Abstract
Although biotin is an essential enzyme cofactor found in all three domains of life, our knowledge of its biosynthesis remains fragmentary. Most of the carbon atoms of biotin are derived from pimelic acid, a seven carbon dicarboxylic acid, but the mechanism whereby Escherichia coli assembles this intermediate remains unknown. Genetic analysis identified only two genes of unknown function required for pimelate synthesis, bioC and bioH. We report in vivo and in vitro evidence that the pimeloyl moiety is synthesized by a modified fatty acid synthetic pathway in which the ω-carboxyl group of a malonyl-thioester is methylated by BioC which allows recognition of this atypical substrate by the fatty acid synthetic enzymes. The malonyl-thioester methyl ester enters fatty acid synthesis as the primer and undergoes two reiterations of the fatty acid elongation cycle to give pimeloyl-acyl carrier protein (ACP) methyl ester which is hydrolyzed to pimeloyl-ACP and methanol by BioH.
INTRODUCTION
Biotin (also known as vitamin H) is a covalently bound enzyme cofactor required by all forms of life. Although biotin was discovered over 70 years ago and is an essential nutrient for animals, its detailed biosynthetic pathway is not completely understood in any organism1,2. Biotin consists of two fused heterocyclic rings plus a valeric acid side-chain (Fig. 1). The late steps of the pathway are responsible for assembly of the rings whereas the early steps are concerned with the synthesis of pimelic acid, a seven carbon dicarboxylic acid. The origins of the biotin carbon atoms in E. coli are known from 13C NMR analysis of products labeled in vivo3,4. The C-3, C-5, and C-7 carbons (Fig. 1) are derived from C-1 of acetate whereas C-2 of acetate contributes the C-2, C-4 and C-6 carbons. The labeling pattern indicates that the pimeloyl moiety is formed by head to tail incorporation of three intact acetate units as is the case in fatty acid (or polyketide) synthesis with CO2 providing C-1. Since the biotin C-1 and C-7 atoms show different labeling patterns, free pimelic acid (a symmetrical molecule) cannot be an intermediate. Hence, the pimeloyl moiety must be assembled with one of the carboxyl groups covalently linked. This is likely via a thioester since pimeloyl-CoA is thought to be the activated form of pimelic acid1,2.
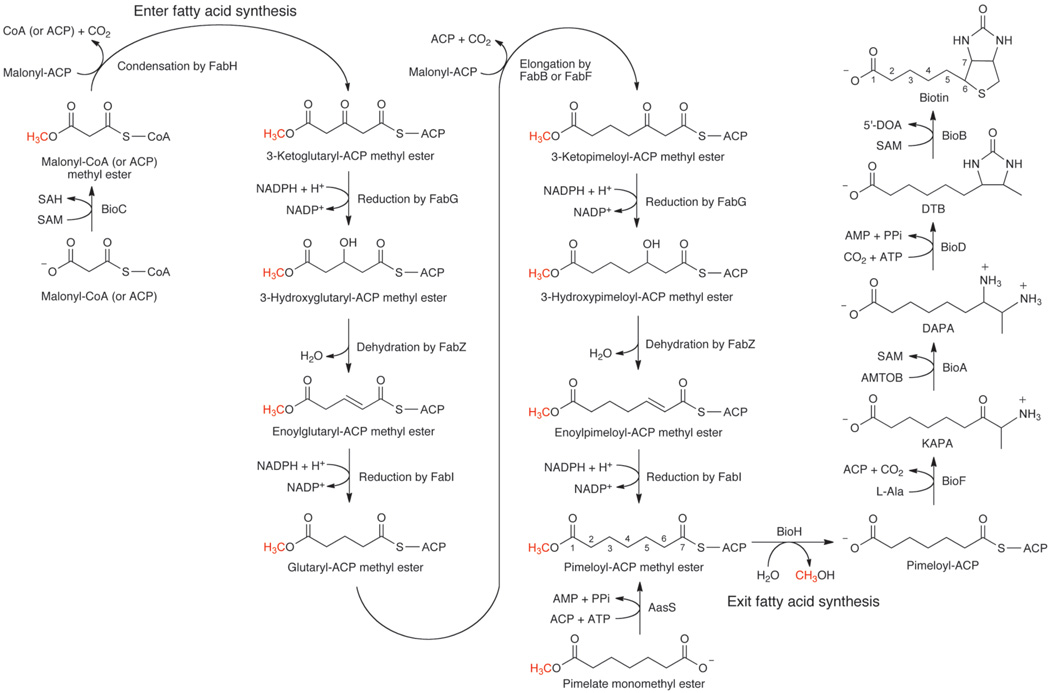
Fig. 1. Scheme of the proposed biotin synthetic pathway.
Pimeloyl-ACP methyl ester synthesis proceeds either artificially by acylation of ACP with the monomethyl ester of pimelate or physiologically by elongation of a primer molecule of malonyl-CoA methylated at its ω-carboxyl group by BioC. In the physiological pathway the committed step is the BioC-catalyzed SAM-dependent methylation of malonyl-CoA (or malonyl-ACP) . Malonyl-CoA methyl ester enters the fatty acid synthetic pathway as a dedicated biotin precursor and acts as primer (in place of the acetyl-CoA normally used in fatty acid synthesis) for the FabH catalyzed condensation reaction. The product of the proposed FabH reaction would be 3-ketoglutaryl-ACP methyl ester, a five-carbon dicarboxylate carrying the methyl ester moiety that allows the substrate to interact with the other fatty acid synthetic enzymes. 3-Ketoglutaryl-ACP methyl ester would be reduced by 3-ketoacyl-ACP reductase (FabG) to 3-hydroxylglutaryl-ACP methyl ester which would be dehydrated by 3-hydroxyacyl-ACP dehydratase (FabZ) to the enoyl-ACP methyl ester species. The double bond would be reduced by enoyl-ACP reductase (FabI) to give glutaryl-ACP methyl ester. The product of the first elongation cycle would then be elongated by a second decarboxylative Claisen condensation with malonyl-ACP (probably by FabB or FabF) to give 3-ketopimeloyl-ACP methyl ester which would be reduced and dehydrated as above to give pimeloyl-ACP methyl ester. BioH cleavage of the ester prevents further elongation and the resulting pimeloyl-ACP utilized by BioF to make KAPA which would begin synthesis of biotin. Abbreviations: DAPA, 7,8-diaminopelargonic acid; DTB, dethiobiotin; AMTOD, S-adenosyl-2-oxo-4-methylthiobutyric acid; SAH, S-adenosylhomocysteine; 5’-DOA, 5’-deoxyadenosine.
