polymorphisms.
Each group is represented by one or several SNPs that are in high LD with each other (R2>0.7), meaning that one SNP in each group can be used to represent the entire group. The first marker from each group was used because of genotyping convenience. Marker PD1.9 was omitted in current analyses because it defined the same haplotype as PD1.5 and PD1.6 together and was difficult to genotype (by sequencing). Haplogroup 7 defined by markers C_56215, C_56216 was infrequent (<2%) in all ethnic groups and was not analyzed further.
Table 3.
‘Tag’ SNPs and corresponding haplotypes within the PDCD1 gene
| ‘Tag’ SNPs | ||||
|---|---|---|---|---|
| PD1.1 | PD1.7 | PD1.3 | PD1.5 | PD1.6 |
| G | C | A | C | G |
| G | C | G | C | G |
| G | T | G | C | G |
| G | T | G | T | G |
| G | T | G | T | A |
| A | T | G | C | A |
Of note, Haplotype 5 shows an interesting gradient of frequency in unaffected individuals, from 63% in Mexican Amerindians (Mazateco area in the State of Guerrero kindly provided by Dr Julio Granados Arriola) to 49% in Chinese,11 30% in Mexicans,6 19% in Hispanics, 5% in African Americans and 1% in Europeans (current data).
LD patterns and blocks in the PDCD1 gene
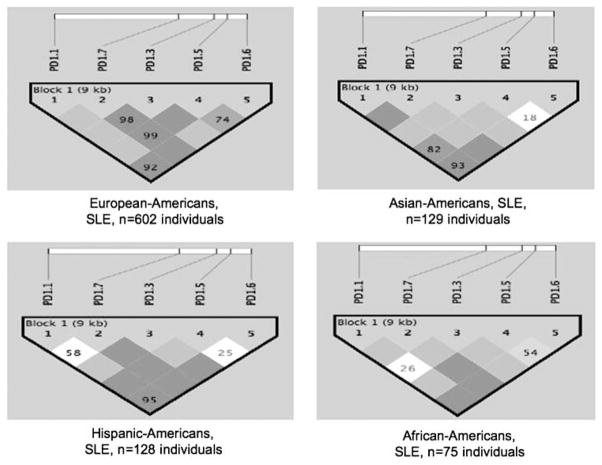
As we have established, five SNPs within 10 kb of the PDCD1 gene are needed to detect six major haplotypes in any population studied (Caucasian, African American, Asian or Hispanic) (see Figure 2). This means that LD blocks in the PDCD1 gene are very short, only 1–2 kb each, and that a very dense SNP map would be needed in this region to detect a predisposing genetic variant.
Figure 2.
LD structure of PDCD1 gene using six tag SNPs in SLE patients from four populations studied. Figure generated using the Haploview program.
Alleles and haplotypes of the PDCD1 gene in a large multi-ethnic group of SLE families
Table 4 displays the demographic and clinical characteristics of the 1036 American SLE patients (from 1011 nuclear families). Our cohort was 90% female subjects and the ethnic breakdown was 61% Caucasian, 13% Hispanic, 13% Asian, 8% African American and 5% others, reflecting the San Francisco population demographics, where the majority of our cohort was sampled from. The percentages of patients with LN, dsDNA antibodies, APLAs and arthritis are comparable with those reported for other general SLE cohorts.12–14 Table 5 presents the PDCD1 SNP minor allele and haplotype frequencies for the SLE patients by ethnic subgroup. These results reveal substantial ethnic variation in PDCD1 allele and haplotype frequencies. Both 4-marker and 5-marker haplotypes were evaluated. The 5-marker haplotypes include marker PD1.7, which was less effectively typed and, therefore, many cases or family members had missing data for this marker. For our ethnic subgroups with fewer family members typed for PD1.7, the related 4-marker haplotypes are reported instead.
Table 4.
