Abstract
Systemic lupus erythematosus (SLE), an autoimmune disease, develops at a female-to-male ratio of 10:1. Increased serum levels of type I interferons (IFN-α/β) and induction of “IFN-signature” genes are associated with an active SLE disease in patients. Moreover, SLE patients exhibit three- to four-fold increase in the risk of developing malignancies involving B cells, including non-Hodgkin lymphoma (NHL) and Hodgkin's lymphoma (HL). Interestingly, homozygous mice expressing a deletion mutant (the proline-rich domain deleted) of the p53 develop various types of spontaneous tumors, particularly of B-cell origin upon aging. The deletion is associated with defects in transcriptional activation of genes by p53 and inhibition of DNA damage-induced apoptosis. Notably, increased levels of the p202 protein, which is encoded by the p53-repressible interferon-inducible Ifi202 gene, in B cells of female mice are associated with defects in B cell apoptosis, inhibition of the p53-mediated transcription of pro-apoptotic genes, and increased lupus susceptibility. In this review we discuss how increased levels of the p202 protein (and its human functional homologue IFI16 protein) in B cells increase lupus susceptibility and are likely to increase the risk of developing certain B cell malignancies. A complete understanding of the molecular mechanisms that regulate B cell homeostasis is necessary to identify SLE patients with an increased risk to develop B cell malignancies.
Keywords: SLE, interferons, sex bias, B cell malignancies, p53, apoptosis, p200-family
1. Introduction
Defects in innate and adaptive immune responses are associated with the development of systemic lupus erythematosus (SLE), an autoimmune disease with strong sex bias [1-3]. SLE patients develop pathogenic autoantibodies against nuclear antigens and the disease involves multiple organs, including the kidneys [2]. Moreover, consistent with a role for the innate immune responses in SLE, increased serum levels of type I interferons (IFN-α/β) and induction of “IFN-signature” genes are associated with active SLE disease in patients [4-6]. The IFNs are a family of cytokines, which inhibit cell proliferation and modulate cell survival [7]. In particular, IFNs inhibit apoptosis induced by signaling through the B-cell receptor [8]. Therefore, it is interesting that individuals with SLE exhibit an increased susceptibility to develop certain types of cancers [9,10]. Of particular concern are three- to four-fold increases in the risk of developing malignancies involving B cells, including non-Hodgkin lymphoma (NHL) and Hodgkin's lymphoma (HL) [9,10].
Mouse models of SLE have provided important insight into the pathogenesis of this disease and defects in elimination of self-reactive cells and increased expression of the IFN-inducible genes (ISGs) are also associated with the development of lupus-like disease in certain strains of mice [1,4,5]. Moreover, mice that are defective in the type I IFN-signaling do not develop lupus-like disease [11]. Furthermore, certain lupus-prone strains of mice are reported to develop malignancies, including the malignancies involving the B cells [12,13,14].
In this review we focus on the role of the IFN-inducible p200-family proteins, such as the murine p202 protein, in the modulation of B cell survival and its role in lupus susceptibility. In addition, we also discuss the relevance of findings concerning the p202 protein in human SLE and an increased risk to develop B cell malignancies.
2. p53 in B cell survival and SLE
p53 protein is a sequence-specific transcription factor, which is conserved (78% amino acid residues unchanged) between mouse and humans [15,16]. Upon activation by various stress stimuli, the p53 protein binds to the regulatory region of target genes as a tetramer and regulates transcription [16]. The binding of p53 to the regulatory region of the target genes can either activate or repress transcription of genes. Therefore, transcriptional activation of pro-apoptotic genes by p53 as well as transcriptional repression of the anti-apoptotic genes is thought to contribute to the cell growth regulatory and apoptotic functions of the p53 [16]. The cell growth-inhibitory activities of p53 are important for tissue homeostasis as well as in protection against the development of cancers. Notably, mutations in the TP53 gene are associated with a poorer prognosis in NHL, diffuse large B-cell lymphoma (DLBCL), and follicular lymphoma [17,18]. Furthermore, restoration of the p53 function in certain human lymphoma cell lines inhibits cell proliferation and induces apoptosis [19].
Studies have indicated that the p53 induces apoptosis through transcription-dependent and independent mechanisms [20,21]. The transcription-dependent mechanisms include binding of p53 protein to the regulatory region of target genes in a sequence-specific manner, resulting in transcriptional activation of pro-apoptotic genes (for example, PUMA and gadd45) and transcriptional repression of anti-apoptotic genes, such as survivin and Ifi202 [22,23]. The transcription-independent mechanisms include the mitochondrial pathway, which involves translocation of p53 protein to mitochondria and release of pro-apoptotic proteins, such as cytochrome c, from the mitochondria, resulting in activation of the effector caspases [24,25]. Moreover, a study has revealed that the translocation of p53 to mitochondria in response to death stimuli triggers a rapid wave of caspase-3 activation and cell death during the physiologic death response of radiosensitive organs, such as spleen and thymus, in vivo [26].