The conundrum is how to assemble a seven-carbon dicarboxylic acid in E. coli. Genetic analyses implicate only two genes, bioC and bioH, in pimeloyl moiety synthesis, but neither gene product appears able to play a direct role in assembling the carbon chain5–7. BioC is annotated as an S-adenosyl-L-methionine (SAM)-dependent methyltransferase whereas BioH has been shown to have esterase activity on short and medium chain acyl p-nitrophenyl esters8–10 and on the methyl ester of dimethylbutyryl-S-methyl mercaptopropionate10. The BioC annotation was especially puzzling because all of the pimeloyl moiety carbon atoms are derived from acetate and CO2 3,4. The remaining bio genes encode enzymes that function late in the pathway and thus it seemed that assembly of the pimeloyl moiety must require additional enzymes belonging to another biosynthetic pathway that are somehow assisted in this task by BioC and BioH. In 196 a pathway was proposed in which pimeloyl-CoA synthesis could be formed by the enzymes of fatty acid synthesis11. The proposal was that three malonyl-CoA molecules would be condensed with the primer malonyl moiety retaining the carboxyl group introduced by acetyl-CoA carboxylase fixation of CO2. The other two malonyl-CoA molecules would lose their free carboxyl groups in the course of the two decarboxylative Claisen reactions required to give the C7 dicarboxylate, a scheme consistent with the 13C labeling studies and the precedent of type III polyketide synthases12,13. However, in fatty acid synthesis the growing chains are attached to ACP rather than CoA and unlike polyketides, where the keto groups are either retained or consumed in rearrangements of the carbon chain (e.g., cyclization), pimelate synthesis requires that the keto groups be converted to methylene groups. Although the enzymes of fatty acid synthesis could in principle perform this conversion, it seemed most unlikely that the fatty acid synthetic enzymes could accept substrates having a carboxyl group in place of the usual terminal methyl group because the fatty acid synthetic enzymes sequester the growing fatty acyl chains in strongly hydrophobic tunnels or clefts14. It occurred to us that BioC and BioH could circumvent this conundrum.
In our model (Fig. 1) the role of BioC is to convert the free carboxyl group of a malonyl-thioester to its methyl ester by transfer of a methyl group from SAM. Methylation would both cancel the charge of the carboxyl group and provide a methyl carbon to mimic the methyl ends of normal fatty acyl chains. The esterified malonyl-thioester would enter the fatty acid synthetic pathway as in the 1963 proposal11. Two reiterations of the elongation cycle would produce pimeloyl-ACP methyl ester. BioH would then cleave the methyl ester to give pimeloyl-ACP which BioF would utilize to make 7-keto-8-aminopelargonic acid (KAPA), the first intermediate in biotin ring assembly. In this scenario, introduction of the methyl ester disguises the biotin synthetic intermediates such that they become substrates for the fatty acid synthetic pathway. When synthesis of the pimeloyl moiety is complete and disguise is no longer needed, the methyl group is removed to free the carboxyl group that will eventually be used to attach biotin to its cognate metabolic enzymes15. We report that the monomethyl esters of malonic, glutaric and pimelic acid allow growth of a ΔbioC strain in the absence of biotin, but fail to allow growth of ΔbioC ΔbioH strains. An in vitro system was developed in which dialyzed cell extracts converted malonyl-CoA to dethiobiotin (DTB, the last intermediate of the pathway) which defined the proposed pathway by allowing the precursor requirements of the pathway and the effects of inhibitors of fatty acid synthesis and methyl transfer on DTB synthesis to be determined.
RESULTS
Dicarboxylate monoesters allow growth of a ΔbioC strain
We propose that the intermediates of pimelate synthesis are acyl carrier protein (ACP) thioesters. To obtain such substrates we tested malonic, glutaric and pimelic acids and their monomethyl esters as substrates for acyl-ACP synthetase (AasS) from Vibrio harveyi16. AasS was completely inactive with the diacids probably due to the inability of the terminal carboxyl groups to enter the hydrophobic tunnel where the substrate acyl chain resides16. In contrast, the three monomethyl esters were readily converted to their ACP thioesters (Supplementary Fig. 1). Given these data and prior success in using this enzyme to insert exogenous fatty acids into the fatty acid synthetic pathway in vivo17, we tested whether or not expression of AasS in E. coli ΔbioC, ΔbioH or ΔbioC ΔbioH strains would allow growth in the absence of biotin when the medium was supplemented with one of the three monomethyl esters. Note that essentially the entire coding sequences were deleted in construction of the ΔbioC and ΔbioH strains (Supplementary Methods).
Supplementation of biotin-free medium with any of the three monomethyl esters allowed AasS-dependent growth of the ΔbioC strain whereas the ΔbioH and ΔbioC ΔbioH strains failed to grow under these conditions (Supplementary Fig. 2). Supplementation with malonic, glutaric and pimelic acids also failed to support growth, although the last of these could be shown to enter E. coli (Supplementary Fig. 3). This was demonstrated by expression of the BioW pimeloyl-CoA synthetase of Bacillus subtilis in a ΔbioC ΔbioH strain which upon supplementation with pimelic acid allows growth of these mutant strains in the absence of biotin18. Putative intermediates in the pathway, the enoyl, 3-keto and 3-hydroxy derivatives of the monomethyl ester of glutarate and the 3-keto and 3-hydroxy derivatives of the monomethyl ester of pimelate allowed growth whereas the 2-keto, 2-hydroxy and 4-keto derivatives did not (Supplementary Table 1). The specificity of the in vivo system was also demonstrated by the failure of the monomethyl esters of C4, C6, C8, C9 and C11 dicarboxylates to support growth of the ΔbioC strain. The compounds active as monomethyl esters were also active as their monoethyl, monopropyl and monobutyl esters (Supplementary Table 1). In the case of malonic acid, monoesterification with the longer alcohols resulted in better growth than seen with the monomethyl ester, perhaps due to increased activity with AasS. Growth with the longer esters indicated that the enzyme catalyzing cleavage of the ester linkages tolerates the larger alcohol moieties (see below). The fact that either AasS plus the monomethyl ester of pimelate or BioW plus pimelate allowed growth of ΔbioC strains argues that BioF can accept either thioester. This is not unprecedented, a number of fatty acid synthetic enzymes accept acyl-CoA substrates in vitro, albeit these are generally less efficient substrates than the cognate acyl-ACPs.
Fatty acid synthesis-dependent DTB production in vitro
To further test the proposed pathway we developed an in vitro system in which DTB synthesis was coupled to the fatty acid synthetic pathway. DTB (the last intermediate of the pathway) was assayed rather than biotin because BioB (biotin synthase), the last enzyme of the pathway, is a notoriously unstable and oxygen-sensitive protein that has not been demonstrated to be catalytic in vitro19. The in vitro system consisted of cell-free extracts of wild type or Δbio strains that had been subjected to ammonium sulfate precipitation (where ACP remains soluble) followed by dialysis; manipulations designed to remove small molecules and deplete the extracts of ACP. The strains sometimes carried extra copies of the bio operon genes on a multicopy plasmid. The in vitro system allowed us to test which precursors are required for DTB synthesis, show that SAM is required for KAPA synthesis, and that the methyl group is required for elongation of the C5 species.
