Abstract
Background
Martian regolith (unconsolidated surface material) is a potential medium for plant growth in bioregenerative life support systems during manned missions on Mars. However, hydrated magnesium sulfate mineral levels in the regolith of Mars can reach as high as 10 wt%, and would be expected to be highly inhibitory to plant growth.
Methodology and Principal Findings
Disabling ion transporters AtMRS2-10 and AtSULTR1;2, which are plasma membrane localized in peripheral root cells, is not an effective way to confer tolerance to magnesium sulfate soils. Arabidopsis mrs2-10 and sel1-10 knockout lines do not mitigate the growth inhibiting impacts of high MgSO4·7H2O concentrations observed with wildtype plants. A global approach was used to identify novel genes with potential to enhance tolerance to high MgSO4·7H2O (magnesium sulfate) stress. The early Arabidopsis root transcriptome response to elevated concentrations of magnesium sulfate was characterized in Col-0, and also between Col-0 and the mutant line cax1-1, which was confirmed to be relatively tolerant of high levels of MgSO4·7H2O in soil solution. Differentially expressed genes in Col-0 treated for 45 min. encode enzymes primarily involved in hormone metabolism, transcription factors, calcium-binding proteins, kinases, cell wall related proteins and membrane-based transporters. Over 200 genes encoding transporters were differentially expressed in Col-0 up to 180 min. of exposure, and one of the first down-regulated genes was CAX1. The importance of this early response in wildtype Arabidopsis is exemplified in the fact that only four transcripts were differentially expressed between Col-0 and cax1-1 at 180 min. after initiation of treatment.
Conclusions/Significance
The results provide a solid basis for the understanding of the metabolic response of plants to elevated magnesium sulfate soils; it is the first transcriptome analysis of plants in this environment. The results foster the development of Mars soil-compatible plants by showing that cax1 mutants exhibit partial tolerance to magnesium sulfate, and by elucidating a small subset (500 vs. >10,000) of candidate genes for mutation or metabolic engineering that will enhance tolerance to magnesium sulfate soils.
Introduction
Long duration human missions to Mars must rely on more than just stored supplies and physico-chemical means to regenerate air and clean water. The Advanced Life Support (ALS) scenarios envisioned for extended manned missions will depend upon the efficient use of local planetary resources and the recycling of limited materials such as water, pressurized atmosphere, and organic matter, while producing food to augment supplies [1]. The use of in situ regolith for plant growth in a future bioregenerative life support system on Mars may have several advantages over hydroponic systems [2], [3]. These include the immediate bioavailability of plant essential ions, low-tech mechanical support for plants, and easy access to in situ materials once on the surface. However, plant growth may be reduced or inhibited by phytotoxic substances in the regolith, such as high levels of soluble magnesium sulfate minerals. Hydrated forms of magnesium sulfate such as MgSO4·7H2O (epsomite) and MgSO4·H2O (kieserite) have been detected in several regions by the Mars Express Satellite [4], [5], [6]. Analyses by the Mars Exploration Rover landers at Meridiani Planum and Gusev crater have also indicated the presence of high levels of magnesium sulfate minerals (up to 10 wt%) in outcrops and soils [7], [8], [9].
High levels of hydrated sulfate minerals in regolith on Mars used in bioregenerative life support systems will lead to exposure of plant roots to supra-optimal concentrations of both Mg2+ and SO4 2− ions in the soil solution. Plants may have evolved to cope with relatively high levels of elements in the soil environment by limiting internal accumulation or tolerating high internal concentrations [10]. In a potential bioregenerative life support system on Mars, an excess of a particular element in the crew's diet could affect the availability of other required elements. This study therefore first determined whether knockout mutant lines for genes encoding certain transporters responsible for uptake of Mg2+ and SO4 2− ions in roots could enhance plant tolerance to high levels of magnesium sulfate in the growth medium and then moved to a molecular analysis of the responses to Mg2+ and SO4 2− in order to increase the potential pool of candidate genes.
Various efforts have previously illustrated that the disabling of transporter genes can indeed improve tolerance to certain elements. For example, a line of transgenic wheat plants expressing an antisense construct of the high affinity K+ transporter TaHKT2;1 showed reduced sodium uptake by roots and enhanced growth relative to unstressed plants compared to a control line at high levels of NaCl in the growth medium [11].
Several plasma membrane localized proteins in the outer cell layers of Arabidopsis roots are known to be responsible for magnesium or sulfate ion uptake. Two specific genes, AtMRS2-10 and AtSULTR1;2, were chosen for this study, in part because of their localization and because of availability of knockout lines. AtMRS2-10 ( = AtMGT1) functionally complemented a bacterial mutant lacking Mg2+ transport capability and AtMRS2-10-GFP–expressing plants showed fluorescence in the periphery of root cells, suggesting a plasma membrane association [12]. AtMRS2-10 is, to date, the only member of the AtMRS2 family that is known to be associated to the plasma membrane of root cells in addition to having Mg2+ ion transport capability. Therefore, a knockout mutant line of the AtMRS2-10 gene (mrs 2-10) was characterized for this study based on the SALK_100361.41.30.x T-DNA insertion line [13]. AtSULTR1;2 is a constitutively expressed sulfate transporter gene whose product AtSULTR1;2 is localized in root hair, epidermal and cortical cells where it ensures sulfate uptake into plants under sulfur-replete conditions [14]. A knockout mutant line of the AtSULTR1;2 gene (sel1-10) was shown to take up 80% less sulfate than wildtype when grown for 12 days on agar with optimal levels (1.7 mM MgSO4·7H2O) of sulfate [15].
Knockout lines of genes encoding vacuolar H+/Ca2+ transporters CAX1 and CAX3 were also analyzed to present perspective of other, non-plasma membrane, cellular transporter locations. The cax1-1 and cax1/cax3 lines were previously shown to be more tolerant of high Mg2+ levels in the form of MgCl2 when grown on agar [16], [17]. In a separate study, a CAX1 knockout mutant was identified through a mutant screen on nutrient solutions reflecting low Ca∶Mg ratios characteristic of serpentine soils [18]. When high MgSO4·7H2O concentrations are applied, the Ca∶Mg ratio of the nutrient solution decreases, which therefore makes the cax1-1 and cax1/cax3 mutants candidates for high magnesium sulfate tolerance. This study determined whether enhanced tolerance to low Ca∶Mg ratios is maintained when cax1-1 and cax1/cax3 mutants are exposed to elevated SO4 2− in addition to elevated Mg2+ in soil medium.
These candidate genes for enhanced magnesium sulfate tolerance that are presented above were selected based on previous literature describing the results of individual gene or gene family analyses. Genome-wide transcriptome analyses of plants exposed to elevated concentrations of magnesium, sulfate, or magnesium sulfate in the growth medium have not been reported to date. In order to increase the number of candidate genes that could play a role in enhancing growth performance of plants exposed to this abiotic stress, root transcriptome remodeling in Col-0 was analyzed after short-term (45 min.) hydroponic exposure to a non-lethal, high concentration of MgSO4·7H2O. This early time point was chosen to capture part of the primary stress responses. The differential expression of transporter genes was analyzed after 90 min. and 180 min. in addition to 45 min., as this group in particular may represent a metabolic strategy for tolerance to an elevated magnesium sulfate environment. The root transcriptome of the transporter gene knockout mutant cax1-1 was furthermore contrasted with that of Col-0 after both genotypes were exposed to MgSO4·7H2O treatment for 180 min. Based on their previously identified tolerance to high MgCl2 and low Ca∶Mg ratios, cax1 mutants are likely to show enhanced growth compared to wildtype when exposed to high magnesium sulfate levels. Furthermore, ionome analyses of cax1 mutants showed that they have significantly reduced levels of Mg2+ in their leaves compared to wildtype [18].The cax1/cax3 double knockout mutant, while having the potential to show tolerance to elevated MgSO4·7H2O based on its tolerance to MgCl2 and its reduced shoot levels of Mg2+, showed significant differences in concentration of multiple elements, and grew more slowly than wildtype under regular nutrient conditions [17]. Since cax1-1 mutants did not display negative effects on growth under regular nutrient conditions, cax1-1 rather than cax1/cax3 was included in the root transcriptome analysis. Genes differentially regulated between cax1-1 and Col-0 grown under stress conditions could reveal those molecular processes possibly leading to significant differences in growth performance at the whole plant level. A hydroponic medium was chosen to mimic root exposure to a specific soil solution in order to directly link transcriptome responses to the level of bioavailable ions that roots may experience in a hypothetical root zone of a regolith profile on Mars [19]. The treatment concentration of MgSO4·7H2O was based on the low Ca∶Mg ratio that can occur in serpentine soils on Earth. Because of their high amount of bioavailable magnesium, Serpentine soils can be seen as a partial analogue for regolith high in magnesium sulfate on Mars [20].