Defects in p53-mediated apoptosis are linked to the development of SLE. In blood lymphocytes from SLE patients with an active disease, relatively higher levels of the p53 protein are detected [21]. Given that type I IFNs increase p53 levels [27], which remain transcriptionally inactive, it remains unclear whether p53 in lymphocytes of SLE patients remains transcriptionally active or not. Moreover, p53 is required for spontaneous autoantibody production in B6/lpr lupus-prone mice [28]. In contrast, mice null for the Gadd45a gene develop lupus-like disease [29] and loss of expression of p21WAF1 (encoded by the p21 gene), a known transcriptional target of p53, is also linked to the development of autoimmunity [30]. p53 inhibits autoimmune diabetes and innate immune responses through down-regulating the expression of STAT1, a transcription factor that activates transcription of the IFN-inducible genes, and production of proinflammatory cytokines [31]. These observations make it likely that, depending upon the system, defects in p53-mediated transcriptional activation and repression of genes that regulate cell survival contribute to the development of autoimmune diseases.
Protein-protein interactions are central to the regulation of the p53-mediated functions, including apoptosis [15,16]. It has been proposed that the proline-rich domain (PRD; amino acid residues 63-85; Fig. 1) of p53 is important for the p53-mediated apoptosis that is induced by DNA damage involving transcription-dependent and independent apoptosis [16,32]. Based on in vitro studies involving cultured cells, it has been noted that the PRD domain is indispensible for p53-mediated transcriptional repression of the target genes [33]. However, based on in vivo studies involving generation of mice with mutations in the PRD domain of p53, the role of PRD domain in the regulation of p53-mediated apoptosis remains unresolved. A recent study by Slatter et al [34] investigated the role of PRD domain in p53-mediated apoptosis by generating p53 homozygous (p53 mΔpro/mΔpro) and heterozygous (p53+/mΔpro) mice (on C57BL/6 background) that lacked the amino acid residues 58-88 within the PRD domain in p53 (Fig. 1). Characterization of these mice revealed that deletion of amino acid residues 55-88 within the PRD domain in the murine p53 is associated with inhibition of DNA damage-induced apoptosis. However, the deletion mutant could induce an arrest of cell-cycle progression. Notably, cells from the mutant mice were defective in the p53-mediated transcriptional activation of genes that encode the pro-apoptotic proteins. However, the expression of the p53-repressible genes was not analyzed. The study also noted that defects in expression of pro-apoptotic proteins were associated with the development of various types of spontaneous tumors, particularly of B-cell origin in aged (7 months old) mice [34]. The prominence of B-cell malignancies in the homozygous (p53 mΔpro/mΔpro) rather than the heterozygous (p53+/mΔpro) mice suggests a relatively complex role of p53 as a transcriptional regulator during the B cell homeostasis.
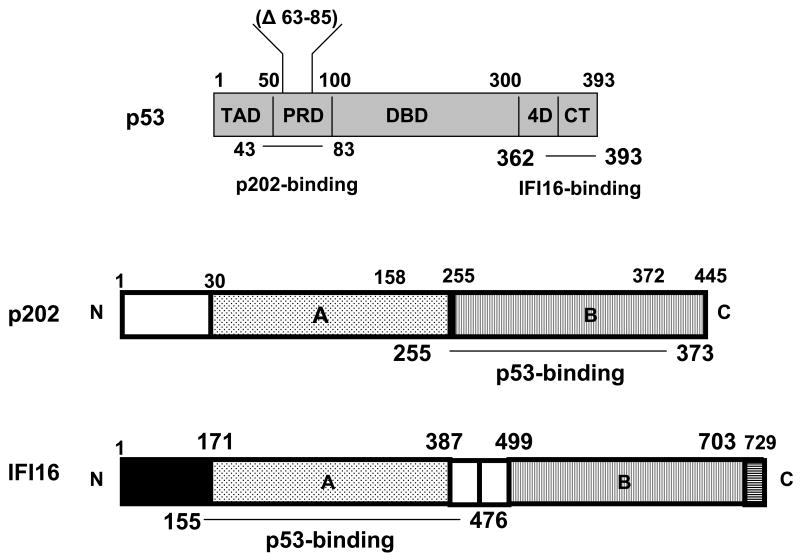
Fig. 1.
Top panel: p53 protein and its structural domains: the transactivation domain (TAD), proline-rich domain (PRD), specific DNA-binding domain (DBD), tetramerization domain (4D), and C-terminal regulatory (CT) domain. The p202-binding region (amino acids residues 43-83) within the proline-rich domain of the p53 protein is indicated [46]. The amino acid residues (Δ63-85) that were deleted in the murine p53 protein in the study by Slatter et al. [34] are also indicated. The IFI16-binding region (amino acids residues 362-393) within the proline-rich domain of the human p53 protein is indicated [63]. Middle panel: p202 protein and its structural domains. The p53-binding region (amino acid residues 255-273) in the p202 protein is indicated. Lower panel: IFI16 protein and its structural domains. The p53-binding region (amino acid residues 155-476) in the IFI16 protein is indicated.