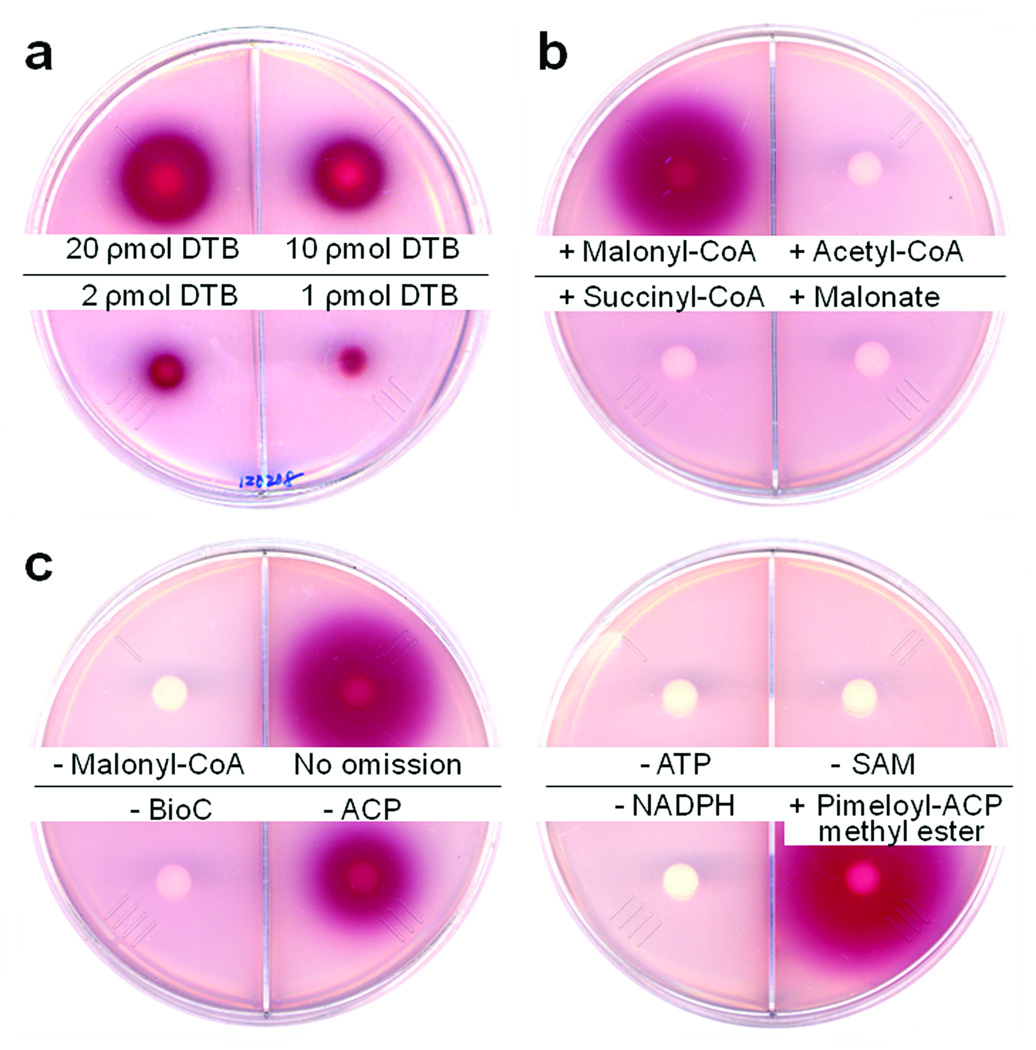
DTB synthesis was determined by bioassay with E. coli strain ER90 (ΔbioF bioC bioD) which carries an insertion-deletion mutation within bioF that also inactivates the downstream genes, bioC and bioD, by transcriptional polarity5,20. Hence, strain ER90 is defective in synthesis of KAPA, DAPA and DTB, but proficient in conversion of DTB to biotin20. Detection by bioassay was required because biotin synthesis is an extremely low capacity biosynthetic pathway (E. coli requires only about 100 biotin molecules per cell). In the bioassay, which can reliably detect 1 ρmol of DTB (Fig. 2a), the test solution diffuses from a filter disk into biotin-free minimal medium agar seeded with an appropriate DTB (or biotin)-requiring E. coli strain. If grow proceeds a redox indicator becomes reduced and forms a bright red, insoluble deposit with the area of the red spot being proportional to the concentration of the biotin pathway intermediate21.
Fig. 2. Requirements for DTB synthesis from malonyl-CoA.
The cell free extract was prepared from the ΔbioC strain STL96 which carries multiple copies of all the bio operon genes except bioC. Purified and refolded BioC was added to all reactions except as shown. Bioassays were done using strain ER90 (ΔbioF) as described in Methods. (a) Standard DTB was bioassayed and reliably detected in levels as low as 1 pmol. (b) DTB synthesis specifically required malonyl-CoA as substrate. Structurally related compounds such as acetyl-CoA, succinyl-CoA and malonate were inactive. (c) DTB synthesis required malonyl-CoA, BioC, NADPH and all other reactions components as given for in vitro DTB synthesis in Methods. When pimeloyl-ACP methyl ester was added, both BioC and malonyl-CoA were omitted.
DTB synthesis was obtained upon supplementation of extracts of a wild type strain with the cofactors (malonyl-CoA, NADPH and ACP) required for in vitro fatty acid synthesis22 plus those required for the late steps of the biotin synthetic pathway (L-alanine, ATP and SAM). In contrast, parallel assays of extracts of ΔbioC, ΔbioH or ΔbioA strains produced no detectable DTB (Figs. 2 and 3). DTB synthesis was observed when malonyl-CoA was added to the reaction whereas the structurally related compounds, acetyl-CoA, succinyl-CoA and malonate, were inactive (Fig. 2b). The requirements for DTB synthesis were then examined in extracts of the ΔbioC strain STL96 (Fig. 2c). The extracts were inactive, but activity was restored upon addition of BioC purified from inclusion bodies and refolded into a soluble form (Fig. 2c). NADPH was required in addition to malonyl-CoA and thus two essential components of the fatty acid synthetic pathway were required for DTB synthesis (Fig. 2c). Omission of ACP from the reaction reduced, but did not abolish DTB synthesis, probably due to residual ACP in the extracts. As expected DTB synthesis also required ATP and SAM, the respective substrates of BioD and BioA (Fig. 2c). In the absence of BioC, pimeloyl-ACP methyl ester (synthesized using V. harveyi AasS, Supplementary Figs. 1 and 4) was converted to DTB (Fig. 2c). Bypass of the BioC requirement was also observed when malonyl-ACP methyl ester and glutaryl-ACP methyl ester were used as substrates (Supplementary Fig. 1). However, unlike pimeloyl-ACP methyl ester, DTB synthesis from these substrates required the presence of malonyl-CoA (Supplementary Fig. 1). In agreement with the in vivo results (Supplementary Fig. 2) the methyl esters of succinyl-ACP, adipyl-ACP, suberyl-ACP and azelayl-ACP lacked activity in vitro (Supplementary Fig. 1).