Materials and Methods
Mrs2-10 T-DNA Insertion Line Characterization
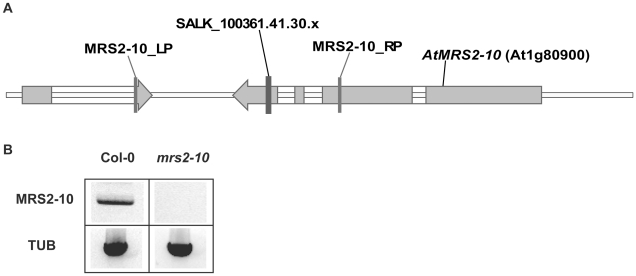
A homozygous knockout T-DNA insertion line for AtMRS2-10 (At1g80900) was identified by PCR and RT-PCR using the original SALK_100361.41.30.x line (Fig. 1a,b). For PCR, oligonucleotide primers MRS2-10_LP (5′-CAGGATCAAAGCATCGTTCTC-3′) and MRS2-10_RP (5′-TAGGAGCTCAGAAGACGCAAC-3′) were designed using software available on the Salk Institute website (http://signal.salk.edu/tdnaprimers.html). In addition, the T-DNA specific primer LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) was included as designed and recommended by the Salk Institute. Combinations of the primers were used to identify plants for which the T-DNA insertion was present in both AtMRS2-10 alleles. Genomic DNA was extracted from T-DNA insertion mutant leaves using the Shorty method. For this method, 500 uL Shorty Buffer (0.2 M Tris-HCL pH 9.0, 0.4 M LiCl, 25 mM EDTA, and 1% SDS) was added to ground leaf tissue (∼0.25 cm2) in a microcentrifuge tube. The tube was then spun for 5 min. at high speed in a microcentrifuge. Supernatant (350 uL) was transferred to a new tube containing isopropanol (350 uL). The two volumes were mixed by inversion and the tube was spun for 10 min. at top speed in a microcentrifuge. After this, the liquid was discarded and the pellet was air-dried before being resuspended in 100 uL TE buffer. Genomic DNA was extracted from Col-0 leaves using the DNeasy Plant Mini Kit (Qiagen). PCR was carried out using JumpStart Taq DNA polymerase (Sigma). PCR products were separated in agarose gels and stained with SYBR Safe DNA gel stain (Invitrogen). A confirmed homozygous T-DNA insertion mutant was backcrossed to Col-0 three times before allowing self-fertilization. Homozygous plants backcrossed 3× were identified by PCR and used for seed generation. For RT-PCR, RNA was extracted from leaves of Col-0 and a homozygous T-DNA insertion line for AtMRS2-10 (At80900) with the RNeasy Plant Mini Kit (Qiagen). Gene specific oligonucleotide primers MRS2-10_LP1 (5′-AGGGTTACTTTGTCGGAGA-3′) and MRS2-10_RP1 (5′-TACACGGGGTTTTATCTTG-3′) were designed based on Arabidopsis genomic DNA sequence information (NCBI). Alpha-(α)-Tubulin was used as a constitutive control with primers according to Yoshimoto (2002). RT-PCR was carried out using the OneStep RT-PCR kit (Qiagen). PCR products were separated in agarose gels and stained with SYBR Safe DNA gel stain (Invitrogen). An ionome analysis of the backcrossed homozygous AtMRS2-10 knockout mutants (mrs2-10) was performed as part of the Purdue Ionomics project and results are available online through the Purdue Ionomics Information Management System (http://www.ionomicshub.org/home/PiiMS) [21].
Figure 1. Homozygous knockout mutant line mrs2-10.
(a) A homozygous T-DNA insertion mutant for AtMRS2-10 (At1g80900) was identified by PCR based on the SALK_100361.41.30.x line. (b) RNA extracted from mrs2-10 and Col-0 leaves was tested for the presence of MRS2-10 mRNA with gene-specific primers. Alpha-(α)-Tubulin (TUB) was used as a constitutive control.
Plant Growth Experiments on Soil Medium
Seeds from the Ws, Col-0, mrs2-10, sel1-10 [15], cax1-1 [16] and cax1/cax3 [17] lines were sterilized by soaking them for 15 minutes in a 40% (v/v) bleach solution with a drop of Tween 20 (polyoxyethylenesorbitanmonolaureate). The seeds were then washed six times with sterilized (autoclaved) water. To control for environmental variation when comparing plant growth per concentration, each tray was divided in 8 sections of 6 wells each, with either a mutant line or its associated wildtype background line planted in one section, in alternating fashion. This method resulted in 4 sections per seed line per tray. The seeds were evenly planted (5 seeds per tray well) on the soil medium (sphagnum peat moss, vermiculite and perlite) that was wetted with nutrient solution. The basic and control nutrient solution consisted of 2.2 g/L Murashige and Skoog salts (Sigma-Aldrich) and 0.5 g/L MES buffer (Sigma-Aldrich). For the treatments, MgSO4·7H2O (Sigma-Aldrich) was added to the basic solution at 20, 60, 80 or 100 mM concentration. The pH of the solutions was adjusted to 5.70–5.75 with KOH. The trays with soil were covered with plastic wrap to maintain sufficient humidity and placed in a cold room at 4°C for three days. After stratification, the trays were moved to a growth room at 23°C where they were randomly placed on the growth benches. There, the seeds germinated, the plastic wrap was removed after 3–5 days, and the surviving plants were thinned to two plants per well (maximum of 12 plants per section) after one week to maintain sufficient spacing. The plants were sub-irrigated twice a week, alternatively with tap water and the appropriate nutrient solution. The plants were analyzed at week 4 and the experiment was repeated three times.
Growth Performance Measurements and Statistical Analysis
Fresh weight biomass of plants grown on soil was measured by grouping and weighing whole shoots of plants per tray section. Leaf chlorophyll content of plants grown on soil medium was measured with a Minolta SPAD-502 meter. One leaf measurement per plant per tray well was taken. Biomass or chlorophyll levels of wildtype and knockout mutant Arabidopsis lines were evaluated in a mixed analysis of variance (ANOVA) model using SAS 9.2 and JMP Genomics 7 software. Genotype and concentration were included in the ANOVA model as fixed effects, while batch (replication set) was included as a random effect. Unbiased estimates of biomass or chlorophyll levels (least-square means) were generated for each effect (genotype, concentration, or genotype and concentration) per experiment. The estimated levels of biomass or chlorophyll were then compared using a series of t-tests, generating estimated differences and corresponding p-values. Differences with associated p-values<0.05 were considered significant. Results of the ANOVA analysis are presented in Table S1.
Hydroponic Root Growth
The hydroponic set-up for Arabidopsis that was used in this study is based on two previously described systems [22], [23]. Glass containers (2.6 L) were covered with opaque black plastic sheeting along the glass, and with opaque plastic lids across the top. In each opaque plastic lid, 18 holes (d = 1.43 cm) were made at regular intervals with a hole puncher. A 2-cm-long Rockwool (Grodan) plug (d = 1.59 cm) was placed in each hole. Containers were filled with a nutrient solution adjusted to pH 5.7 by KOH that was replaced once a week. It consisted of distilled water, 0.25 g/L MES buffer, and 1/32x MS salts during week 1 and 2, while the concentration of MS salts increased to 1/16x during week 3. Arabidopsis seeds were sterilized and stratified before planting about ten seeds per Rockwool plug. Col-0 seeds were planted on 18 containers while cax1-1 seeds were planted on 6. Containers were covered with transparent plastic wrap during the first 3–5 days of germination. Holes were made in the plastic wrap after 1–3 days depending on the general atmospheric humidity. The containers were randomly placed over 3 benches (8 containers/bench). Air was supplied to the roots by one 3W air pump per 8 containers; each container had an airstone made from glass beads. After one week, seedlings were thinned to one seedling per Rockwool plug, so that every container supported 18 plants (Fig. 2a). At day 21, Col-0 and cax1-1 roots were exposed to the basic nutrient solution (0.25 g/L MES, 1/16x MS, pH 5.7) with an additional 2.08 mM magnesium sulfate (total Ca∶Mg ratio = 1∶15). Col-0 was exposed for 45 min., 90 min., or 180 min., while cax1-1 was exposed for 180 min. only. The Col-0 control set received no extra magnesium sulfate and was harvested at 45 min. together with the first Col-0 treatment set. Four replicate containers were harvested for the control and each of the treatment sets (Fig. 2b). Roots were cut below the Rockwool plug and pooled per container before being flash frozen in liquid nitrogen and stored at −80°C.
Figure 2. Overview of hydroponic Arabidopsis growth and root harvest.
(a) Set-up for the microarray experiment. (b) Example of Arabidopsis roots after 21 days of growth, and of root harvest per container for the microarray and Q-PCR experiments.