3. p202 protein in lupus susceptibility
The p202a protein (encoded by Ifi202a gene) is a member of the interferon-inducible p200-protein family [35,36]. The family includes structurally and functionally-related murine (for example, p202a, p202b, p204, and Aim2) and human (for example, IFI16 and AIM2) proteins [35]. The protein p202a differs from the p202b protein (encoded by Ifi202b gene) in the N-terminus by seven amino acids residues [37]. Both proteins contain two repeat of 200-amino acids residues [35]. The repeat contains two consecutive oligosaccharide/ oligonucleotide binding (OB-fold) folds [38], which allow the p202a protein to bind single and double stranded DNA and interact with other proteins, including the p53 and NF-κB (p50 and p65) [35,39].
Expression levels of the p202 protein depend on the mouse strain and gender [36,40]. Moreover, constitutive basal levels of the p202 protein are detected in the cytoplasm of embryonic fibroblasts (MEFs) derived from the B6.Nba2 congenic (congenic for the NZB derived Nba2 interval on C57BL/6 background) mice [36,41]. Furthermore, IFN-α treatment of MEFs, which induces the p202 expression, potentiates the nuclear accumulation of the p202 protein [42]. Similarly, p202 protein is detected in the cytoplasm of bone marrow-derived macrophages (BMM) and in splenic B cells from C57BL/6 female mice [43,44]. In contrast, in mouse fibroblast cell lines and in splenic B cells from the B6.Nba2 female mice, the p202 protein is detected both in the cytoplasm and in the nucleus [44,45]. Consistent with the presence of a mitochondrial targeting sequence in the p202a protein in the N-terminus, in the cytoplasm, a significant fraction of the p202a protein is associates with the mitochondria [42]. Considering the above observations, it seems likely that sub-cellular (cytoplasmic versus nuclear) localization of the p202 protein depends on several factors, including the mouse strain, gender, cell types, and IFNs.
Increased levels of the p202 protein in B6.Nba2 splenic B cells are associated with defects in apoptosis and accumulation of B cells in spleen, thus, resulting in splenomegaly [46]. We have noted that increased expression of p202 in B6.Nba2 splenocytes is associated with up-regulation of p53 protein levels and a significant (>70%) reduction in steady-state levels of Gadd45a and PUMA mRNAs (PUMA is BH3-only pro-apoptotic protein), and a 50-60% decrease in levels of p21 and Mdm2 mRNAs [46]. Moreover, increased levels of the p202 in B6.Nba2 fibroblasts are associated with increases in survivin protein, an antiapopototic protein, expression of which is transcriptionally repressed by the p53 [22]. Furthermore, p202 protein binds to p53 in the PRD domain (Fig. 1) [46]. These observations suggest that increased levels of p202 in B6.Nba2 B cells inhibit the p53-mediated transcriptional activation of genes that encode pro-apoptotic proteins as well as transcriptional repression of genes that encode anti-apoptotic proteins (Fig. 2).
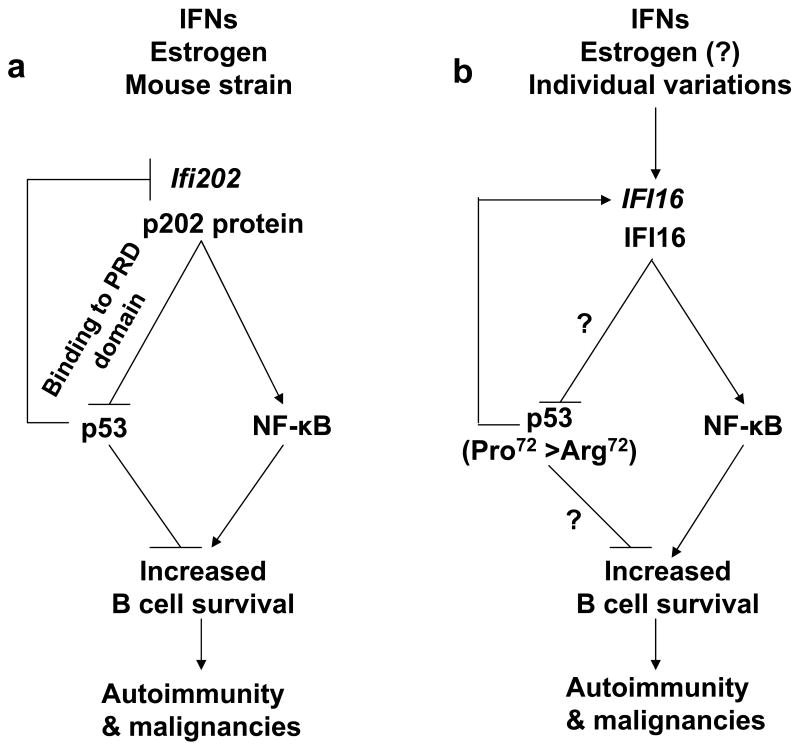
Fig. 2.