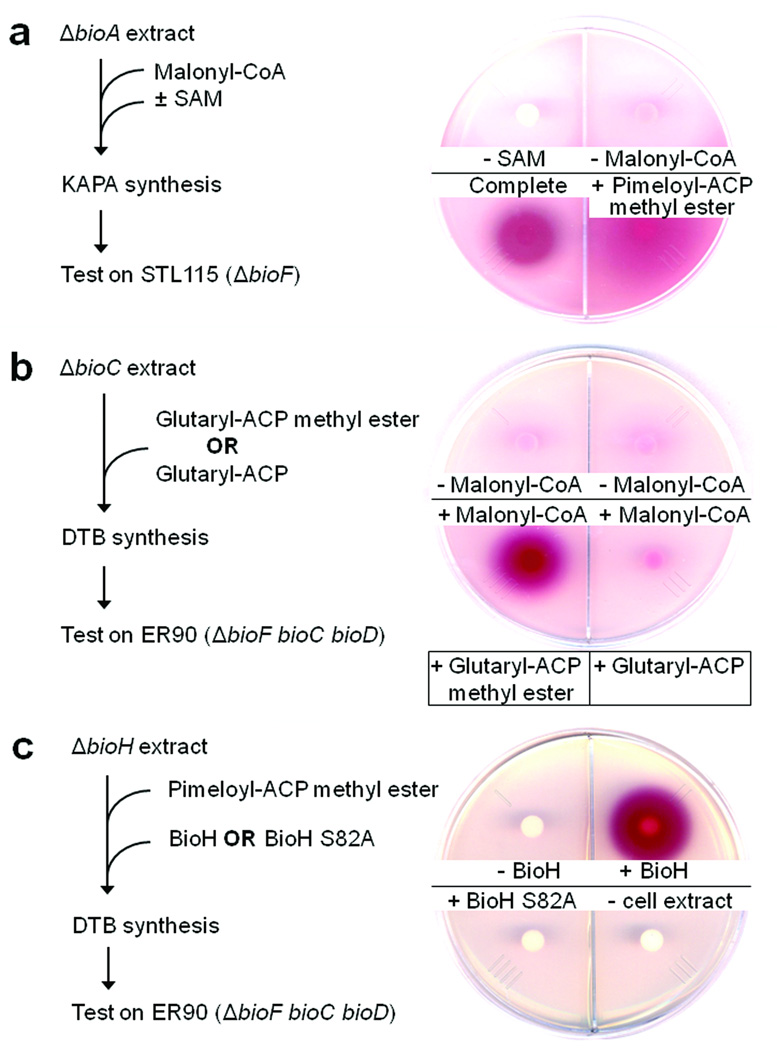
Figure 3. SAM and the methyl ester moiety are both required for synthesis of the pimeloyl moiety.
(a) KAPA synthesis in an extract lacking BioA with malonyl-CoA as substrate. The product was detected by bioassay on the ΔbioF strain STL115. When pimeloyl-ACP methyl ester was added both SAM and malonyl-CoA were omitted. (b) Requirement for the methyl ester moiety in conversion of glutaryl-ACP to DTB. The extract lacked BioC and the bioassay was performed on strain ER90 (ΔbioF bioC bioD). Glutaryl-ACP methyl ester was added to the reactions assayed in the two left hand quadrants whereas the reactions of the two right hand quadrants received glutaryl-ACP. (c) The requirement for active BioH in conversion of pimeloyl-ACP methyl ester to DTB. The extract lacked BioH with pimeloyl-ACP methyl ester as the substrate and the bioassay was performed using strain ER90. Either purified wild type BioH or the mutant BioH S82A protein (which lacks the active site nucleophile) was added to 20 µg/ml. The reaction of the lower right quadrant lacked extract, but contained both the wild type and mutant BioH proteins.
The methyl ester moiety is essential for chain elongation
We postulated that methylation of the ω-carboxyl group of the malonyl thioester was catalyzed by BioC and was essential for accessing the fatty acid elongation cycle enzymes. To test this hypothesis the assay was modified to specifically ask whether or not KAPA synthesis requires SAM. The in vitro KAPA assay used malonyl-CoA as the starting substrate and an extract from the ΔbioA strain STL112. Since in the absence of BioA KAPA cannot be converted to DAPA, KAPA would accumulate in the reaction. KAPA was detected by bioassay using the ΔbioF strain, STL115 that required KAPA (or a later intermediate) for growth. SAM was clearly required for in vitro KAPA synthesis (Fig. 3a). Moreover, the requirement for SAM was bypassed when pimeloyl-ACP methyl ester was added in place of malonyl-CoA (Fig. 3a).
To directly test whether or not the methyl ester was required for chain elongation by the fatty acid synthetic pathway, we synthesized glutaryl-ACP methyl ester using AasS and removed the ester group by BioH treatment to obtain glutaryl-ACP (Supplementary Fig. 4). Purified samples of glutaryl-ACP methyl ester and glutaryl-ACP were tested for the ability to support DTB synthesis in a ΔbioC extract. In the presence of glutaryl-ACP (which lacked the methyl ester moiety) no DTB was produced (Fig. 3b) whereas DTB was synthesized in the presence of glutaryl-ACP methyl ester, but only when malonyl-CoA was available for elongation (Fig. 3b). Therefore, the methyl ester moiety was essential for the conversion of the glutaryl moiety to DTB.