Microarray Procedures, Statistical Analysis and Data display
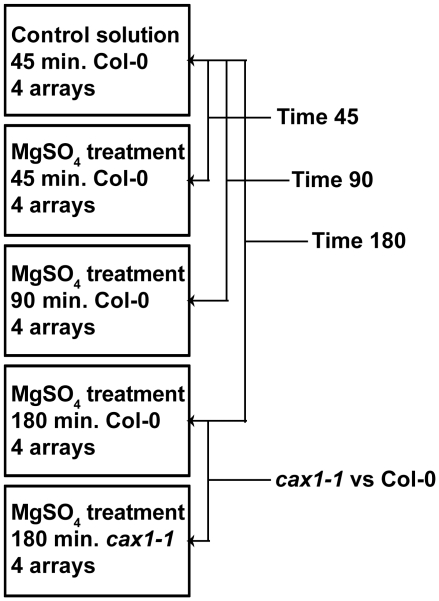
RNA was extracted from root samples with the RNeasy Plant Mini Kit (Qiagen). Samples were weighed while still frozen and reagents were adjusted to the total measured weight for the grinding and lysing step. Two replicate extractions were completed per sample with lysate volumes corresponding to 100 mg frozen wet weight each. The remainder of a sample was stored at −80°C as cleared lysate according to the Qiagen protocol. The quality and quantity of the extracted RNA was checked with denaturing agarose gels stained with ethidium bromide and the NanoDrop 1000 Spectrophotometer (Thermo Scientific). RNA from each sample was amplified and labeled with cy3 dye by using Agilent Quick Amp labeling kit, one color. A total of 20 samples were hybridized to five 4×44k Arabidopsis microarray slides (Agilent), which were then washed and scanned before data was extracted. Microarray handling and data extraction was done at the Interdisciplinary Center for Biotechnology Research (ICBR) at the University of Florida according to the One-Color Microarray-Based Gene Expression Analysis (Quick Amp Labeling) Protocol (version 5.7). Median signal intensities were quantile normalized using R software. Log2 transformed normalized data were evaluated using a mixed analysis of variance (ANOVA) model [24], [25] implemented in SAS 9.2 that included the fixed effect of each genotype and time of tissue harvest. Unbiased, least square means estimates of transcript abundance were generated and gene expression measured in each treatment was then compared to a control using a series of t-tests; Col-0 sets treated for 45 min., 90 min., or 180 min were compared to the Col-0 set exposed to a control solution for 45 min., and these three comparisons are referred to in the text, tables and figures as Time 45, Time 90, and Time 180 respectively (Fig. 3). In addition, the cax1-1 set treated for 180 min. was compared to the Col-0 set treated for 180 min. (Fig. 3). The p-values generated in these comparisons were corrected for multiple testing by controlling the False Discovery Rate (FDR) with the Q-value procedure in the Q-value 1.0 package (default settings) of the R software [26]. A default q-value threshold of 0.05 was used to declare a significant difference in gene expression between treatment and control. The results of the ANOVA analysis are presented in Table S2. Lists of genes with statistically significant changes in expression were further organized, analyzed and displayed in tables and figures using JMP Genomics 7, R software, GeneVenn, GOstat, Cluster 3.0, Java TreeView 1.1.4r5, the National Center for Biotechnology Information (NCBI) BLAST tool, and the Arabidopsis Information Resource (TAIR) Motif analysis tool.
Figure 3. Overview of the microarray experiment.
The experiment included 20 arrays in total, each with a single cy3-labeled sample hybridized to it. Each sample set consisted of 4 biological replicates. Gene expression was compared among the sets as indicated by the arrows; Col-0 sets treated for 45 min., 90 min., or 180 min. were compared to the Col-0 set exposed to a control solution for 45 min., and these three comparisons are referred to in the text, tables and figures as Time 45, Time 90, and Time 180 respectively. The comparison of cax1-1 with Col-0 treated for 180 min. is not referred to in abbreviated form.
Quantitative Real-time PCR
Wildtype Arabidopsis plants (Col-0) were grown hydroponically for 21 days as described above. At day 21, four containers received a replacement of the basic nutrient solution (0.25 g/L MES, 1/16x MS, pH5.7), while another four containers received the basic solution with an additional 2.08 mM magnesium sulfate (total Ca∶Mg ratio = 1∶15). Roots of treatment and control plants were exposed for 180 min. before being harvested per container. RNA was extracted and checked for quality and quantity as described above. RNA from these independently grown plants exposed or unexposed for 180 min., as well as RNA from plants exposed for 180 min. and unexposed for 45 min. in the microarray experiment, were used for the quantitative PCR (Q-PCR) analyses. Q-PCR was performed using TaqMan reverse transcription reagents and Power SYBR Green PCR Master Mix (Applied Biosystems). 1 ug of total RNA was reverse transcribed per sample for a total of three control and three treatment samples. Primers were designed for 5 genes using Primer Express (Applied Biosystems) (Table 1). At2g32170 was chosen as a stably expressing reference gene appropriate for abiotic stress treatment [27]. Each template/primer pair combination was run in triplicate. The relative increase or decrease of mRNA abundance between the two sample sets was calculated by using the Pfaffl method, and statistical analysis of the results was done with the REST 2008 2.0.7 software.
Table 1. Genes and related primer sequences selected for Q-PCR analysis.
| Gene ID | Primer sequence |
| At2g32170 (reference gene) | FW: 5′-GTTAAATCATGACCATGGCAGTGT-3′ |
| RV: 5′-CTACATCAACCAGAGGAACATGTGT-3′ | |
| At2g38170 (CAX1) | FW: 5′-GCGACTCAGATTGGCTTATTCG-3′ |
| RV: 5′-GATCCATATTAATTCCCAAAATCCA-3′ | |
| At1g80900 (MRS2-10) | FW: 5′-TTCTCTGTCTGCGCCAGTTTC-3′ |
| RV: 5′-GGCTCCTTACAATGCTCAAGCT-3′ | |
| At3g15990 (SULTR3;4) | FW: 5′-GGTGAAGCTGTGGCTGATCTC-3′ |
| RV: 5′-GCTCCATCTTCAGAAACAGTCTCTCT-3′ | |
| At1g80830 (NRAMP1) | FW: 5′-ACAGGATCTGGACGGTCTCAA-3′ |
| RV: 5′-GATGAGTGGAGAATTGGAGAAGCT-3′ |
Results
Plant Growth Performance
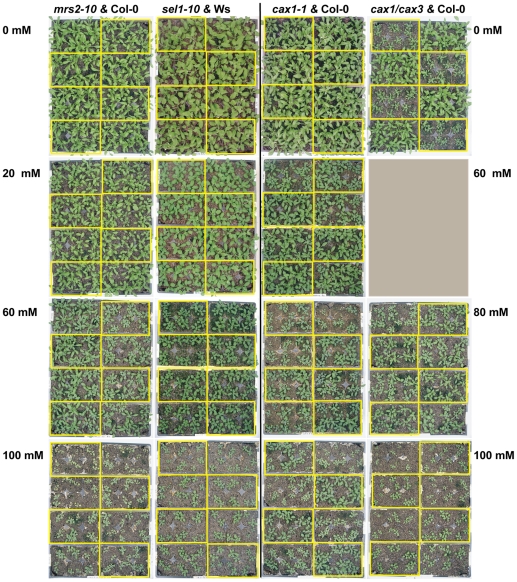
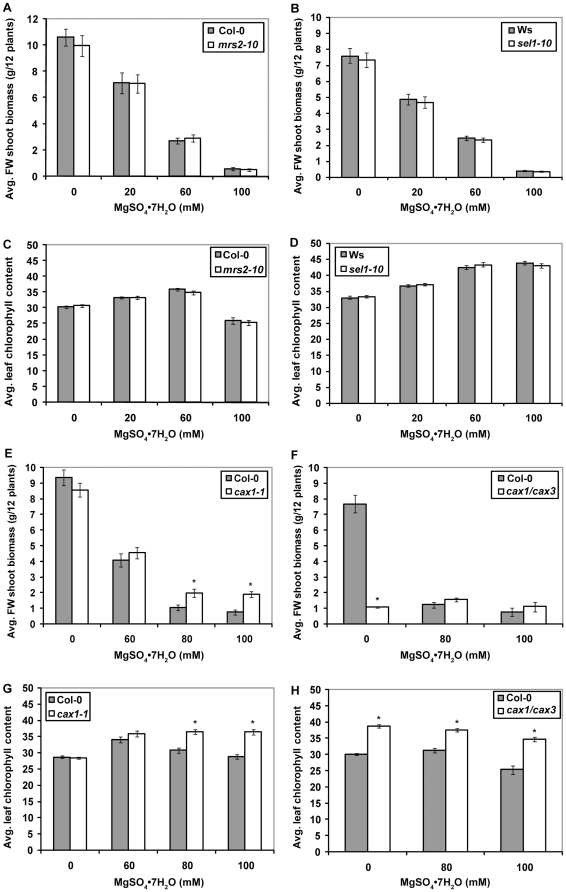
The growth performance of transporter gene knockout mutant lines and their respective wildtype backgrounds grown for four weeks on soil treated with MgSO4·7H2O in solution (0–100 mM) was compared on the basis of shoot fresh weight (FW) biomass and leaf chlorophyll levels. Figure 4 shows the growth habit of mrs2-10 and sel1-10 plants compared to wildtype in the left hand panels, and of cax1-1 and cax1/cax3 compared to wildtype in the right-hand panels. Statistical analysis of the FW shoot biomass of wildtype Arabidopsis (Ws, Col-0) with ANOVA confirmed the significant phytotoxic effects of increasing concentrations of dissolved MgSO4·7H2O on wildtype plant growth (Table S1). Subsequent ANOVA analyses assessed whether selected mutant lines could alleviate the growth-limiting effects seen in wildtype. Results showed that FW shoot biomass and leaf chlorophyll content of mrs2-10 and sel1-10 lines were indistinguishable from that of wildtype at the tested concentrations (Fig. 5a–d, Table S1). Ionome analysis of the mrs2-10 mutant exposed to regular nutrient conditions furthermore did not reveal a statistically different leaf Mg content compared to Col-0 (http://www.ionomicshub.org/home/PiiMS). Although there was little difference between the mrs2-10 and sel1-10 lines and their respective wild-type backgrounds, the cax1-1 and cax1/cax3 lines showed relative improvement in their ability to grow on MgSO4·7H2O enriched soil. Cax1-1 plants had significantly higher FW shoot biomass and leaf chlorophyll content than Col-0 grown at 80 and 100 mM MgSO4·7H2O (Fig. 5e,g, Table S1). The increase in cax1-1 shoot biomass over that of Col-0 was 89% and 149% at 80 and 100 mM respectively. The CAX1 knockout mutation did not fully eliminate the effects of high magnesium sulfate on plant growth performance; the absolute FW shoot biomass of cax1-1 grown on soil for four weeks was still low (20%) compared to untreated Col-0 (Fig 5e). Cax1/cax3 plants showed significantly higher leaf chlorophyll content than Col-0 grown at 80, and 100 mM, but the average FW shoot biomass increases of 26.8% and 33.2%, at 80 and 100 mM, were not found to be statistically significant (Fig. 5f,h, Table S1). The significant differences in FW shoot biomass and leaf chlorophyll content between cax1/cax3 and Col-0 grown at 0 mM MgSO4·7H2O were described in detail by Cheng et al. (2005). In this study, 0 mM was only included as a point of reference for the high concentrations of MgSO4·7H2O.