Model for the proposed role of the p202 and IFI16 proteins in the regulation of B cell survival. (a) Increased levels of p202 in B cells from the B6.Nba2 congenic female mice are associated with defects in apoptosis [46]. The defect is associated with inhibition of the p53-mediated transcriptional activation of pro-apoptotic genes and inhibition of p53-mediated transcriptional repression of anti-apoptotic genes. The p202 protein binds to the PRD domain of the p53 [46]. The p202 protein also modulates the transcriptional activity of NF-κB in a cell type-dependent manner [57-59]. Increased levels of the p202 protein in splenic B cells from B6.Nba2 female mice are associated with the activation of NF-κB transcriptional activity. (b) Increased levels of IFI16 protein in immune cells are predicted to increase B cell survival by inhibiting the p53-mediated transcription of pro-apoptotic genes. The Pro/Pro genotype (as compared to the Arg/Arg genotype) of the p53 in SLE patients may increase the risk of developing B cell malignancies by up-regulating the expression of the IFI16 gene. The IFI16 protein activates the NF-κB transcriptional activity in endothelial cells [68].
Given that: (i) the p53 protein negatively regulates expression of the Ifi202 gene [23]; (ii) the p202 protein can bind the PRD domain of p53 [46]; (iii) increased levels of p202 protein in B6.Nba2 lupus-prone female mice are associated with an inhibition of p53-mediated transcriptional activation of pro-apoptotic genes [46]; and (iv) p202 protein inhibits the p53-induced apoptosis [46], it is likely that increased levels of p202 in B cells contribute to increased risk to develop B cell malignancies (Fig. 2).
4. NF-κB and p202 in lupus susceptibility
The NF-κB family of transcription factors includes p50, p52 (NF-κB1 and NF-κB2, respectively), p65 (RelA), c-Rel (Rel) and RelB proteins [47,48]. The p65, c-Rel, and RelB proteins contain C-terminal transcriptional activation domains. Members of the NF-κB family contain a Rel homology domain, which contains a nuclear localization sequence and is involved in DNA binding, dimerization and interaction with κB inhibitory proteins (I κB). The NF-κB proteins can form homo- or heterodimers in response to various stimuli. In response to an activating signal, the p50 and p65 heterodimer is rapidly activated in cells to activate the transcription of the target genes. In contrast, the p50/p50 and p52/p52 homodimers that lack transcriptional activation domains act as transcriptional repressors [48].
SLE patients exhibit high plasma levels of the B cell activating factor (BAFF) [49], which activates the NF-κB activity [50]. Like SLE patients, high serum levels of BAFF are also noted in the murine models of the lupus disease and mice that overproduce BAFF develop SLE- like disease and exhibit B cell activation [51]. The BAFF enhances the long-term B cell survival primarily through the NF-κB activation [52]. The activation of NF-κB results in integrin up-regulation, thereby retention of the autoreactive B cells in the splenic marginal zone, a compartment that contributes to the survival of the B cells. Although, limited information is available on the role of the NF-κB in lupus disease, a recent study compared the NF-κB status in B cells between lupus-prone and non lupus-prone mice [53]. The study noted that the NF-κB pathway is significantly up-regulated in B cells from unmanipulated and anti-IgM-stimulated bicongenic mice as well as in BXSB and MRL/lpr mice but not in B6.Sle1z monocongenic mice [53]. Since NF-κB is upregulated only in the strains that develop severe lupus (but not the B6.Sle1z congenic strain), this study concluded that NF-κB activation may be necessary for the development of lupus disease. Similarly, in a mouse model of SLE, in which deletion of the inhibitory Fc receptor FcγRIIb results in the production of anti-nuclear antibodies and glomerulonephritis, the expression of the inhibitory molecule IκB-α is significantly decreased in mice that developed SLE [54]. Thus, these studies revealed that the occurrence of SLE in patients and certain mouse models of the disease are significantly associated with the activation of NF-κB.
Although studies using various mouse models of the lupus disease have suggested a role for NF-κB in B cell survival, it remains unclear whether the female sex hormones regulate the NF-κB activity in B cells. A study has reported that treatment of total splenic cells with the female sex hormone estrogen activates the NF-κB activity [56]. Moreover; IFN induces NF-κB activation to mediate IFN-dependent cell survival signals [56].
The Lengyel laboratory has demonstrated that increases (∼50% increase) in levels of p202 protein in cultured murine fibroblast cell lines inhibit NF-κB-mediated transcription of reporter genes and certain target genes [57,58]. The increased levels of p202 protein in cells inhibit the specific DNA-binding activity of the transcriptionally active p65 homodimers and p50/65 heterodimers [58]. However, the specific DNA-binding activity of the transcriptional repressive p50 homodimers is increased by p202 protein. The p202 protein binds to the p50 and p65 proteins in vitro and in vivo, and the p202 protein is detected in the p50 homodimer complex that binds to DNA [58]. However, increased expression of p202 in (NZB × NZW) F1 dendritic cells has been reported to stimulate the NF-κB-mediated transcription of target genes, such as IL-6 and IL-12p40 [59]. Similarly, increased levels of p202 protein in splenic B cells from B6.Nba2 congenic female mice (as compared with the age and gender-matched C57BL/6 mice) are associated with increased basal and TNF-α-induced transcriptional activity of the NF-κB (Shen and Choubey, unpublished data). Therefore, it is likely that increased levels of p202 protein in cells regulate the NF-κB activity in a cell type-dependent manner.