Demographic and clinical characteristics of 1036 SLE patients
| Characteristic | N (%) |
|---|---|
| Sex, female | 937 (90.4) |
| Ethnicity | |
| Caucasian | 628 (60.7) |
| Hispanic | 138 (13.3) |
| Asian | 137 (13.2) |
| African American | 82 (7.9) |
| Other | 50 (4.8) |
| Age at diagnosis of SLE, mean in years (s.d.) | 31.0 (12.6) |
| Disease duration, mean in years (s.d.) | 8.5 (8.0) |
| Clinical phenotype | |
| Lupus nephritis | 304 (29.3) |
| dsDNA antibodies | 529 (51.1) |
| APLA | 314 (30.4) |
| Arthritis | 635 (61.3) |
Abbreviations: APLA, antiphospholipid antibodies; dsDNA, double-stranded DNA; s.d., standard deviation; SLE, systemic lupus erythematosus.
Table 5.
PDCD1 SNP minor allele and haplotype frequencies among SLE patients, according to ethnicity. Cells show proportions and frequencies (counts)
| SNP minor allele frequency | Caucasian (N = 1222)a | Hispanic (N = 268) | African American (N = 160) | Asian (N = 274) |
|---|---|---|---|---|
| PD1.1A | 0.01 (15) | 0.19 (50) | 0.06 (9) | 0.36 (98) |
| PD1.3A | 0.10 (120) | 0.06 (14) | 0.02 (3) | 0.008 (2) |
| PD1.5T | 0.41 (482) | 0.44 (110) | 0.43 (67) | 0.35 (90) |
| PD1.6A | 0.09 (107) | 0.42 (103) | 0.26 (37) | 0.59 (150) |
| PD1.7C | 0.34 (299) | 0.15 (25) | 0.20 (20) | 0.10 (17) |
| 4-Marker haplotypeb | Caucasian (N = 1086) | Hispanic (N = 212) | African American (N = 128) | Asian (N = 216) |
| 1 | 0.10 (113) | 0.05 (11) | 0.02 (3) | 0.009 (2) |
| 2 | 0.47 (512) | 0.32 (67) | 0.51 (65) | 0.25 (55) |
| 3 | 0.33 (358) | 0.18 (38) | 0.21 (27) | 0.11 (23) |
| 4 | 0.08 (90) | 0.26 (55) | 0.20 (26) | 0.22 (48) |
| 5 | 0.01 (11) | 0.18 (39) | 0.05 (6) | 0.35 (76) |
| 6 | 0.002 (2) | 0.009 (2) | 0.008 (1) | 0.06 (12) |
N is total number of chromosomes. Cells show percentages.
Haplotype 1 (PD1.1G/PD1.3A/PD1.5C/PD1.6G); 2 = (PD1.1G/PD1.3G/PD1.5C/PD1.6G) serves as referent haplotype with all common alleles; 3 = (PD1.1G/PD1.3G/PD1.5T/PD1.6G); 4 = (PD1.1G/PD1.3G/PD1.5T/PD1.6A); 5 = (PD1.1A/PD1.3G/PD1.5C/PD1.6A); 6 = all other haplotypes.
Variants of PDCD1 and SLE risk: Family Based Association Tests
We used the Family Based Association Tests (FBAT) software to evaluate individual PDCD1 SNPs and associated haplotypes with SLE susceptibility. Table 6 displays PDCD1 allelic and haplotypic associations with SLE. When all subjects were analyzed together irrespective of ethnic background, the haplotype that includes the minor allele, PD1.3A, was associated with susceptibility to SLE with borderline significance (P = 0.05). This result probably is driven by the larger number of Caucasian families included. Evaluation of the Caucasian families separately showed the same result with a more significant P-value (P = 0.01). The PD1.3A allele itself showed a trend toward association (P = 0.08) among the Caucasian families. Among the Hispanic families, the SNPs PD1.7T (P = 0.005) and PD1.6A (P = 0.01), and the associated 4-marker haplotype including PD1.6A (P = 0.07), were associated with SLE susceptibility. There were no statistically significant results among the Asian and African American families, but our power for these ethnic subgroups is limited and therefore we cannot rule out false negative results.