Figure 4. Overview of the Arabidopsis growth experiment on soil.
Mutant lines are grown alongside their respective wildtype backgrounds on trays containing soil medium with different levels of dissolved MgSO4·7H2O. To control for environmental variation, each tray was divided in 8 sections, with a mutant line and its associated wildtype background line planted in alternating fashion. Yellow boxes highlight the sections with mutant plants within each flat; the other sections contain wildtype plants. For the comparison between cax1/cax3 and Col-0, 60 mM was not tested. The plants shown here are 3 weeks old and were harvested and analyzed after completing 4 weeks of growth. The experiment was repeated three times.
Figure 5. The shoot fresh weight biomass comparisons of mutant and wildtype lines grown on soil.
Average fresh weight shoot biomass of (a) mrs2-10 and Col-0, (b) sel1-10 and Ws, (e) cax1-1 and Col-0, or (f) cax1/cax3 and Col-0 plants in response to increasing concentrations of MgSO4·7H2O in soil medium. Bars indicate standard error, n = 12. Average leaf chlorophyll content of (c) mrs2-10 and Col-0, (d) sel1-10 and Ws, (g) cax1-1 and Col-0, or (h) cax1/cax3 and Col-0 plants in response to increasing concentrations of MgSO4·7H2O in soil medium. Bars indicate standard error, n = 72. The asterisks indicate statistically significant differences between genotypes (p<0.05) at specific concentrations of MgSO4·7H2O based on ANOVA.
Root Transcriptome
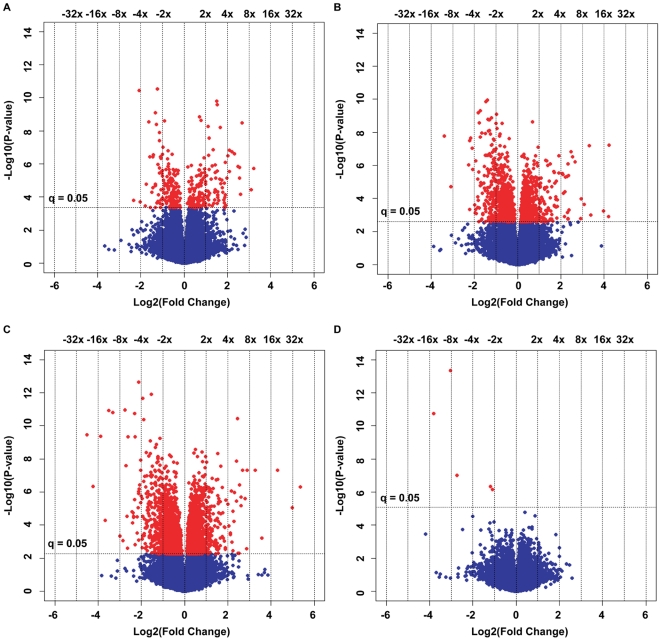
Root transcriptome remodeling was analyzed in Arabidopsis after hydroponic exposure to a non-lethal, high concentration of MgSO4·7H2O (Fig. 2, Fig. 3, Table S2). The number of genes with statistically significant differences in expression between Col-0 exposed to MgSO4·7H2O and Col-0 exposed to control conditions increased from 325 (Time 45) to 1516 (Time 90) and 3265 (Time 180) (Fig. 6). The number of genes with a significant difference in expression of over 2-fold increased accordingly, from 100 to 248 and 445. Between cax1-1 and Col-0 exposed to MgSO4·7H2O for 180 min., only 4 unique transcripts showed a significant difference in expression, and all were over a 2-fold change in abundance (Fig. 6).
Figure 6. Volcano plots of gene expression comparisons at distinct timepoints.
(a) Time 45 (Col-0 treated for 45 min. vs Col-0 exposed to a control solution for 45 min.) (b) Time 90 (Col-0 treated for 90 min. vs Col-0 exposed to a control solution for 45 min.) (c) Time 180 (Col-0 treated for 180 min. vs Col-0 exposed to a control solution for 45 min.) (d) cax1-1 treated for 180 min. vs Col-0 treated for 180 min. The x-axes show log2 values of the fold changes in gene expression between sample sets. Each dot represents one of 37478 transcripts. Vertical lines indicate absolute fold change values as indicated on top of the graphs. The y-axes show the −log10 p-values corresponding to the log2 fold change values. The horizontal line indicates the −log10 p-value where the q-value is 0.05. Transcripts whose expression difference (fold change) corresponds to a p-value for which q<0.05 are above the horizontal line and indicated in red.
Primary MgSO4·7H2O stress response
The primary root transcriptome responses to MgSO4·7H2O stress were analyzed in Col-0 treated for 45 min. versus Col-0 treated with a control solution for 45 min. (Time 45) to reveal genes involved in the metabolic response to this environment as candidate genes that could play a role in high magnesium sulfate tolerance. The resulting set of 325 genes with significant differences in expression was analyzed for over- and under-represented GO molecular function categories. Significantly over-represented categories were polygalacturonase activity, calcium ion binding, and transcription factor activity (Table 2). Many of the transcripts represented at Time 45 encode phytohormone associated proteins, transcription factors, Ca2+-binding proteins, kinases, phosphatases, disease resistance proteins, cell wall related proteins, membrane-based transporters, as well as proteins of unknown functional category. The better characterized of these genes are listed by category in Table 3. For example, phytohormone associated genes encoding enzymes involved in ABA (9-cis-epoxycarotenoid dioxygenase), ethylene (1-aminocyclopropane-1-carboxylate synthase) and jasmonic acid (lipoxygenase ) biosynthesis pathways show up-regulated expression. The gene encoding C2H2 type zinc finger family transcription factor ZAT7 is up-regulated, while the gene encoding transcription factor WRKY70 is down-regulated at Time 45. The expression of the gene encoding CBL-interacting protein kinase CIPK9 is up-regulated. Genes encoding cell-wall and disease-resistance associated proteins are also well represented; fasciclin-like arabinogalactan proteins, UDP-glucose 4-epimerase (RHD1), xyloglucan endotransglucosylase (XTH9) and beta-expansin (EXPB1) show down-regulated expression, while the expression of several genes encoding alpha-expansin proteins is up-regulated at Time 45. Well-characterized genes encoding membrane based transporters that are differentially expressed include those encoding calcium exchanger CAX1, calcium-transporting ATPase ACA2, cyclic nucleotide-gated channels CNGC19 and CNGC1, potassium transporter HAK5 and inorganic phosphate transporter PHT1. The 500 bp upstream regions of the down-regulated and up-regulated genes at Time 45 were statistically analyzed for over-represented six-mer sequences (motifs), which may correspond to cis-regulatory elements. Table 4 lists the ten motifs with the highest significance (lowest p-values) for both the up-regulated and down-regulated subsets of genes. The motif with the highest significance in the subset of up-regulated genes is an ABRE coupling element (ABRE-CE). The 325 differentially expressed genes at Time 45 were furthermore compared with a cluster of 197 genes that are known to be differentially expressed in response to a broad range of stress conditions, including cold, osmotic stress, salinity, wounding, and biotic stresses [28]. The comparison showed that at least 18 of the 325 genes differentially expressed at Time 45 appear to be universally responsive to stress conditions (Table 5). These include for example genes encoding Ca2+-binding, kinase, zinc finger, and disease resistance proteins.
Table 2. Significantly over-represented GO molecular function categories within the set of genes with significant differences in expression at Time 45.
| GO category | Molecular function | Count | P-Value |
| Over-represented | |||
| GO:0004650 | Polygalacturonase activity | 6 | 0.0176 |
| GO:0005509 | Calcium ion binding | 8 | 0.0176 |
| GO:0003700 | Transcription factor activity | 27 | 0.0443 |
The set of 325 genes with significant differences in expression at Time 45 was analyzed for over- and under-represented GO molecular function categories. The results were corrected for multiple testing using False Discovery Rate and the p-value threshold was set to 0.05.