DTB synthesis from pimeloyl-ACP methyl ester requires BioH
Our in vivo results indicated that BioH activity was required for conversion of the pimeloyl methyl ester moiety to biotin (Supplementary Fig. 2). To explore this in more detail, a cell-free extract of the ΔbioH strain STL98 was prepared and was found to be unable to convert pimeloyl-ACP methyl ester to DTB unless the extract was supplemented with purified BioH (Fig. 3c). We synthesized various ω-carboxyl acyl-ACP esters and tested the ability of BioH in vitro to cleave the ester moieties of these substrates. BioH hydrolyzed the methyl, ethyl, propyl and butyl esters of pimeloyl-ACP (Supplementary Fig. 5), confirming our in vivo bypass assay results. BioH also readily hydrolyzed the methyl esters of adipyl-, pimeloyl-, suberyl- and azelayl-ACPs, but failed to cleave malonyl-ACP methyl ester and succinyl-ACP methyl ester whereas glutaryl-methyl ester seemed to cleaved less efficiently (Supplementary Fig. 5). As a control we expressed and purified a BioH S82A mutant protein. This protein was expected to be inactive because in the BioH crystal structure9 Ser82 had been modified by reaction with phenylmethylsulfonyl fluoride added as a proteolysis inhibitor. Since this inhibitor inhibits serine proteases and esterases by forming a covalent adduct with the active site nucleophile, there seemed little doubt that the S82A substitution would inactivate BioH. As expected the mutant protein was inactive in the in vitro hydrolysis assay (Supplementary Fig. 5) and failed to support DTB synthesis in a ΔbioH extract (Fig. 3c). Moreover, expression of BioH S82A in a ΔbioH strain failed to allow growth on biotin-free medium whereas expression of wild type BioH supported robust growth (Supplementary Fig. 6). These results, together with the biotin requirement of ΔbioH strains, indicate that the methyl ester moiety is specifically cleaved by BioH and not by other E. coli hydrolases.
Inhibitors of fatty acid synthesis block in vitro DTB synthesis
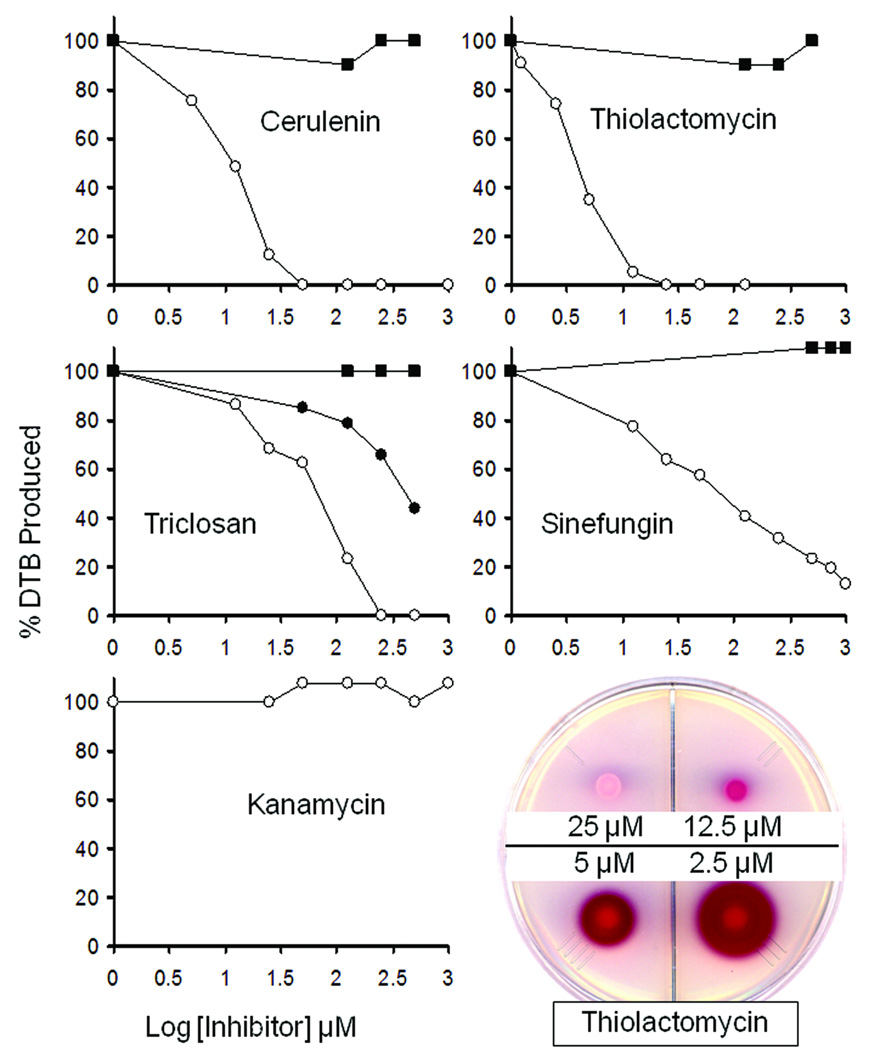
The proposed pimelate synthetic pathway was further tested by use of the antibiotics, cerulenin and thiolactomycin plus the microbiocide, triclosan, to block fatty acid synthesis in vitro (Fig. 4). Cerulenin inhibits the β-ketoacyl-ACP synthases, FabB and FabF, whereas thiolactomycin inhibits FabB and FabF plus the short chain β-ketoacyl-ACP synthase, FabH23. Triclosan inhibits FabI, the sole E. coli enoyl-ACP reductase23. Addition of any of these three inhibitors severely reduced the levels of DTB synthesized from malonyl-CoA in a concentration-dependent manner (Fig. 4). Moreover, the effects of triclosan were shown to be due to inhibition of FabI because addition of purified Vibrio cholerae FabV, a naturally triclosan-resistant enoyl-ACP reductase24, largely restored DTB synthesis to triclosan-treated extracts. The fatty acid synthesis inhibitors had no effect on DTB synthesis when pimeloyl-ACP methyl ester was the substrate indicating that their effects were due to inhibition of synthesis of the pimeloyl moiety.
Figure 4. Inhibition of DTB synthesis by inhibitors of fatty acid synthesis and SAM-dependent methyltransferases.
The microbiocide, triclosan, or the antibiotics cerulenin, thiolactomycin and sinefungin were added to the in vitro DTB synthesis with either malonyl-CoA (○) or pimeloyl-ACP methyl ester (■) as substrate. The level of DTB produced was estimated based on the area of the bioassay red zone and was standardized to the control reactions containing no inhibitor (100 % DTB synthesis was approximately 30 ρmol). The lack of inhibition seen when pimeloyl-ACP methyl ester was the substrate demonstrated that the amounts of inhibitor transferred to the assay disks were insufficient to inhibit growth of the bioassay strain ER90 (ΔbioF bioC bioD). Inhibition by triclosan was reduced when FabV, a triclosan resistant enoyl-ACP reductase, was added to 50 µg/ml in the reaction (●). The protein synthesis inhibitor, kanamycin, was tested with malonyl-CoA as the substrate and gave no inhibition over the same range of concentrations. A bioassay plate showing the effects of thiolactomycin on DTB synthesis is given at the lower right of the figure. Type of file: figure
Sinefungin, a SAM analog and a potent inhibitor of SAM-dependent methyltransferases25, also inhibited DTB synthesis when malonyl-CoA was the substrate. No inhibition was seen when pimeloyl-ACP methyl ester was the substrate (Fig. 4) indicating that the blockage of DTB synthesis was not due to inhibition of BioA, the step later in the pathway where SAM is used as an amino donor.