Table 6.
PDCD1 allelic and haplotypic associations with SLE based on FBAT
| Subgroup | Allele/haplotypea | Number of informative familiesb | Z score | P-value | Model type |
|---|---|---|---|---|---|
| All subjects | |||||
| SLE | GCACG | 93 | 1.99 | 0.05 | Additive |
| Caucasian | |||||
| SLE | GCACG | 76 | 2.47 | 0.01 | Additive |
| PD1.3A | 102 | 1.74 | 0.08 | Dominant | |
| Hispanic | |||||
| SLE | PD1.6A | 41 | 2.46 | 0.01 | Additive |
| PD1.7T | 26 | 2.78 | 0.005 | Additive | |
| AGCA | 27 | 1.82 | 0.07 | Dominant | |
Abbreviations: FBAT, Family Based Association Testing; SLE, systemic lupus erythematosus.
5-Marker haplotype listed in order: PD1.1/PD1.7/PD1.3/PD1.5/PD1.6; 4-marker haplotypes exclude the PD1.7 allele.
Total number of families analyzed = 844 (575 Caucasian, 108 Hispanic, 105 Asian, 56 African American).
Variants of PDCD1 and clinical manifestations of SLE
We chose to evaluate associations between clinical subphenotypes and PDCD1 variants among all SLE cases regardless of the availability of family members, as that would allow for the largest possible sample size and therefore maximize our power for these analyses. Each clinical phenotype was characterized as a dependent bivariate variable. As the variables sex, age at diagnosis and disease duration were associated with one or more of the clinical outcomes (data not shown), separate logistic regression models for each ethnic group were created to characterize the association between each of the minor alleles (PD1.1A, PD1.3A, PD1.5T and PD1.6A) with the clinical phenotype, controlling for these variables. Similar logistic regression models were evaluated with the haplotypes included as a multiple categorical variable, with the referent haplotype containing all the major alleles.
There was no particular SNP that was consistently associated with one or more clinical outcomes across all ethnic groups (data not shown). Table 7 presents the statistically significant haplotypic associations with SLE clinical subtypes stratified by ethnic group. There were no allele-level associations that were not captured by the haplotypic associations, and in all cases, the haplotypic associations were stronger despite having less power to detect them. Notably, the PD1.3A allele and its associated haplotype were not associated with any clinical phenotype among any of the ethnic subgroups (data not shown).
Table 7.
Significant PDCD1 haplotypic associations with SLE clinical subtypes in multivariate logistic regression analyses
| Subgroup | Haplotypea | ORb | 95% CI |
|---|---|---|---|
| Caucasian | |||
| APLA | GGTG | 1.50 | 1.09–2.06 |
| Hispanic | |||
| dsDNA | GGTA | 0.37 | 0.16–0.84 |
| Asian | |||
| LN | AGCA | 0.34 | 0.11–0.99 |
| Arthritis | GGTA | 2.35 | 1.03–5.36 |
| African American | |||
| APLA | GGTA | 3.49 | 1.04–11.74 |
Abbreviations: APLA, antiphospholipid antibody positive; 95% CI, 95% confidence interval; dsDNA, double-stranded DNA; LN, lupus nephritis; OR, odds ratio; SLE, systemic lupus erythematosus.
4-marker haplotypes evaluated (PD1.1/PD1.3/PD1.5/PD1.6).
Covariates include sex, age at diagnosis and disease duration in years.
Among Caucasians, Haplotype 3 (PD1.1G/PD1.3G/PD1.5T/PD1.6G) was associated with the presence of APLAs (odds ratio (OR) = 1.5, 95% confidence interval (CI): 1.09–2.06). Even though we had more than 90% power to detect an OR of 2.0 or greater among the Caucasian subgroup (n = 628), no other haplotypes were associated with clinical outcomes in these patients.