Table 3. Well-characterized genes with significant differences in expression at Time 45 (Col-0 treated for 45 min. vs Col-0 exposed to a control solution for 45 min.).
| Transporters: | ||
| Gene | Description | Log2 |
| At2g38170 | calcium exchanger (CAX1) | −0.5676 |
| At4g13420 | potassium transporter (HAK5) | 1.5548 |
| At5g43350 | inorganic phosphate transporter (PHT1) (PT1) | 0.3541 |
| At4g37640 | calcium-transporting ATPase 2 (ACA2) PM-type | 0.3525 |
| At3g17690 | cyclic nucleotide-binding transporter 2 (CNGC19) | 0.9209 |
| At5g53130 | cyclic nucleotide-regulated ion channel (CNGC1) | −0.3754 |
| Transcription factors: | ||
| Gene name | Description | Log2 |
| At3g46090 | zinc finger (C2H2 type) family protein (ZAT7) | 0.9947 |
| At3g56400 | WRKY family transcription factor (WRKY 70) | −0.9454 |
| Kinases: | ||
| Gene name | Description | Log2 |
| At1g01140 | CBL-interacting protein kinase 9 (CIPK9) | 1.7794 |
| phytohormone associated: | ||
| Gene name | Description | Log2 |
| At1g78390 | 9-cis-epoxycarotenoid dioxygenase | 2.554 |
| At5g65800 | 1-aminocyclopropane-1-carboxylate synthase | 1.8605 |
| At1g72520 | lipoxygenase | 1.3473 |
| Cell wall related: | ||
| Gene name | Description | Log2 |
| At2g23130 | arabinogalactan-protein (AGP17) | −1.149 |
| At2g23130 | arabinogalactan-protein (AGP17) | −0.7107 |
| At1g55330 | arabinogalactan-protein (AGP21) | −1.0069 |
| At5g10430 | arabinogalactan-protein (AGP4) | −0.4932 |
| At5g65390 | arabinogalactan-protein (AGP7) | −0.7772 |
| At4g12730 | fasciclin-like arabinogalactan-protein (FLA2) | −0.3949 |
| At1g03870 | fasciclin-like arabinogalactan-protein (FLA9) | −0.501 |
| At1g64440 | UDP-glucose 4-epimerase (RHD1) | −0.5767 |
| At4g03210 | xyloglucan endotransglycosylase (XTH9) | −0.4144 |
| At2g20750 | beta-expansin (EXPB1) | −1.0351 |
| At1g20190 | alpha-expansin (EXP11) | 0.7098 |
| At3g15370 | alpha-expansin (EXP12) | 3.2099 |
| At4g01630 | alpha-expansin (EXP17) | 2.4955 |
| At5g02260 | alpha-expansin (EXP9) | 0.793 |
Table 4. Motif analysis of 500bp upstream region of genes with significant differences in expression at Time 45 (Col-0 treated for 45 min. vs Col-0 exposed to a control solution for 45 min.).
| oligomer | motif name | query set | genomic set | p-value |
| up-regulated genes | ||||
| ACGCGG/CCGCGT | ABRE-CE | 33/156 | 1155/33518 | 5.11E-17 |
| CGCGTA/CGCGTA | 28/156 | 1163/33518 | 9.30E-13 | |
| AACGCG/CGCGTT | 32/156 | 1731/33518 | 1.72E-11 | |
| ACGCGT | 24/156 | 1053/33518 | 1.36E-10 | |
| ACACGG/CCGTGT | 31/156 | 1927/33518 | 1.04E-09 | |
| CGCGTC/GACGCG | 23/156 | 1148/33518 | 3.74E-09 | |
| CACGCG/CGCGTG | D-box | 26/156 | 1525/33518 | 8.63E-09 |
| AACACG/CGTGTT | 48/156 | 4731/33518 | 5.07E-08 | |
| ACACGT/ACGTGT | 51/156 | 5594/33518 | 4.64E-07 | |
| ACCGCG/CGCGGT | 17/156 | 940/33518 | 1.72E-06 | |
| down-regulated genes | ||||
| ACAGCT/AGCTGT | 40/153 | 3998/33518 | 7.44E-07 | |
| AATAGA/TCTATT | 40/153 | 13809/33518 | 3.79E-05 | |
| GACAGC/GCTGTC | 23/153 | 2068/33518 | 4.57E-05 | |
| AATATG/CATATT | 87/153 | 13955/33518 | 5.03E-05 | |
| AGCCTG/CAGGCT | 20/153 | 1727/33518 | 8.48E-05 | |
| AATAAT/ATTATT | 120/153 | 21856/33518 | 1.33E-04 | |
| ATATTA/TAATAT | 106/153 | 18637/33518 | 1.72E-04 | |
| ATTAGC/GCTAAT | 47/153 | 6439/33518 | 2.17E-04 | |
| CAGCTG | 16/153 | 1317/33518 | 2.54E-04 | |
| AAGACA/TGTCTT | 72/153 | 11483/33518 | 3.17E-04 |
The 500 bp upstream regions of the significantly down-regulated and up-regulated genes at Time 45 were statistically analyzed for over-represented six-mer sequences (motifs), which may correspond to cis-regulatory elements. Table 4 lists the ten motifs with the highest significance for both the up-regulated and down-regulated subsets of genes.
Table 5. Genes with significant expression differences between Col-0 treated for 45 min. and Col-0 exposed to a control solution for 45 min.
| Gene | Description | Log2 |
| At1g02400 | gibberellin 2-oxidase, putative | 0.6555 |
| At1g18740 | expressed protein | 0.706 |
| At1g19180 | expressed protein | 0.8927 |
| At1g73540 | MutT/nudix family protein | 0.7802 |
| At1g74450 | expressed protein | 1.1068 |
| At2g27080 | harpin-induced protein-related | 0.5201 |
| At2g30040 | protein kinase family protein | 0.9232 |
| At2g34930 | disease resistance family protein | 1.3017 |
| At2g41410 | calmodulin, putative | −0.78 |
| At2g46600 | calcium-binding protein, putative | −1.0632 |
| At3g10300 | calcium-binding EF hand family protein | 1.1562 |
| At3g16720 | zinc finger (C3HC4-type RING finger) family protein | −0.4137 |
| At4g24570 | mitochondrial substrate carrier family protein | 1.1447 |
| At4g27652 | expressed protein | 0.8255 |
| At4g29780 | expressed protein | 1.4961 |
| At4g35985 | senescence/dehydration-associated protein-related | 0.6918 |
| At5g12010 | expressed protein | 0.6498 |
| At5g16830 | syntaxin 21 (SYP21)/PEP12 homolog | 0.3345 |
(Time 45) that are known to be universally responsive to abiotic stress.
Membrane transporters Col-0 time series
Of special interest is the differential expression of transporter genes as they may represent a metabolic strategy for tolerance to an elevated magnesium sulfate environment. Over 200 different genes encoding membrane-based transporters were differentially expressed across the Col-0 time series (Table S2). Since the Time 90 and 180 comparisons are not fully controlled for diurnal effects, gene expression differences for several transporter genes of interest to this study were analyzed by Q-PCR using diurnally controlled samples. The number of differentially expressed transporter genes increased from 13 at Time 45, to 74 at Time 90 and 189 at Time 180 (Table 2). The expression of the 217 unique genes encoding transporters across the Col-0 time series was analyzed by cluster algorithms to reveal subsets of genes with corresponding patterns of expression (Fig. 7 and Fig. S1).
Figure 7. Hierarchical average linkage cluster analysis of transporter gene expression using uncentered correlation.
The cluster analysis is based on transporter genes with significant expression at Time 45, 90 or 180. Yellow denotes a higher, and blue a lower expression of a gene in the treated plants versus the control. The figure shows that distinct clusters of expression patterns can be distinguished within the group of transporter genes across the three comparisons. The full cluster set is shown on the left; subsets of the clusters are expanded to the right to allow closer inspection of the differential expression patterns. The first 10 letters of the annotation are provided in the expanded sections. The fully annotated figure can be found in the Supplemental material (Fig. S1).
The differential expression of genes encoding known magnesium, sulfate and calcium/proton transporters is summarized in Table 6. The summary shows that the expression of the genes encoding magnesium transporters MRS2-10 and MRS2-7 is slightly up-regulated. Genes encoding sulfate transporters SULTR3;4, SULTR3;1 and SULTR4;1 (a vacuolar H+/SO4 2− cotransporter) show down-regulated expression. The down-regulated expression of the gene encoding SULTR3;4 was confirmed by Q-PCR at 180 min., although its down-regulated expression was less pronounced when controlled for diurnal effects (Table 7, Fig. 8). The gene encoding the vacuolar Mg2+/H+ antiporter (MHX) shows up-regulated expression, while the vacuolar Ca2+/H+ antiporters CAX1, CAX2 and CAX3 show down-regulated expression. The down-regulated expression of CAX1 was confirmed by Q-PCR after 180 min. of exposure when controlled for diurnal effects (Table 7, Fig. 8). Besides the genes described above, there were many examples of differentially expressed genes encoding transporters of unknown function belonging to several large transporter gene families. Represented families include the MATE efflux family, the ABC transporter family, the integral membrane family, the major intrinsic protein family, the cation efflux family, the cation-chloride cotransporter family, the anion exchange family, and the ATPase E1–E2 type family. In addition, several genes encoding transporter-related proteins and putative transporters were differentially expressed.