Interestingly, p53 dependent apoptosis also requires the transcriptional activity of NF-κB in some cell types [60]. Moreover, analogous to the p53 function during the B cell development, the NF-κB also significantly contributes to the regulation of B cell homeostasis and inhibition of NF-κB results in accumulation of immature B cells [48,61]. Therefore, it is likely that p53 and NF-κB transcription factors cooperate with each other in regulating the B cell development. Given that the p202 protein can also modulate the transcriptional activity of NF-κB in cell-type dependent manner [57-59], it is likely that increased levels of the p202 protein in B cells, which are associated with increased NF-κB activity, contribute to increased B cell survival (Fig. 2). The increased B cell survival is known to contribute to B cell malignancies and autoimmunity [48].
5. IFI16 protein, a functional homologue of the p202
The p202 protein does not have any known human homologue [36,62]. However, the IFI16 protein, a member of the p200-protein family, is functionally similar protein in certain aspects [62]. The IFI16 protein is detected primarily in the nucleus and increased expression of IFI16 protein in normal human cells is associated with cellular senescence-associated permanent cell growth arrest. Notably, up to 29% SLE patients develop high titer autoantibodies to the IFI16 protein [62]. Although, the role of IFI16 protein in B cell survival remains unknown, the following observations make the IFI16 protein a good candidate for an increased risk to develop B cell malignancies in SLE patients: (i) IFI16 protein binds to p53 (Fig. 1) [63]; (ii) expression of IFI16 protein was reported to inhibit the p53-mediated transcription of the p21 gene in human osteosarcoma cell line U2OS [62], thus, raising the possibility that the IFI16-mediated regulation of p53-mediated transcription depends on cell type [62]; (iii) expression of the IFI16 gene is reported to vary among individuals [62], thus, raising the possibility that basal and IFN-induced increased levels of IFI16 protein in immune cells of certain SLE patients may inhibit the p53-mediated transcription of target genes; (iv) increased levels of IFI16 mRNA are associated with several aging-associated epithelial cancers [62]; and (v) functional p53 up-regulates expression of the IFI16 gene in human normal diploid fibroblasts [64]. Given that re-expression of p53 (Pro72) polymorphic form in Saos-2 cells up-regulates the expression of IFI16 protein relatively more than the p53 (Arg72) form [65], it is conceivable that the Pro/Pro genotype (as compared to the Arg/Arg genotype) of the p53 in SLE patients may increase the risk of developing B cell malignancies. Accordingly, it is known that the individuals who carry the Pro72 allele of the p53 have a higher risk of developing SLE than those who carry the Arg72 allele [66]. Moreover, mutations in the p53 encoding gene (TP53) are unfavorable prognostic factor in lymphomas in autoimmune diseases [67]. Therefore, it would be important to determine whether a particular genotype of the p53 in SLE patients is associated with increased levels of the IFI16 protein and, more importantly, with an increased risk to develop B cell malignancies (Fig. 2).
Persistent activation of the NF-κB transcription factor is associated with the development of B cell lymphoma [48]. Moreover, the IFI16 protein activates the NF-κB transcription factor and potentiates the transcription of the target genes in human endothelial cells [68]. Therefore, it is conceivable that IFN-increased levels of the IFI16 protein in B cells activate NF-κB transcription factor and increase transcription of the target genes. In addition, NF-κB induces IL-6 expression in response to a variety of signals and IL-6 induces the IFI16 expression in human cell lines [62]. These observations along with the observations that SLE patients exhibit increased levels of type I IFNs and IL-6 may suggest that the IFI16-NF-κB-IL-6 axis could contribute to the B cell pathologies, including the increased risk to develop B cell malignancies (Fig. 1).
6. Conclusions
Studies have revealed that development of SLE in patients and certain mouse models of the disease have several common risk factors, including being female, increased serum levels of type I IFNs, and induced expression of “IFN signature” genes in immune cells [2,3,7]. Therefore, an increased risk to develop B cell malignancies among SLE patients raise several questions, including: Do the female sex hormones (for example, estrogen) and increased levels of the type I IFNs contribute to an increased risk to develop B cell malignancies in SLE patients? Do signaling pathway(s) that are activated by the female sex hormones and/or the type I IFNs increase the B cell survival? Do proteins, expression of which is regulated by the female sex hormones and/or type I IFNs, modulate the B cell survival? To begin to address these important questions, it would be important to determine whether increased nuclear levels of the estrogen and IFN-inducible p202 protein, which are associated with increased lupus susceptibility in certain strains of female mice, also contribute to B cell malignancies. The availability of mouse models, such as the one described by Slatter et al. [34] should allow to test whether: (i) levels of the p202 protein increase in the homozygous (p53 mΔpro/mΔpro) mice than the wild type mice; (ii) there are differences between the homozygous and heterozygous (p53+/mΔpro) mice with respect to the p202 expression; and (iii) there is sex bias in the development of various B cell malignancies in the homozygous (p53 mΔpro/mΔpro) mice. In addition, a careful re-examination of the current strains of lupus-prone mice (including the NZB and (NZB/W)F1), which express increased levels of the p202 protein [36], is needed to determine whether increased levels of the p202 protein in B cells are associated with an increased risk to develop B cell malignancies. Furthermore, the role of IFI16 protein needs to be investigated in B cell survival following the activation of p53. A complete understanding of the molecular mechanisms by which the p200-family proteins regulate B cell homeostasis is necessary to identify SLE patients with an increased risk to develop B cell malignancies.