The Hispanic subgroup of patients showed a protective association between Haplotype 4 (PD1.1G/PD1.3G/PD1.5T/PD1.6A) and the presence of dsDNA antibodies (OR = 0.37, CI: 0.16–0.84). Our Hispanic patients are predominantly of Mexican heritage. To our knowledge, this is the first association demonstrated between dsDNA antibodies and any PDCD1 variant among Mexicans.
The same Haplotype 4 was associated with the presence of APLAs among the African American patients (OR = 3.49, CI: 1.04–11.74). As we have the fewest number of African American patients in our cohort, we had less than 50% power to detect associations with ORs smaller than 2.0. Thus, we cannot rule out the possibility that other haplotypes may have weaker associations with other clinical phenotypes among African Americans.
Among our Asian patients, there were two significant haplotypic associations. Haplotype 4 was associated with arthritis (OR = 2.35, CI: 1.03–5.36). Haplotype 5, common among Asian patients, was protective for LN (OR = 0.34, CI: 0.11–0.99). This haplotype includes PD1.1A, which was uncommon in all subgroups except for the Asian patients.
Discussion
The PDCD1 PD1.3A allele has been associated with SLE susceptibility in Swedish, Mexican and European American cohorts,6,7 whereas it has been shown to be protective for SLE among a Spanish cohort,15 and not associated with SLE susceptibility among a biethnic (African American and Caucasian) US cohort.9 The PD1.3A allele has also been positively associated with susceptibility to LN among Swedish Europeans7,8 and negatively associated with APLA presence.9 As it appears that PD1.3A does not explain all the PDCD1 associations in SLE, we evaluated additional polymorphisms within PDCD1 for susceptibility to SLE and clinical sub-phenotypes.
We have analyzed a total of 15 SNPs, covering on average one SNP per 670 bp of the PDCD1 gene. We examined the distribution of these SNPs and their haplotypes in several populations (data not shown). We find that the size of the LD blocks in PDCD1 is rather small (1–2 kb), suggesting that in order to identify susceptibility variants in this gene, a very dense SNP map is required. We also identified five SNPs that can provide non-redundant information about haplotypes in any population and can be used as ‘tags’ for association studies.
In the current study, using family-based association methods, we detected a weak association for the PD1.3A allele among European American families (P = 0.08, dominant model; P = 0.11, additive model). In addition, the 5-marker haplotype containing PD1.3A showed an even stronger association with SLE risk (P = 0.01, additive model). The fact that we have not detected any other associations indicates that there may be no other disease variants of similar frequency/relative risk within this gene in Europeans, but it does not exclude the possible existence of additional rare, weak and/or recessive mutations that we were not able to detect using the current data set.
We can exclude PD1.3A as a significant disease variant in non-European populations because of its extreme rarity and acknowledge that additional studies will be needed to clarify the contribution of PDCD1 to disease in these populations using both families and case/control analyses. However, we did observe an association between two other PDCD1 SNPs (PD1.6A and PD1.7T) and SLE susceptibility among Hispanic families. To our knowledge this is a novel finding and should encourage others to evaluate these variants in Hispanic populations.
In our evaluation of PDCD1 and SLE sub-phenotypes, we have identified several haplotypic associations. Notably, Haplotype 5 containing the PD1.1A allele located in the promoter region of the gene (position −531 from the translation start) was much more frequent among the Asian American patients, and it conferred a protective effect for LN in this subgroup. This haplotype has also been recently demonstrated to be associated with decreased levels of tumor necrosis factor alpha.16 Haplotype 4 was positively associated with arthritis among Asian Americans and with APLA among African American patients, whereas it was negatively associated with dsDNA antibodies among the Hispanic American patients. Among the Caucasian patients, Haplotype 3 was marginally associated with APLAs. These novel findings should be viewed as specific hypotheses to be confirmed in independent cohorts.