Table 6. Genes encoding known magnesium, sulfate and calcium/proton transporters with significant differences in expression in Col-0 at Time 45, 90 and 180.
| Magnesium transporter family: | ||||
| Gene | Description | Log2 Time 45 | Log2 Time 90 | Log2 Time 180 |
| At1g80900 | magnesium transporter (MRS2-10) | 0.2182 | 0.2425 | |
| At5g09690 | magnesium transporter (MRS2-7) | 0.2772 | ||
| Sulfate transporter family | ||||
| Gene | Description | Log2 Time 45 | Log2 Time 90 | Log2 Time 180 |
| At3g51895 | sulfate transporter (SULTR3;1) | −2.6591 | ||
| At5g13550 | sulfate transporter (SULTR4;1) | −0.5451 | ||
| At3g15990 | sulfate transporter (SULTR3;4) | −0.7491 | −0.8085 | |
| Cation/proton antiporter families | ||||
| Gene | Description | Log2 Time 45 | Log2 Time 90 | Log2 Time 180 |
| At2g38170 | calcium exchanger (CAX1) | −0.5676 | −0.8702 | −0.7321 |
| At3g13320 | calcium exchanger (CAX2) | −0.2668 | −0.2707 | |
| At3g51860 | cation exchanger (CAX3) | −1.1699 | −1.1249 | |
| At2g47600 | magnesium/proton exchanger (MHX1) | 0.3887 | 0.4055 | |
| At3g53720 | cation/hydrogen exchanger (CHX20) | 0.4314 | 0.6706 | |
Table 7. Q-PCR analysis results.
| Gene | Type | Relative Expression (Array results) | Std. Error | P-value | Result |
| Q-PCR results of gene expression in Col-0 treated for 180 min. vs Col-0 exposed to a control solution for 45 min. (RNA sources are the same as for the transcriptome analysis): | |||||
| At2g32170 | REF | 1 | |||
| CAX1 | TRG | 0.57 (0.602) | 0.436–0.766 | 0 | DOWN |
| MRS2-10 | TRG | 1.035 (1.183) | 0.758–1.377 | 0.714 | |
| SULTR3;4 | TRG | 0.557 (0.541) | 0.462–0.691 | 0 | DOWN |
| NRAMP1 | TRG | 1.196 (1.317) | 0.886–1.515 | 0.107 | |
| Q-PCR results of gene expression in Col-0 treated for 180 min. vs Col-0 exposed to a control solution for 180 min. (diurnally controlled samples): | |||||
| At2g32170 | REF | 1 | |||
| CAX1 | TRG | 0.641 (0.602) | 0.429–0.966 | 0.004 | DOWN |
| MRS2-10 | TRG | 1.017 (1.183) | 0.876–1.195 | 0.763 | |
| SULTR3;4 | TRG | 0.861 (0.541) | 0.695–1.059 | 0.049 | DOWN |
| NRAMP1 | TRG | 1.128 (1.317) | 0.970–1.381 | 0.156 | |
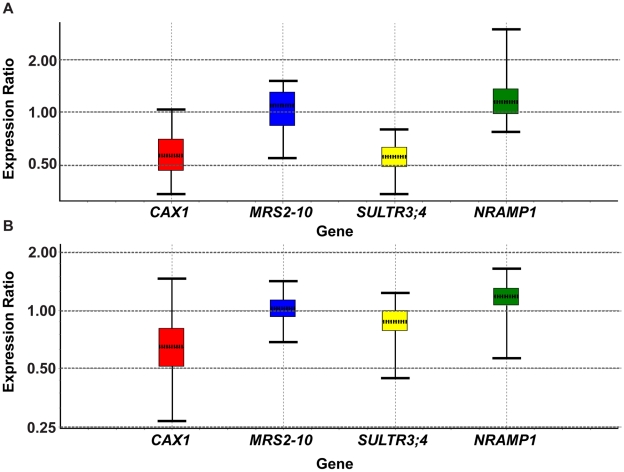
Figure 8. Whisker box plots representing gene expression ratio distributions for the Q-PCR analysis of four transporter genes.
The gene expression ratio distributions of transporter genes that showed significant differences in expression in the root transcriptome analysis of Col-0 treated for 180 min. vs Col-0 exposed to a control solution for 45 min are represented by Whisker box plots. Results show permutated expression data that are calculated by the REST 2008 statistical analysis software, which uses randomization techniques. The graphs give an impression of the expression ratio distribution per gene related to the results presented in Table 7. (a) RNA sources were the same as for the transcriptome analysis (Col-0 treated for 180 min. vs Col-0 exposed to a control solution for 45 min.). (b) RNA was extracted from Col-0 treated for 180 min vs Col-0 exposed to a control solution for 180 min. (diurnally controlled samples).
Differentially regulated genes cax1-1/Col-0 comparison
Only 4 unique transcripts showed a significant difference in expression between the cax1-1 and Col-0 sample sets treated for 180 minutes (Table 8). Among those, the transcript with the smallest q-values and largest differences in expression was CAX1, the gene that was knocked out in the mutant cax1-1, and which is represented on the microarray by two different probes. The other three transcripts included At3g01345, an expressed protein with similarity to beta-galactosidases in Arabidopsis, At4g07526, an unknown protein with similarity to other unknown proteins in Arabidopsis, and chromosomal region CHR2:011819877–011819818, which corresponds to a portion of the mitochondrial photorespiration gene At2g27730.
Table 8. Transcripts with differential expression at q<0.05 between Arabidopsis thaliana cax1-1 and Col-0 treated for 180 min.
| q-value | Log2(FC) | Gene and DNA region description |
| 1.66E-09 | −3.0461 | calcium exchanger (CAX1) [At2g38170.1] |
| 3.28E-07 | −3.8209 | calcium exchanger (CAX1) [At2g38170.3] |
| 0.001201795 | −2.7293 | Expressed protein [At3g01345.1] |
| 0.004291231 | −1.1998 | hypothetical protein [At4g07526.1] |
| 0.005396832 | −1.0903 | Unknown [CHR2:011819877–011819818] |
The table shows that 5 transcripts are identified with a significant difference in expression. Two of the transcripts are identical (At2g38170), which means that 4 unique transcripts show a significant difference in expression between cax1-1 and Col-0 treated for 180 min.
Discussion
The use of regolith on Mars with high levels of hydrated sulfate minerals for bioregenerative life support systems will lead to exposure of plant roots to supra-optimal concentrations of both Mg2+ and SO4 2− ions in the soil solution. The results in this study show that knockout mutant lines of the known genes in Arabidopsis encoding root plasma membrane based uptake transporters of Mg2+ and SO4 2− ions failed to confer an adaptive advantage to a MgSO4·7H2O enriched environment. Because sel1-10 mutants are known to take up 80% less sulfate than their wildtype background Ws, these results mean that phytotoxic effects of magnesium sulfate on overall plant growth are dominated by the Mg2+ cation. Overexpression of AtMRS2-10 (AtMGT1) in Nicotiana benthamiana led to increased accumulation of magnesium (Mg), manganese (Mn), and iron (Fe) per unit dry weight and per plant compared to wildtype plants [29]. However, the potential role of AtMRS2-10 as a major uptake transporter of Mg2+ in Arabidopsis remains uncertain based on the observation that mrs2-10 plants did not show a significant difference in leaf Mg levels compared to Col-0 under standard nutrient conditions. This could explain why, despite the indication that Mg dominantly affects overall plant growth, the mrs2-10 mutant did not show a significant improvement in growth performance compared to Col-0. Interestingly, the dominant role of Mg is also supported by the relative tolerance of the cax1-1 vacuolar H+/Ca2+ transport mutant plants to elevated levels of MgSO4·7H2O. The soil data shown in Figures 5 and 6 are in line with the relative tolerance of cax1-1 on agar to high concentrations of dissolved MgCl2, a mineral in which the Mg2+ cation is the dominant phytotoxic factor [16]. In addition, Bradshaw et al (2005) observed that cax1 mutants are better able to tolerate the low Ca∶Mg ratios characteristic of terrestrial serpentine soils. The CAX1 knockout mutation does not fully eliminate the effects of high magnesium sulfate on plant growth performance; the absolute FW shoot biomass of cax1-1 grown on soil for four weeks was still low (20%) compared to untreated Col-0.
The primary Arabidopsis Col-0 root transcriptome responses to elevated levels of magnesium sulfate suggest a number of candidate genes that could play a role in enhancing the growth performance of plants exposed to this nutrient stress. Transcriptome responses of Arabidopsis Col-0 roots exposed to MgSO4·7H2O versus a control solution for 45 min. revealed over 300 differentially expressed genes. Genes of known functional category include those encoding enzymes involved in hormone metabolism, transcription factors, calcium-binding proteins, kinases, disease resistance proteins, cell wall related proteins, and membrane-based transporters. The biological significance of this seemingly diverse set of gene classes illustrates the comprehensive nature of a primary abiotic stress response. The differentially expressed genes at Time 45 encoding enzymes in the ABA, ethylene and jasmonate biosynthesis pathways point to possible changes in the synthesis of these phytohormones. ABA, ethylene, and jasmonic acid have all been implicated in adaptive mechanisms of osmotic stress, ion-mediated signal transduction and the regulation of transporters [30], [31], [32]. The exact role of many of the differentially expressed genes at Time 45 encoding transcription factors is currently unknown, although some are associated with tolerance to other abiotic stresses. The up-regulation at Time 45 of the gene encoding ZAT7 is for example in line with the finding that ZAT7 renders plants more tolerant to salinity stress when constitutively expressed [33]. The down-regulation at Time 45 of the gene encoding WRKY70 correlates with the observation that WRKY70 functions as a negative regulator of developmental senescence and is involved in plant defense signaling pathways [34]. The majority of transcripts encoding kinases that are differentially expressed at Time 45 encode uncharacterized protein kinases as well as receptor-like protein kinases (RLKs) with extracytoplasmic leucine-rich repeats (LRRs) or with an extracellular lectin-like domain. RLKs are membrane-spanning proteins with a predicted signal sequence and a cytoplasmic kinase domain that have been implicated in a wide range of signal transduction pathways [35]. The up-regulated expression of the gene encoding CBL-interacting protein kinase CIPK9 at Time 45 is consonant with the fact that CIPK9 is required for low-potassium tolerance in Arabidopsis [36]. Genes encoding proteins that belong to the transcription factor or kinase categories might be transcriptional or post-translational regulators of transporters [37].