Acknowledgments
We thank former and current members of the Choubey laboratory for their contributions to our progress. Studies in the Choubey laboratory are supported by the NIH (AI026066, AG025035) and VA Merit Awards.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85:303–6. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- 2.Crispín JC, Liossis SN, Kis-Toth K, Lieberman LA, Kyttaris VC, Juang YT, et al. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends Mol Med. 2010;16:47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pisetsky DS. The role of innate immunity in the induction of autoimmunity. Autoimmun Rev. 2008;8:69–72. doi: 10.1016/j.autrev.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 4.Baccala R, Kono DH, Theofilopoulos AN. Interferons as pathogenic effectors in autoimmunity. Immunol Rev. 2005;204:9–26. doi: 10.1111/j.0105-2896.2005.00252.x. [DOI] [PubMed] [Google Scholar]
- 5.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–36. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 6.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Su L, David M. Inhibition of B cell receptor-mediated apoptosis by IFN. J Immunol. 1999;162:6317–21. [PubMed] [Google Scholar]
- 9.Gayed M, Bernatsky S, Ramsey-Goldman R, Clarke A, Gordon C. Lupus and cancer. Lupus. 2009;18:479–85. doi: 10.1177/0961203309102556. [DOI] [PubMed] [Google Scholar]
- 10.Bernatsky S, Ramsey-Goldman R, Clarke AE. Malignancy in systemic lupus erythematosus: what have we learned? Best Pract Res Clin Rheumatol. 2009;23:539–47. doi: 10.1016/j.berh.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, et al. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197:777–88. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellors RC. Autoimmune disease in NZB-Bl mice. II. Autoimmunity and malignant lymphoma. Blood. 1966;27:435–48. [PubMed] [Google Scholar]
- 13.Sugai S, Palmer DW, Talal N, Witz IP. Protective and cellular immune responses to idiotypic determinants on cells from a spontaneous lymphoma of NZB-NZW F1 mice. J Exp Med. 1974;140:1547–58. doi: 10.1084/jem.140.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaglione BJ, Salerno E, Balan M, Coffman F, Landgraf P, Abbasi F, et al. Murine models of chronic lymphocytic leukaemia: role of microRNA-16 in the New Zealand Black mouse model. Br J Haematol. 2007;139:645–57. doi: 10.1111/j.1365-2141.2007.06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 16.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–42. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 17.Ichikawa A, Kinoshita T, Watanabe T, Kato H, Nagai H, Tsushita K, et al. Mutations of the p53 gene as a prognostic factor in aggressive B-cell lymphoma. N Engl J Med. 1997;337:529–34. doi: 10.1056/NEJM199708213370804. [DOI] [PubMed] [Google Scholar]
- 18.Dolcetti R, Boiocchi M. Cellular and molecular bases of B-cell clonal expansions. Clin Exp Rheumatol. 1996 14:S3–13. [PubMed] [Google Scholar]
- 19.Buttgereit P, Schakowski F, Märten A, Brand K, Renoth S, Ziske C, et al. Effects of adenoviral wild-type p53 gene transfer in p53-mutated lymphoma cells. Cancer Gene Ther. 2001;8:430–39. doi: 10.1038/sj.cgt.7700323. [DOI] [PubMed] [Google Scholar]
- 20.Rathmell JC, Thompson CB. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell. 2002;109(Suppl):S97–107. doi: 10.1016/s0092-8674(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 21.Miret C, Molina R, Filella X, Garcia-Carrasco M, Claver G, Ingelmo M, et al. Relationship of p53 with other oncogenes, cytokines and systemic lupus erythematosus activity. Tumour Biol. 2003;24:185–88. doi: 10.1159/000074428. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–57. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 23.D'Souza S, Xin H, Walter S, Choubey D. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J Biol Chem. 2001;276:298–305. doi: 10.1074/jbc.M007155200. [DOI] [PubMed] [Google Scholar]
- 24.Chipuk JE, Green DR. Cytoplasmic p53: bax and forward. Cell Cycle. 2004;3:429–431. [PubMed] [Google Scholar]
- 25.Baptiste N, Prives C. p53 in the cytoplasm: a question of overkill? Cell. 2004;116:487–89. doi: 10.1016/s0092-8674(04)00164-3. [DOI] [PubMed] [Google Scholar]
- 26.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–41. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, et al. Integration of interferon-alpha/beta signaling to p53 responses in tumor suppression and antiviral defense. Nature. 2003;424:516–23. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 28.Kuan AP, Cohen PL. p53 is required for spontaneous autoantibody production in B6/lpr lupus mice. Eur J Immunol. 2005;35:1653–60. doi: 10.1002/eji.200525982. [DOI] [PubMed] [Google Scholar]