Although strengths of the current study include the ethnically diverse cohort and the relatively large number of non-Caucasians overall, we still had reduced power to detect genetic associations among the smaller ethnic subgroups and for the less frequent alleles/haplotypes. Another strength of the study is the family-based design, which minimizes problems with population admixture.
In conclusion, we have carefully evaluated 15 SNPs in the PDCD1 gene to further define its role in SLE risk and disease expression. The knowledge of haplotype structure in different populations allowed us to select five SNPs that can be used as ‘tags’ for association studies in any population. We have used these ‘tags’ to study a large multi-ethnic set of SLE families. We have shown that a 5-marker haplotype containing PD1.3A is associated with SLE in European Americans using family-based association methods. We also identified haplotypic associations for clinical SLE subphenotypes among Asian Americans, African Americans and Hispanic Americans.
The new haplotypic associations identified herein provide further evidence that PDCD1 is an important gene for expression of SLE. The associations suggest either a true haplotypic effect or alternatively, indicate that another polymorphism within or near PDCD1 may be causal for disease apart from PD1.3A. The fact that haplotype associations are stronger than allele associations and that the haplotype associations vary by ethnic group, suggest there may be additional causal variants that have not yet been identified.
Patients and methods
Patients
For the association studies with SLE and its sub-phenotypes (development of LN, APLA positivity, arthritis and dsDNA antibody positivity), we studied 1036 American SLE patients (from 1011 nuclear families). Four ethnic subgroups were analyzed: European American, Hispanic American, African American and Asian American (Table 4). Ethnicity was determined based on grand-parental countries of origin. Informed consent was obtained from all participants and the study was approved by the institutional review board at the University of California, San Francisco (UCSF). Patients were participants in the UCSF Lupus Genetics Project and were recruited from UCSF Arthritis Clinics and private rheumatology practices in northern California, as well as by nationwide outreach. Study enrollment for SLE patients includes completion of a 10-page questionnaire about her/his family structure and SLE history, review of medical records to document diagnoses and clinical manifestations, and collection of blood or buccal cells for DNA analysis. Medical records are reviewed to verify the presence of APLAs on at least one occasion in the subset of patients with these antibodies, the presence of dsDNA antibodies, and the presence of arthritis diagnosed by a clinician. A diagnosis of LN was made for patients who met the American College of Rheumatology (ACR) criteria for LN17 or who had evidence of LN by kidney biopsy.
Additional subjects were used in our investigation of haplotypes (see below). Specifically, we studied 16 Mexican trios, 96 Mexican controls and 96 Swedish controls. The Swedish controls were blood donors sampled in Uppsala, Sweden and are of Swedish ancestry for two generations and have been described earlier.6 Mexican controls were from the general population of Mexico City and may be considered as Mestizos, or admixed between Caucasians and Mexican Indians.6,18
SNP selection and genotyping
We evaluated 15 SNPs, including 10 that were identified previously by sequencing of DNA samples of patients and controls6 and five additional SNPs located in intron 1 of the PDCD1 gene, not previously genotyped in multiple individuals (Table 1). SNPs were genotyped either by sequencing, Dynamic Allele Specific Hybridization (DASH), or Pyrosequencing as described previously6 or using the TaqMan Allelic Discrimination Assays of ABI as recommended.
Haplotype determination
As the PDCD1 gene is not yet fully represented in public databases such as HapMap,19 Perlegen20 or the UCSC Genome Browser (http://genome.ucsc.edu), we determined the haplotype structure of the gene before performing association analyses. Thus, we first genotyped 17 SNPs in 16 Mexican trios. We chose Mexican trios as both European and Amerindian (partly Asian) haplotypes are present in this population. We also genotyped 96 Swedish and 96 Mexican controls for all SNPs to estimate their allele frequencies and confirm haplotype structure (Table 1).