Within the group of genes encoding membrane based transporters, the Ca2+/H+ antiporter CAX1 is one of the few transporter genes that are already differentially expressed at Time 45. Its down-regulated expression is in line with our finding that the cax1-1 mutant performed better compared to wildtype when grown on soil with elevated levels of magnesium sulfate. The slightly up-regulated expression of the gene encoding magnesium transporter MRS2-10 at Time 90 and 180 agrees with the observation that a knockout mutant of AtMRS2-10 (mrs2-10) does not show improved growth in the form of higher FW shoot biomass compared to Col-0 at elevated levels of magnesium sulfate. MRS2 transporters have been shown to transport other ions in addition to Mg2+ [29], [38]. Schock et al. (2002) speculate that, in line with the large number of AtMRS2 family members, the function of the AtMRS2 gene family may be the maintenance of metal ion homeostasis in different cellular compartments (i.e. over different cellular membrane systems). The expression of the gene encoding SULTR1;2, a high-affinity sulfate transporter, showed no significant differences at the three time points. The fact that expression of the gene is not down-regulated in the early adaptation response of Arabidopsis roots to high levels of magnesium sulfate supports the outcome of the FW shoot biomass comparison between the AtSULTR1;2 knockout mutant sel1-10 and Col-0, which showed no advantage for sel1-10. The gene encoding EIL3, one of the known transcriptional regulators of SULTR1;2 [39], did not show a significant difference in expression at any of the time points either.
Interestingly, two sulfate transporter genes of the same family as SULTR1;2 show down-regulated expression; the gene encoding SULTR3;4 at Time 90 and 180, and the gene encoding SULTR3;1 at Time 180. The spatial and subcellular localization of these transporters is not known, and so far no influence on the expression of Group 3 sulfate transporters by the sulfur status of plants has been reported [40]. This study shows for the first time that genes encoding Group 3 sulfate transporters SULTR3;1 and SULTR3;4 are differentially expressed in roots upon exposure to high levels of sulfate. The expression of the gene encoding another sulfate transporter - the tonoplast-localized H+/SO4 2− cotransporter SULTR4;1 - is also down-regulated at Time 180, pointing at the possible retention of excess SO4 2− in the vacuole. Similarly, the up-regulated expression of the gene encoding the vacuolar MHX transporter indicates the possible storage of excess Mg2+ in the vacuole. Both MHX and SULTR1;4 could be seen as marker genes for excess magnesium sulfate recognition by the plant, since vacuolar storage of excess ions is a well-known defense mechanism against ion stress.
Several members of the CAX (cation exchanger) family show down-regulated expression in response to elevated magnesium sulfate. The Ca2+/H+ antiporter CAX1, which shows down-regulated expression across the time points, is localized to the tonoplast and responsible for 50% of the Ca2+/H+ antiport activity there [16]. CAX2 has been shown to transport Ca2+ and Mn2+ into the vacuole in Arabidopsis and other plant species [41], and CAX3 is known as a weak Ca2+ vacuolar transporter. CAX1 and CAX3 can be combined as “hetero-CAX” complexes to form functional transporters with distinct transport properties [42]. The down-regulated expression of the genes encoding these three vacuolar Ca2+/H+ antiporters indicates a possible shortage of calcium in the cytosol upon high magnesium sulfate exposure.
In total, over 200 differentially expressed genes encoding membrane-based transporters were identified across the Col-0 time series. Among the genes encoding transporters of known and unknown function, candidates for improving plant tolerance to high magnesium sulfate may be found. Transporters of unknown function include for example major intrinsic proteins, cyclic nucleotide gated channels, integral membrane proteins, cation efflux family proteins, ATPase E1–E2 type family proteins, MATE efflux family proteins, transporter-related proteins and putative transporters.
The differential expression of only four unique transcripts between cax1-1 and Col-0 plants after 180 min. of MgSO4·7H2O treatment indicates that the root transcriptome responses are virtually identical for the two genotypes at this time after initiation of treatment. In our transcriptome study we see down-regulation of the gene encoding vacuolar sulfate efflux transporter SULTR4;1 at Time 180, and up-regulation of the gene encoding vacuolar magnesium influx transporter MHX at Time 90 and Time 180 in Col-0 exposed to elevated magnesium sulfate. Both of these responses point to storage of excess magnesium and sulfate ions in the vacuole. This means that the profile is most probably not performed at a time point before plants recognize excess magnesium sulfate, especially since over 3000 differentially expressed genes were detected in Col-0 at Time 180, in addition to the vacuolar marker genes. We could therefore not foresee that after 180 minutes of exposure there were no more than four differentially expressed transcripts between the cax1-1 mutant and Col-0. One of the main reasons for the limited number of differentially regulated transcripts between the genotypes at this time after initiation of treatment may be that CAX1 is one of the few transporter genes that are already differentially expressed after 45 min. of exposure in Col-0, which means that down-regulated expression of CAX1 is an important natural response of Arabidopsis to elevated levels of magnesium sulfate. The response of CAX1 and other CAX family members in the Col-0 time series suggests that a deficiency of calcium caused by high levels of magnesium and possibly by calcium sulfate precipitation could play a major part in the reduction of plant growth performance in the presence of high magnesium sulfate concentrations. The difference between the immediately down-regulated level of CAX1 in Col-0 and the absence of CAX1 in the cax1-1 knockout mutant is apparently not sufficiently large at 180 min. after initiation of treatment to reveal many of the downstream molecular processes eventually leading to enhanced growth performance and reduced leaf Mg content of cax1-1 compared to Col-0 after several weeks of growth. This result is significant in that follow-up studies contrasting cax1-1 with Col-0 can look at profiles after possibly even days of exposure.
Conclusions and perspectives
It was initially thought that the mrs2-10 and sel1-10 mutants would have an advantage over wildtype in elevated magnesium sulfate soils, but this was not the case. However, fresh weight shoot biomass analyses showed that although mrs2-10 and sel1-10 mutants were not more successful than wildtype in this soil environment, the cax1 mutants were. Thus, cax1 mutants are currently the only confirmed genotype with partial tolerance to the phytotoxic levels of magnesium sulfate expected for the regolith on Mars.
To further characterize the response of Arabidopsis to Mars-like levels of magnesium sulfate a set of experiments was conducted to evaluate patterns of gene expression in wildtype (Col-0) and cax1 mutants. The 325 genes differentially expressed in Col-0 roots after 45 min. of exposure to high magnesium sulfate can be seen as candidate genes for enhanced tolerance. This set of genes included CAX1, 18 genes previously associated with enhanced tolerance to broad ranges of abiotic stresses, and several well-characterized genes known to enhance tolerance to other salts like NaCl. Many of the differentially regulated genes contain promoter sequence motifs that are known to play a role in ABA-mediated regulation, such as ABRE-CE. This abundance suggests that ABA plays a role in regulating the stress response to elevated magnesium sulfate. In addition, a rich pool of candidates is found in the 200+ transporter genes that are differentially expressed in Col-0 across the time points. Many genes in this set encode transporters of unknown function. These unknown transporter genes, along with the four differentially regulated transcripts between cax1-1 and Col-0, are of particular interest as they encode proteins of unknown function whose roles are now at least implicated.
This study functions as a solid basis for the development of Mars soil-compatible plants by reducing the number of potential candidate genes from tens of thousands to several hundred. Future research efforts could determine whether any of these candidate genes, or their potential regulators, can be confirmed as genomic loci for (crop) plant growth enhancement in the presence of mars-like levels of magnesium sulfate.
Supporting Information
Plant growth performance ANOVA. ANOVA (t-test) results are shown for specific genotype concentration effects in the growth experiments.
(0.01 MB XLS)
Root transcriptome ANOVA. An ANOVA analysis of differential expression in the root transcriptome.
(0.71 MB XLS)
The fully annotated Figure 7: hierarchical average linkage cluster analysis of transporter gene expression using uncentered correlation. Hierarchical average linkage cluster analysis of transporter gene expression using uncentered correlation. The cluster analysis is based on transporter genes with significant expression at Time 45, 90 or 180. Yellow denotes a higher, and blue a lower expression of a gene in the treated plants versus the control. The figure shows that distinct clusters of expression patterns can be distinguished within the group of transporter genes across the three comparisons.