DISCUSSION
The early steps of biotin synthesis in E. coli have long been an enigma. Indeed, more than 40 years after the discovery of the two genes, our data are the first to demonstrate that BioC precedes BioH in the pathway. It has been argued that BioH is not involved in pimelate synthesis26 and also that it is intimately involved6. BioC and BioH were postulated6 to catalyze Claisen condensations and to transfer the pimeloyl moiety from a BioC active site thiol to CoA, respectively. A more recent proposal was that BioH would somehow condense CoA and pimelic acid to produce pimeloyl-CoA perhaps with the help of BioC9.
We have demonstrated that these hypotheses are incorrect and that the roles of BioC and BioH are to allow E. coli to synthesize pimeloyl-ACP via the fatty acid synthetic pathway (Fig. 1). The pimeloyl-ACP synthetic pathway differs from normal fatty acid synthesis in that the ω-carboxyl group of a malonyl-thioester is converted to its methyl ester by SAM-dependent methyl transfer catalyzed by BioC. We expect that the malonyl-thioester is malonyl-CoA because FabH, which uses acetyl-CoA rather than acetyl-ACP14,23, seems likely to catalyze the first condensation. Moreover, E. coli FabH is known to accept propionyl-CoA, a reasonable analogue of malonyl-CoA methyl ester although it cannot accept bulkier branched-chain acyl-CoAs as substrates27. However, we cannot presently determine whether the methyl acceptor is malonyl-CoA or malonyl-ACP (or both). This is because E. coli BioC (as well as the BioCs of Salmonella enterica, Pseudomonas putida, Bacillus cereus and Kurthia sp.) is an extremely recalcitrant protein. E. coli BioC invariably formed inclusion bodies under a wide range of expression conditions both in native form and as a fusion protein. Others have encountered the same problem with BioC28. Although we have recovered traces of activity by refolding denaturated BioC, the activities are too low to detect methyl transfer by direct means (e.g., transfer of the methyl group from [methyl-3H]-SAM). It should also be noted that native BioC might be an intrinsically poor catalyst. E. coli requires only trace quantities of biotin and thus catalytic efficiency is not required of the biotin pathway enzymes. Indeed, the enzymes late in the pathway are notably poor catalysts (kcat values of 0.1−0.01 s−1). Moreover, a catalytically efficient BioC might well compromise synthesis of the fatty acids required for membrane lipids.
Methyl transfer to a carboxyl group is an unusual reaction for a SAM-dependent methyltransferase and thus it is interesting that data base searches using several BLAST algorithms indicate that the closest relative of BioC is a known carboxyl methyltransferase, the E. coli trans-aconitate methyltransferase (Tam) which converts trans-aconitate to its 6-methyl ester29,30. The E. coli Tam and BioC proteins align with 28% identity over the full lengths of the proteins with only a few small gaps.
The pimeloyl-ACP synthetic pathway also requires BioH. We believe that the physiological role of BioH is to remove the methyl group introduced by BioC when the pimeloyl moiety is complete. If so, this would prevent further chain elongation to azelayl-ACP methyl ester, a physiologically useless product. However, we find that BioH is a rather promiscuous hydrolase that also cleaves various esters of pimeloyl-ACP and the methyl esters of the C5 through C9 acyl-ACPs although the thioester bonds of these substrates are not cleaved. Others report that BioH cleaves an ester10 and acyl p-nitrophenyl esters8,9. One possibility is that the specificity of BioH for pimeloyl-ACP depends on acyl-ACP structure. Early work demonstrated that the first 6–8 carbon atoms of the acyl chain are sequestered from solvent by the protein31. This picture has been borne out by X-ray structures32 and molecular dynamics simulations33 indicating that the protected carbon atoms reside within a hydrophobic tunnel formed by a bundle of four helices. Hence, the ester groups of the shorter intermediates would be largely or completely protected from BioH and only upon chain elongation to the pimeloyl moiety would the ester group become fully exposed to solvent and BioH.
We believe that pimeloyl-ACP rather than pimeloyl-CoA is the physiological substrate of BioF. Pimeloyl-CoA was the first substrate tested based on analogy to the synthesis of δ-aminolevulinic acid from glycine and succinyl-CoA and the ability of pimelate to bypass the biotin requirements of certain bacteria and fungi34. However, pimeloyl-CoA seems the only acyl donor tested and it is possible that it acts as a mimic of pimeloyl-ACP or that the enzyme can accept either pimeloyl-thioester. The suggestion that pimeloyl-ACP is the physiological acyl donor for BioF is consistent with recent reports on the BioI of Bacillus subtilis, a bacterium that makes the pimeloyl moiety by a mechanism entirely different from that of E. coli. BioI, which appears restricted to this organism and its close relatives, is a cytochrome P450 family member that binds long chain acyl-ACPs both in vivo and in vitro and cleaves the acyl chains to generate pimeloyl-ACP35,36. B. subtilis BioI functionally replaces both BioC and BioH in E. coli18 which argues strongly that pimeloyl-ACP is a physiological BioF acyl donor and eliminates models in which BioH is involved in transfer of the pimeloyl moiety from ACP to CoA.
METHODS
Bacterial strains, plasmids and primers are given in Supplementary Table 2
Materials
The minimal medium was M937 supplemented (final concentrations) with 0.2 % glucose, 0.1 % Vitamin Assay Casamino Acids, 1 mM MgSO4 and 1 µg/ml thiamine except when 0.2 % arabinose replaced glucose. Agar was added to 1.5 %. Antibiotics were used at the following concentrations (in µg/ml): sodium ampicillin, 100; kanamycin sulfate, 50; chloramphenicol, 30. Assay Buffer is 50 mM 3-(N-morpholino)propanesulfonic acid (pH 7.5), 200 mM NaCl, 10 % glycerol and 5 mM 2-mercaptoethanol. ACP (holo form) and AasS were prepared from strains DK574 and YFJ239, respectively, strictly according to the detailed protocols given previously16,39. The hexahistidine-tagged BioC, BioH and BioH S82A proteins were purified as described in Supplemental Methods.