- 29.Salvador JM, Hollander MC, Nguyen AT, Kopp JB, Barisoni L, Moore JK, et al. Mice lacking the p53-effector gene Gadd45a develop a lupus-like syndrome. Immunity. 2002;16:499–508. doi: 10.1016/s1074-7613(02)00302-3. [DOI] [PubMed] [Google Scholar]
- 30.Balomenos D, Martín-Caballero J, García MI, Prieto I, Flores JM, Serrano M, et al. The cell cycle inhibitor p21 controls T-cell proliferation and sex-linked lupus development. Nat Med. 2000;6:171–76. doi: 10.1038/72272. [DOI] [PubMed] [Google Scholar]
- 31.Zheng SJ, Lamhamedi-Cherradi SE, Wang P, Xu L, Chen YH. Tumor suppressor p53 inhibits autoimmune inflammation and macrophage function. Diabetes. 2005;54:1423–28. doi: 10.2337/diabetes.54.5.1423. [DOI] [PubMed] [Google Scholar]
- 32.Murphy ME. The thousand doors that lead to death: p53-dependent repression and apoptosis. Cancer Biol Ther. 2003;2:381–82. doi: 10.4161/cbt.2.4.439. [DOI] [PubMed] [Google Scholar]
- 33.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–79. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slatter TL, Ganesan P, Holzhauer C, Mehta R, Rubio C, Williams G, et al. p53-mediated apoptosis prevents the accumulation of progenitor B cells and B-cell tumors. Cell Death Differ. 2010;17:540–50. doi: 10.1038/cdd.2009.136. [DOI] [PubMed] [Google Scholar]
- 35.Choubey D, Kotzin BL. Interferon-inducible p202 in the susceptibility to systemic lupus. Front Biosci. 2002;7:e252–62. doi: 10.2741/A921. [DOI] [PubMed] [Google Scholar]
- 36.Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythematosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Chatterjee G, Meyer JJ, Liu CJ, Manjunath NA, Bray-Ward P, Lengyel P. Characteristics of three homologous 202 genes (Ifi202a, Ifi202b, and Ifi202c) from the murine interferon-activatable gene 200-cluster. Genomics. 1999;60:281–94. doi: 10.1006/geno.1999.5923. [DOI] [PubMed] [Google Scholar]
- 38.Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB-folds. Biochem Biophys Res Commun. 2005;327:679–87. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 39.Choubey D, Duan X, Dickerson E, Ponomareva L, Panchanathan R, Shen H, et al. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010 Feb 28; doi: 10.1089/jir.2009.0096. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panchanathan R, Shen H, Bupp MG, Gould KA, Choubey D. Female and male sex hormones differentially regulate expression of Ifi202, an interferon-inducible lupus susceptibility gene within the Nba2 interval. J Immunol. 2009;183:7031–38. doi: 10.4049/jimmunol.0802665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, et al. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–43. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- 42.Choubey D, Pramanik R, Xin H. Sub-cellular localization and mechanisms of nucleocytoplasmic distribution of p202, an interferon-inducible candidate for lupus susceptibility. FEBS Lett. 2003;553:245–49. doi: 10.1016/s0014-5793(03)01006-8. [DOI] [PubMed] [Google Scholar]
- 43.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 44.Choubey D, Panchanathan R, Shen H, Duan X. Comment on “Development of Murine Lupus Involves the Combined Genetic Contribution of the SLAM and FcγR Intervals within the Nba2 Autoimmune Susceptibility Locus”. J Immunol. 2010;184:4051–52. doi: 10.4049/jimmunol.1090015. [DOI] [PubMed] [Google Scholar]
- 45.Choubey D, Lengyel P. Interferon action: cytoplasmic and nuclear localization of the interferon-inducible 52-kD protein that is encoded by the Ifi200-gene from the gene 200-cluster. J Interferon Res. 1993;13:43–52. doi: 10.1089/jir.1993.13.43. [DOI] [PubMed] [Google Scholar]
- 46.Xin H, D'Souza S, Jorgensen TN, Vaughan AT, Lengyel P, Kotzin BL, et al. Increased expression of Ifi202, an IFN-activatable gene, in B6.Nba2 lupus susceptible mice inhibits p53-mediated apoptosis. J Immunol. 2006 May 15;176:5863–70. doi: 10.4049/jimmunol.176.10.5863. [DOI] [PubMed] [Google Scholar]
- 47.Huxford T, Malek S, Ghosh G. Structure and mechanism in NF-kappa B/I kappa B signaling. Cold Spring Harb Symp Quant Biol. 1999;64:533–40. doi: 10.1101/sqb.1999.64.533. [DOI] [PubMed] [Google Scholar]
- 48.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 49.Do RK, Chen-Kiang S. Mechanism of BLyS action in B cell immunity. Cytokine Growth Factor Rev. 2002;13:19–25. doi: 10.1016/s1359-6101(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 50.Laâbi Y, Strasser A. Immunology. Lymphocyte survival--ignorance is BLys. Science. 2000;289:883–84. doi: 10.1126/science.289.5481.883. [DOI] [PubMed] [Google Scholar]