(0.54 MB PDF)
Acknowledgments
We would like to thank Dr. K. Hirschi for providing the cax1-1 and cax1/cax3 lines and Dr. H. Takahashi for providing the sel1-10 line. We gratefully acknowledge the SALK Institute for the distribution of the original T-DNA insertion line from which we characterized the mrs2-10 line. We also wish to thank B. Laughner for assisting with the harvest of hydroponically grown Arabidopsis roots, and Dr. N. Denslow for providing control RNA for the Agilent one color 4×44k microarrays. We likewise acknowledge the Interdisciplinary Center for Biotechnology Research at the University of Florida and, in particular, Dr. G. Ostrow, for the microarray hybridization, scanning, and data extraction. We must also thank Dr. C. Benedict for assistance with the microarray data normalization, and S. Norton and J. McPherson for assistance with the Q-PCR data extraction.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Prins Bernard Cultuurfonds of the Netherlands. NASA NNX07AH27G. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Barta DJ, Henninger DL. Regenerative life support systems: why do we need them? Adv Space Res. 1994;14:403–410. doi: 10.1016/0273-1177(94)90329-8. [DOI] [PubMed] [Google Scholar]
- 2.Ming DW, Henninger DL, editors. Lunar Base Agriculture: Soils for Plant Growth. American Society of Agronomy, Inc; 1989. [Google Scholar]
- 3.Schuerger AC, Ming DW, Newsom HE, Ferl RJ, McKay CP. Near-term lander experiments for growing plants on Mars: requirements for information on chemical and physical properties of Mars regolith. Life Support Biosph Sci. 2002;8:137–147. [PubMed] [Google Scholar]
- 4.Bibring JP, Langevin Y, Mustard JF, Poulet F, Arvidson R, et al. Global mineralogical and aqueous mars history derived from OMEGA/Mars Express data. Science. 2006;312:400–404. doi: 10.1126/science.1122659. [DOI] [PubMed] [Google Scholar]
- 5.Gendrin A, Mangold N, Bibring JP, Langevin Y, Gondet B, et al. Sulfates in Martian layered terrains: the OMEGA/Mars Express view. Science. 2005;307:1587–1591. doi: 10.1126/science.1109087. [DOI] [PubMed] [Google Scholar]
- 6.Arvidson RE, Poulet F, Bibring JP, Wolff M, Gendrin A, et al. Spectral reflectance and morphologic correlations in eastern Terra Meridiani, Mars. Science. 2005;307:1591–1594. doi: 10.1126/science.1109509. [DOI] [PubMed] [Google Scholar]
- 7.Christensen PR, Wyatt MB, Glotch TD, Rogers AD, Anwar S, et al. Mineralogy at Meridiani Planum from the Mini-TES experiment on the Opportunity Rover. Science. 2004;306:1733–1739. doi: 10.1126/science.1104909. [DOI] [PubMed] [Google Scholar]
- 8.Ming DW, Mittlefehldt DW, Morris RV, Golden DC, Gellert R, et al. Geochemical and mineralogical indicators for aqueous processes in the Columbia Hills of Gusev crater, Mars. J Geophys Res. 2006;111 [Google Scholar]
- 9.Haskin LA, Wang A, Jolliff BL, McSween HY, Clark BC, et al. Water alteration of rocks and soils on Mars at the Spirit rover site in Gusev crater. Nature. 2005;436:66–69. doi: 10.1038/nature03640. [DOI] [PubMed] [Google Scholar]
- 10.Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1995. 889 [Google Scholar]
- 11.Laurie S, Feeney KA, Maathuis FJ, Heard PJ, Brown SJ, et al. A role for HKT1 in sodium uptake by wheat roots. Plant J. 2002;32:139–149. doi: 10.1046/j.1365-313x.2002.01410.x. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Tutone AF, Drummond RS, Gardner RC, Luan S. A novel family of magnesium transport genes in Arabidopsis. Plant Cell. 2001;13:2761–2775. doi: 10.1105/tpc.010352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonso JM, Stepanova AN. T-DNA mutagenesis in Arabidopsis. Methods Mol Biol. 2003;236:177–188. doi: 10.1385/1-59259-413-1:177. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K. Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 2002;29:465–473. doi: 10.1046/j.0960-7412.2001.01231.x. [DOI] [PubMed] [Google Scholar]
- 15.Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 2003;132:597–605. doi: 10.1104/pp.102.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng NH, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD. The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell. 2003;15:347–364. doi: 10.1105/tpc.007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng NH, Pittman JK, Shigaki T, Lachmansingh J, LeClere S, et al. Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 2005;138:2048–2060. doi: 10.1104/pp.105.061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradshaw HD., Jr Mutations in CAX1 produce phenotypes characteristic of plants tolerant to serpentine soils. New Phytol. 2005;167:81–88. doi: 10.1111/j.1469-8137.2005.01408.x. [DOI] [PubMed] [Google Scholar]
- 19.Barber SA. Soil nutrient bioavailability: a mechanistic approach. John Wiley and Sons, Inc; 1995. 414 [Google Scholar]
- 20.Brooks RR. Serpentine and its vegetation. Dioscorides Press; 1987. 454 [Google Scholar]
- 21.Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, et al. Purdue ionomics information management system: an integrated functional genomics platform. Plant Physiol. 2007;143:600–611. doi: 10.1104/pp.106.092528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tocquin P, Corbesier L, Havelange A, Pieltain A, Kurtem E, et al. A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol. 2003;3:2. doi: 10.1186/1471-2229-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smeets K, Ruytinx J, Van Belleghem F, Semane B, Lin D, et al. Critical evaluation and statistical validation of a hydroponic culture system for Arabidopsis thaliana. Plant Physiol Biochem. 2008;46:212–218. doi: 10.1016/j.plaphy.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, et al. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 25.Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, et al. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- 26.Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma S, Bohnert HJ. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 2007;8:R49. doi: 10.1186/gb-2007-8-4-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng W, Luo K, Li D, Zheng X, Wei X, et al. Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. J Exp Bot. 2006;57:4235–4243. doi: 10.1093/jxb/erl201. [DOI] [PubMed] [Google Scholar]
- 30.Roberts SK, Snowman BN. The effects of ABA on channel-mediated K(+) transport across higher plant roots. J Exp Bot. 2000;51:1585–1594. doi: 10.1093/jexbot/51.350.1585. [DOI] [PubMed] [Google Scholar]
- 31.Ma S, Gong Q, Bohnert HJ. Dissecting salt stress pathways. J Exp Bot. 2006;57:1097–1107. doi: 10.1093/jxb/erj098. [DOI] [PubMed] [Google Scholar]
- 32.Koiwa H, Bressan RA, Hasegawa PM. Identification of plant stress-responsive determinants in Arabidopsis by large-scale forward genetic screens. J Exp Bot. 2006;57:1119–1128. doi: 10.1093/jxb/erj093. [DOI] [PubMed] [Google Scholar]
- 33.Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, et al. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem. 2007;282:9260–9268. doi: 10.1074/jbc.M611093200. [DOI] [PubMed] [Google Scholar]
- 34.Ulker B, Shahid Mukhtar M, Somssich IE. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta. 2007;226:125–137. doi: 10.1007/s00425-006-0474-y. [DOI] [PubMed] [Google Scholar]
- 35.Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004;18:2545–2556. doi: 10.1101/gad.1245904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey GK, Cheong YH, Kim BG, Grant JJ, Li L, et al. CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res. 2007;17:411–421. doi: 10.1038/cr.2007.39. [DOI] [PubMed] [Google Scholar]
- 37.Shaul O. Magnesium transport and function in plants: the tip of the iceberg. Biometals. 2002;15:309–323. doi: 10.1023/a:1016091118585. [DOI] [PubMed] [Google Scholar]
- 38.Mao DD, Tian LF, Li LG, Chen J, Deng PY, et al. AtMGT7: An Arabidopsis Gene Encoding a Low-Affinity Magnesium Transporter. J Integr Plant Biol. 2008;50:1530–1538. doi: 10.1111/j.1744-7909.2008.00770.x. [DOI] [PubMed] [Google Scholar]
- 39.Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchner P, Takahashi H, Hawkesford MJ. Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J Exp Bot. 2004;55:1765–1773. doi: 10.1093/jxb/erh206. [DOI] [PubMed] [Google Scholar]
- 41.Edmond C, Shigaki T, Ewert S, Nelson MD, Connorton JM, et al. Comparative analysis of CAX2-like cation transporters indicates functional and regulatory diversity. Biochem J. 2009;418:145–154. doi: 10.1042/BJ20081814. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Shigaki T, Mei H, Guo YQ, Cheng NH, et al. Interaction between Arabidopsis Ca2+/H+ exchangers CAX1 and CAX3. J Biol Chem. 2009;284:4605–4615. doi: 10.1074/jbc.M804462200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plant growth performance ANOVA. ANOVA (t-test) results are shown for specific genotype concentration effects in the growth experiments.
(0.01 MB XLS)
Root transcriptome ANOVA. An ANOVA analysis of differential expression in the root transcriptome.
(0.71 MB XLS)
The fully annotated Figure 7: hierarchical average linkage cluster analysis of transporter gene expression using uncentered correlation. Hierarchical average linkage cluster analysis of transporter gene expression using uncentered correlation. The cluster analysis is based on transporter genes with significant expression at Time 45, 90 or 180. Yellow denotes a higher, and blue a lower expression of a gene in the treated plants versus the control. The figure shows that distinct clusters of expression patterns can be distinguished within the group of transporter genes across the three comparisons.
(0.54 MB PDF)