Enzymatic synthesis of pimeloyl-ACP methyl ester
Synthesis of the monomethyl ester of pimelate is described in Supplemental Information. Acyl-ACP synthesis and purification was adapted from the method of Jiang et al.16. Briefly, the acylation reaction contained 50 mM Tris-HCl (pH 8.5), 10 mM MgCl2, 0.5 mM dithiothreitol, 2 mM ATP, 2 mM pimelate methyl ester, 100 µg/ml ACP and 10 µg/ml AasS. The reactions were incubated at 37°C for 2 h. Pimeloyl-ACP methyl ester was purified by ion exchange chromatography using Vivapure D spin columns (GE Healthcare Life Sciences). The reaction mixtures were loaded in Binding Buffer (25 mM 4-morpholineethanesulfonic acid, pH 6, 10 % glycerol and 1 mM dithiothreitol) containing 100 mM LiCl and the column was washed with Binding Buffer containing 250 mM LiCl. Pimeloyl-ACP was eluted in Binding Buffer containing 500 mM LiCl, desalted and analyzed in a conformationally-sensitive electrophoretic mobility assay39 in 20 % polyacrylamide gels containing 2.5 M urea at 130 V for 3 h (Supplementary Fig. 1). Glutaryl-ACP methyl ester and other acyl-ACPs were prepared by the same method. The molecular masses of the products were verified by mass spectrometry with MALDI/MS (Supplementary Fig. 4) at the Mass Spectrometry Laboratory of University of Illinois at Urbana-Champaign. The ACP products were dialyzed in 3,500 molecular weight cut-off membrane against 2 mM ammonium acetate at 4 °C for 15 h. Samples were mixed with α-cyano-4-hydroxycinnamic acid as matrix and analysis was performed using a Voyager-DE STR mass spectrometer (Applied Biosystems) using a UV laser (337 nm N2 laser). All measurements were made using the linear mode and positive ions were recorded (Supplementary Fig. 4).
Preparation of ΔbioC cell-free extracts
Strain STL96 was grown at 37°C to OD at 600 nm of 0.8 in 250 ml of minimal medium containing 2 nM biotin. The cells were washed in M9 salts medium to remove biotin and subcultured into 1 L of minimal medium at 37°C for 5 h to derepress bio operon transcription by starvation for biotin. The cells were lysed in Assay Buffer by French Press treatment at 17,500 psi followed by centrifugation at 20,000 g for 20 min to obtain the soluble fraction of the cell extract. Ammonium sulfate was added slowly to 85 % of saturation to the soluble cell extract in a beaker on ice under constant stirring until completely dissolved. This step was intended to remove small molecules and delete the extracts of ACP which remains soluble in such ammonium sulfate solutions38. The protein precipitant was collected by centrifugation at 10,000 g and stored at −80°C. The precipitant was solubilized before use by dialysis in 7,000 molecular weight cut-off membranes against Assay Buffer at 4°C for 3 h to remove ammonium sulfate and any remaining small molecules. Extracts of the other deletion strains were prepared by the same procedure.
In vitro DTB synthesis
This assay allows in vitro conversion of ACP-bound substrate into DTB using enzymes in cell-free extracts. A 100 µl reaction in Assay Buffer contained 2.5 mg cell-free extract protein, 1 µmol MgCl2, 0.5 µmol dithiothreitol, 0.01 µmol pyridoxal-5'-phosphate, 50 µg ACP, 2 µg BioC, 0.1 µmol L-alanine, 0.1 µmol KHCO3, 0.1 µmol NADPH, 0.1 µmol ATP, 0.1 µmol glucose-6-phosphate and 0.1 µmol SAM. Malonyl-CoA was used at 0.2 µmol whereas pimeloyl-ACP methyl ester or another acyl-ACP substrate was added at 50 µg. The reactions were incubated at 37°C for 3 h and quenched by immersion in boiling water for 10 min. DTB production was bioassayed as follows. Strain ER90 (ΔbioF bioC bioD) was grown in 5 ml of M9 minimal medium containing 2 nM biotin at 30°C overnight. The cells were washed with M9 medium and subcultured in 100 ml of minimal medium at 37°C for 5 h to starve the cells for biotin. The cells were collected by centrifugation, washed again in M9 medium and mixed into 150 ml of minimal agar containing the redox indicator 2,3,5 triphenyl tetrazolium chloride (0.1 %) to a final OD at 600 nm of approximately 0.1. Six ml of the mixture was poured into petri dishes sectored with plastic walls to prevent cross-feeding. A 6 mm paper disk (BBL) was placed upon the agar and the disk was spotted with 10 µl of a reaction to be tested. After incubation of the plates at 30°C overnight, growth of strain ER90 was visualized as a red deposit of formazan. KAPA synthesis was performed similarly using extracts of strain STL112 followed by bioassay on strain STL115.
AasS-mediated bypass assay
The chemical structures and sources of dicarboxylates and derivatives used in this study are summarized in Table S2. Strains STL32, STL33 and STL34 were constructed by transformation with pYFJ84. The strains were grown at 37°C on minimal agar containing 0.1 mM of a dicarboxylate or a derivative plus 0.1 mM isopropyl β-D-1-thiogalactopyranoside to induce aasS expression. Bypass of bio gene function was observed as growth in the absence of biotin. Avidin (0.1 units/ml, Calbiochem) was also added to the medium to prevent cross-feeding and neutralize any biotin or DTB contamination. To verify the permeability of E. coli to pimelate, strain STL74 carrying a plasmid encoding B. subtilis bioW was streaked on M9 minimal agar supplemented with 0.1 mM of sodium pimelate and 0.2 % arabinose for induction (Supplementary Fig. 3).
Supplementary Material
Acknowledgements
This work was supported by NIH grant AI15650 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Author Contributions. S.L., R.E.H. and J.E.C. performed experiments whereas S.L and J.E.C designed experiments and wrote the manuscript.
The authors declare they have no competing financial interests.