Two SNPs were excluded owing to poor genotyping performance (assay C_26891641_10, Applied Biosystems, Foster City, CA, USA) or unresolved genotyping errors (assay C_56214_10, rs7603052). Haplotypes were constructed with Haploview 3.219 and confirmed using PHASE 2.021,22 and by consistency of transmission in nuclear families. Tag SNPs were identified using the ‘Tagger’ application of Haploview 3.2.19
Statistical analyses
Disease risk association
To study the risk of SLE, the transmission of alleles or haplotypes from parents to affected offspring was analyzed using the FBAT.23 These analyses were performed both using all families (n = 844) and among ethnic specific strata (European American, Hispanic American, African American and Asian American). One advantage of the FBAT compared with many other transmission disequilibrium tests is that it can include multiple family types, in addition to complete trios.
Association with specific SLE phenotypes
To study the risk of each clinical sub-phenotype, we performed a nested case–control study with the SLE patients only (n = 1036 cases from 1011 nuclear families). Initially, univariate analyses were performed to examine the association between the individual PDCD1 alleles and the risk of each clinical sub-phenotype. Further analyses utilized multivariate logistic regression to examine the association among haplotypes and the risk of each clinical phenotype, adjusting for sex, age at diagnosis and disease duration (in years), and stratified by ethnicity. For the allele-level analyses, the major allele was the referent group. For the haplotype-level analyses, the haplotype PD1.1G/PD1.3G/PD1.5C/PD1.6G represented by all major alleles was the referent. The PD1.7 SNP was excluded from these analyses, because it was not genotyped in all samples. All logistic regression analyses were performed using STATA software (College Station, TX, USA). Hardy–Weinberg equilibrium was evaluated with Haploview.19
Power calculations
Power calculations for family based analyses were performed using the PBAT program.24 These calculations considered allele frequencies of 5, 10 and 20%, a population prevalence of 0.005. for SLE, a genetic attributable fraction of 0.1 and a significance level of 0.05. Results were not sensitive to variation in the estimated population prevalence between 0.05 and 0.0005.
Acknowledgments
Mexican Amerindians from the Mazateco area in the State of Guerrero, South Eastern Mexico were kindly provided by Dr Julio Granados Arriola (Department of Immunology and Rheumatology, Instituto Nacional de Ciencias Médicas y Nutrición ‘Salvador Zubirán’, México City, INCMNSZ). This work was supported by NIH R01 AR44804, K24 AR02175, ES09911, T32 DK07219, the Arthritis Foundation and the Mary Kirkland Center for Lupus Research. This work was also supported by grants from the Alliance for Lupus Research (to MEAR), The Swedish Research Council, the Swedish Association Against Rheumatism, the Magnus Bergwalls Foundation, The Gustaf V:e 80-Year Jubilee Foundation, the Marcus Borgströms Foundation and the Torsten and Ragnar Söderbergs Foundation. MEAR is a Research Fellow from the Royal Swedish Academy of Sciences supported by a grant from the Knut and Alice Wallenberg Foundation, Sweden. These studies were performed in part in the General Clinical Research Center, Moffitt Hospital, University of California, San Francisco, with funds provided by the National Center for Research Resources, 5 M01 RR-00079, US Public Health Service.