- 51.Mackay F, Woodcock SA, Lawton P, Ambrose C, Baetscher M, Schneider P, et al. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–1710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enzler T, Bonizzi G, Silverman GJ, Otero DC, Widhopf GF, Anzelon-Mills A, et al. Alternative and classical NF-kappa B signaling retain autoreactive B cells in the splenic marginal zone and result in lupus-like disease. Immunity. 2006;25:403–15. doi: 10.1016/j.immuni.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Wu T, Qin X, Kurepa Z, Kumar KR, Liu K, Kanta H, et al. Shared signaling networks active in B cells isolated from genetically distinct mouse models of lupus. J Clin Invest. 2007;117:2186–96. doi: 10.1172/JCI30398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalergis AM, Iruretagoyena MI, Barrientos MJ, González PA, Herrada AA, Leiva ED, et al. Modulation of nuclear factor-kappaB activity can influence the susceptibility to systemic lupus erythematosus. Immunology. 2009;128(1 Suppl):e306–14. doi: 10.1111/j.1365-2567.2008.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai R, Phillips RA, Ahmed SA. Despite inhibition of nuclear localization of NF-kappa B p65, c-Rel, and RelB, 17-beta estradiol up-regulates NF-kappa B signaling in mouse splenocytes: the potential role of Bcl-3. J Immunol. 2007;179:1776–83. doi: 10.4049/jimmunol.179.3.1776. [DOI] [PubMed] [Google Scholar]
- 56.Yang CH, Murti A, Pfeffer LM. Interferon induces NF-kappa B-inducing kinase/tumor necrosis factor receptor-associated factor-dependent NF-kappa B activation to promote cell survival. J Biol Chem. 2005;280:31530–36. doi: 10.1074/jbc.M503120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min W, Ghosh S, Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-kappa B, c-Fos, and c-Jun activities. Mol Cell Biol. 1996;16:359–68. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma XY, Wang H, Ding B, Zhong H, Ghosh S, Lengyel P. The interferon-inducible p202a protein modulates NF-κB activity by inhibiting the binding to DNA of p50/p65 heterodimers and p65 homodimers while enhancing the binding of p50 homodimers. J Biol Chem. 2003;278:23008–19. doi: 10.1074/jbc.M302105200. [DOI] [PubMed] [Google Scholar]
- 59.Yamauchi M, Hashimoto M, Ichiyama K, Yoshida R, Hanada T, Muta T, et al. Ifi202, an IFN-inducible candidate gene for lupus susceptibility in NZB/W F1 mice, is a positive regulator for NF-kappaB activation in dendritic cells. Int Immunol. 2007;19:935–42. doi: 10.1093/intimm/dxm054. [DOI] [PubMed] [Google Scholar]
- 60.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–97. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 61.Feng B, Cheng S, Pear WS, Liou HC. NF-kB inhibitor blocks B cell development at two checkpoints. Med Immunol. 2004;3:1–16. doi: 10.1186/1476-9433-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choubey D, Deka R, Ho SM. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci. 2008;13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- 63.Johnstone RW, Wei W, Greenway A, Trapani JA. Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene. 2000;19:6033–42. doi: 10.1038/sj.onc.1204005. [DOI] [PubMed] [Google Scholar]
- 64.Kwak JC, Ongusaha PP, Ouchi T, Lee SW. IFI16 as a negative regulator in the regulation of p53 and p21(Waf1) J Biol Chem. 2003;278:40899–40904. doi: 10.1074/jbc.M308012200. [DOI] [PubMed] [Google Scholar]
- 65.Song LL, Alimirah F, Panchanathan R, Xin H, Choubey D. Expression of an IFN-inducible cellular senescence gene, IFI16, is up-regulated by p53. Mol Cancer Res. 2008;6:1732–41. doi: 10.1158/1541-7786.MCR-08-0208. [DOI] [PubMed] [Google Scholar]
- 66.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG. The functional p53 codon 72 polymorphism is associated with systemic lupus erythematosus. Lupus. 2005;14:842–45. doi: 10.1191/0961203305lu2224oa. [DOI] [PubMed] [Google Scholar]
- 67.Hoshida Y, Hongyo T, Xu JX, Sasaki T, Tomita Y, Nomura T, et al. TP53 gene mutation, an unfavorable prognostic factor for malignant lymphomas in autoimmune diseases. Oncology. 2005;69:175–83. doi: 10.1159/000087980. [DOI] [PubMed] [Google Scholar]
- 68.Caposio P, Gugliesi F, Zannetti C, Sponza S, Mondini M, Medico E, et al. A novel role of the interferon-inducible protein IFI16 as inducer of proinflammatory molecules in endothelial cells. J Biol Chem. 2007;282:33515–29. doi: 10.1074/jbc.M701846200. [DOI] [PubMed] [Google Scholar]