References
- 1.Marquet A, Bui BTS, Florentin D. In: Vitamins and Hormones. Litwack G, Begley T, editors. Vol. 61. Academic Press; 2001. pp. 51–101. [Google Scholar]
- 2.Webb ME, Marquet A, Mendel RR, Rebeille F, Smith AG. Elucidating biosynthetic pathways for vitamins and cofactors. Nat Prod Rep. 2007;24:988–1008. doi: 10.1039/b703105j. [DOI] [PubMed] [Google Scholar]
- 3.Ifuku O, et al. Origin of the carbon atoms of biotin. Eur. J. Biochem. 1994;220:585–591. doi: 10.1111/j.1432-1033.1994.tb18659.x. [DOI] [PubMed] [Google Scholar]
- 4.Sanyal I, Lee S, Flint DH. Biosynthesis of pimeloyl-CoA, a biotin precursor in Escherichia coli, follows a modified fatty acid synthesis pathway: 13C-labeling studies. J. Am. Chem. Soc. 1994;116:2637–2638. [Google Scholar]
- 5.Cleary PP, Campbell A. Deletion and complementation analysis of the biotin gene cluster of Escherichia coli. J. Bacteriol. 1972;112:830–839. doi: 10.1128/jb.112.2.830-839.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemoine Y, Wach A, Jeltsch JM. To be free or not: the fate of pimelate in Bacillus sphaericus and in Escherichia coli. Mol Microbiol. 1996;19:645–647. doi: 10.1046/j.1365-2958.1996.t01-4-442924.x. [DOI] [PubMed] [Google Scholar]
- 7.Rolfe B, Eisenberg MA. Genetic and biochemical analysis of the biotin loci of Escherichia coli K-12. J. Bacteriol. 1968;96:515–524. doi: 10.1128/jb.96.2.515-524.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon MA, Kim HS, Oh JY, Song BK, Song JK. Gene cloning, expression, and characterization of a new carboxylesterase from I sp. SES-01: comparison with IBioHe enzyme. J Microbiol Biotechnol. 2009;19:147–154. doi: 10.4014/jmb.0804.287. [DOI] [PubMed] [Google Scholar]
- 9.Sanishvili R, et al. Integrating structure, bioinformatics, and enzymology to discover function. J. Biol. Chem. 2003;278:26039–26045. doi: 10.1074/jbc.M303867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie X, Wong WW, Tang Y. Improving simvastatin bioconversion in Escherichia coli by deletion of bioH. Metab Eng. 2007;9:379–386. doi: 10.1016/j.ymben.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Lezius A, Ringelmann E, Lynen F. Zur biochemischen Funktion des Biotins. IV. Die Biosynthese des Biotins. Biochem Z. 1963;336:510–525. [PubMed] [Google Scholar]
- 12.Austin MB, et al. Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J Biol Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 13.Tseng CC, McLoughlin SM, Kelleher NL, Walsh CT. Role of the active site cysteine of DpgA, a bacterial type III polyketide synthase. Biochemistry. 2004;43:970–980. doi: 10.1021/bi035714b. [DOI] [PubMed] [Google Scholar]
- 14.White SW, Zheng J, Zhang YM, Rock The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 15.Chapman-Smith A, Cronan JE., Jr The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem Sci. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Y, Chan C, Cronan JE. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain Acyl-CoA synthetase family. Biochemistry. 2006;45:10008–10019. doi: 10.1021/bi060842w. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, Morgan-Kiss RM, Campbell JW, Chan CH, Cronan JE. Expression of Vibrio harveyi acyl-ACP synthetase allows efficient entry of exogenous fatty acids into the Escherichia coli fatty acid and lipid A synthetic pathways. Biochemistry. 2010;49:718–726. doi: 10.1021/bi901890a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bower S, et al. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J. Bacteriol. 1996;178:4122–4130. doi: 10.1128/jb.178.14.4122-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi-Rhee E, Cronan JE. Biotin synthase is catalytic in vivo, but catalysis engenders destruction of the protein. Chem Biol. 2005;12:461–468. doi: 10.1016/j.chembiol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Choi-Rhee E, Cronan JE. A nucleosidase required for in vivo function of the S-adenosyl-L-methionine radical enzyme, biotin synthase. Chem Biol. 2005;12:589–593. doi: 10.1016/j.chembiol.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 21.del Campillo-Campbell A, Dykhuizen D, Cleary PP. Enzymic reduction of d-biotin d-sulfoxide to d-biotin. Methods Enzymol. 1979;62:379–385. doi: 10.1016/0076-6879(79)62244-9. [DOI] [PubMed] [Google Scholar]
- 22.Lennarz WJ, Light RJ, Bloch K. A fatty acid synthetase from E. coli. Proc Natl Acad Sci U S A. 1962;48:840–846. doi: 10.1073/pnas.48.5.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell JW, Cronan JE. Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu. Rev. Microbiol. 2001;55:305–332. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- 24.Massengo-Tiasse RP, Cronan JE. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J Biol Chem. 2008;283:1308–1316. doi: 10.1074/jbc.M708171200. [DOI] [PubMed] [Google Scholar]
- 25.Pugh CS, Borchardt RT, Stone HO. Sinefungin, a potent inhibitor of virion mRNA(guanine-7-)-methyltransferase, mRNA(nucleoside-2'-)-methyltransferase, and viral multiplication. J. Biol. Chem. 1978;253:4075–4077. [PubMed] [Google Scholar]
- 26.O'Regan M, et al. Nucleotide sequence of the bioH gene of Escherichia coli. Nucleic Acids Res. 1989;17:8004. doi: 10.1093/nar/17.19.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi KH, Heath RJ, Rock CO. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J Bacteriol. 2000;182:365–370. doi: 10.1128/jb.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomczyk NH, et al. Purification and characterisation of the BIOH protein from the biotin biosynthetic pathway. FEBS Lett. 2002;513:299–304. doi: 10.1016/s0014-5793(02)02342-6. [DOI] [PubMed] [Google Scholar]
- 29.Cai H, Clarke S. A novel methyltransferase catalyzes the methyl esterification of trans-aconitate in Escherichia coli. J Biol Chem. 1999;274:13470–13479. doi: 10.1074/jbc.274.19.13470. [DOI] [PubMed] [Google Scholar]
- 30.Cai H, Strouse J, Dumlao D, Jung ME, Clarke S. Distinct reactions catalyzed by bacterial and yeast trans-aconitate methyltransferases. Biochemistry. 2001;40:2210–2219. doi: 10.1021/bi0022902. [DOI] [PubMed] [Google Scholar]
- 31.Cronan JE., Jr Molecular properties of short chain acyl thioesters of acyl carrier protein. J Biol Chem. 1982;257:5013–5017. [PubMed] [Google Scholar]
- 32.Roujeinikova A, et al. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J Mol Biol. 2007;365:135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 33.Chan DI, Stockner T, Tieleman DP, Vogel HJ. Molecular dynamics simulations of the Apo-, Holo-, and acyl-forms of Escherichia coli acyl carrier protein. J Biol Chem. 2008;283:33620–33629. doi: 10.1074/jbc.M805323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenberg MA, Star C. Synthesis of 7-oxo-8-aminopelargonic acid, a biotin vitamer, in cell-free extracts of Escherichia coli biotin auxotrophs. J. Bacteriol. 1968;96:1291–1297. doi: 10.1128/jb.96.4.1291-1297.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cryle MJ, Schlichting I. Structural insights from a P450 carrier protein complex reveal how specificity is achieved in the P450 BioI ACP complex. Proc. Natl. Acad. Sci. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stok JE, De Voss J. Expression, purification, and characterization of BioI: a carboncarbon bond cleaving cytochrome P450 involved in biotin biosynthesis in Bacillus subtilis. Arch Biochem Biophys. 2000;384:351–360. doi: 10.1006/abbi.2000.2067. [DOI] [PubMed] [Google Scholar]
- 37.Miller J. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 38.Rock CO, Cronan JE., Jr Acyl carrier protein from Escherichia coli. Methods Enzymol. 1981;71:341–351. doi: 10.1016/0076-6879(81)71043-7. [DOI] [PubMed] [Google Scholar]
- 39.Cronan JE, Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.