References
- 1.Tsao BP. Update on human systemic lupus erythematosus genetics. Curr Opin Rheumatol. 2004;16:513–521. doi: 10.1097/01.bor.0000132648.62680.81. [DOI] [PubMed] [Google Scholar]
- 2.Lindqvist A-KB, Steinsson K, Johanneson B, Kristjansdottir H, Arnasson A, Grondal G, et al. A susceptibility locus for human systemic lupus erythematosus (hSLE1) on chromosome 2q. J Autoimmun. 2000;14:169–178. doi: 10.1006/jaut.1999.0357. [DOI] [PubMed] [Google Scholar]
- 3.Magnusson V, Lindqvist AK, Castillejo-Lopez C, Kristjansdottir H, Steinsson K, Grondal G, et al. Fine mapping of the SLEB2 locus involved in susceptibility to systemic lupus erythematosus. Genomics. 2000;70:307–314. doi: 10.1006/geno.2000.6374. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura H, Minato N, Nakano T, Honjo T. Immunological studies on PD-1 deficient mice: implication of PD-1 as a negative regulator for B cell responses. Int Immunol. 1998;10:1563–1572. doi: 10.1093/intimm/10.10.1563. [DOI] [PubMed] [Google Scholar]
- 6.Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet. 2002;32:666–669. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 7.Prokunina L, Gunnarsson I, Sturfelt G, Truedsson L, Seligman VA, Olson JL, et al. The systemic lupus erythematosus-associated PDCD1 polymorphism PD1.3A in lupus nephritis. Arthritis Rheum. 2004;50:327–328. doi: 10.1002/art.11442. [DOI] [PubMed] [Google Scholar]
- 8.Johansson M, Arlestig L, Moller B, Rantapaa-Dahlqvist S. Association of a PDCD1 polymorphism with renal manifestations in systemic lupus erythematosus. Arthritis Rheum. 2005;52 :1665–1669. doi: 10.1002/art.21058. [DOI] [PubMed] [Google Scholar]
- 9.Sanghera DK, Manzi S, Bontempo F, Nestlerode C, Kamboh MI. Role of an intronic polymorphism in the PDCD1 gene with the risk of sporadic systemic lupus erythematosus and the occurrence of antiphospholipid antibodies. Hum Genet. 2004;115:393–398. doi: 10.1007/s00439-004-1172-0. [DOI] [PubMed] [Google Scholar]
- 10.Nath SK, Kelly JA, Harley JB, Scofield RH. Mapping the systematic lupus erythematosus susceptibility genes. Methods Mol Med. 2004;102:11–29. doi: 10.1385/1-59259-805-6:011. [DOI] [PubMed] [Google Scholar]
- 11.Kong EK, Prokunina-Olsson L, Wong WH, Lau CS, Chan TM, Alarcon-Riquelme M, et al. A new haplotype of PDCD1 is associated with rheumatoid arthritis in Hong Kong Chinese. Arthritis Rheum. 2005;52:1058–1062. doi: 10.1002/art.20966. [DOI] [PubMed] [Google Scholar]
- 12.Pollak VE, Kant KS. Systemic lupus erythematosus and the kidney. In: Lahita RG, editor. Systemic Lupus Erythematosus. John Wiley & Sons; New York: 1987. [Google Scholar]
- 13.Schur P. Clinical features of SLE. In: Kelley WN, Harris ED, Ruddy S, Sledge CB, editors. Textbook of Rheumatology. 3. WB Saunders Company; Philadelphia, PA, USA: 1989. [Google Scholar]
- 14.Merrill JT. Antibodies and clinical features of the antiphospholipid syndrome as criteria for systemic lupus erythematosus. Lupus. 2004;13:869–876. doi: 10.1191/0961203304lu2026oa. [DOI] [PubMed] [Google Scholar]
- 15.Ferreiros-Vidal I, Gomez-Reino JJ, Barros F, Carracedo A, Carreira P, Gonzalez-Escribano F, et al. Association of PDCD1 with susceptibility to systemic lupus erythematosus: evidence of population-specific effects. Arthritis Rheum. 2004;50:2590–2597. doi: 10.1002/art.20436. [DOI] [PubMed] [Google Scholar]
- 16.Bennet AM, Alarcon-Riquelme M, Wiman B, de Faire U, Prokunina-Olsson L. Decreased risk for myocardial infarction and lower tumor necrosis factor-alpha levels in carriers of variants of the PDCD1 gene. Hum Immunol. 2006;67:700–705. doi: 10.1016/j.humimm.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 18.Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21 :263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Hinds DA, Stokowski RP, Patil N, Konvicka K, Kershenobich D, Cox DR, et al. Matching strategies for genetic association studies in structured populations. Am J Hum Genet. 2004;74:317–325. doi: 10.1086/381716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype – phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 24.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]