Abstract
Dictyostelium discoideum is a unicellular eukaryote that, when starved, aggregates to form multicellular structures. In this report, we identified the proteins secreted by developing Dictyostelium cells using mass spectrometry based proteomics. A total of 349 different secreted proteins were identified, indicating that at least 2.6% of the 13600 predicted proteins in the Dictyostelium genome are secreted. Gene ontology (GO) analysis suggests that many of the secreted proteins are involved in protein and carbohydrate metabolism, and proteolysis.
Keywords: Dictyostelium, development, secreted molecules, proteome
Dictyostelium discoideum is a unicellular eukaryote which lives in soil and proliferates by feeding on bacteria. Upon starvation, cells stop proliferating, communicate with each other by secreting signal molecules, and aggregate to form multicellular groups [1]. Each group consists of ~ 20,000 cells that form a fruiting body, which consists of a mass of spores held up by a column of stalk cells. These spores are dispersed by the wind and germinate to amoeboid cells to continue the life cycle. Because of a wide variety of molecular biology, biochemistry, and cell biology tools to study Dictyostelium, this system is used to elucidate many different mechanisms.
The Dictyostelium genome encodes 13600 predicted proteins [2, 3]. Transcriptome studies have been performed to understand the complex network of signal pathways that regulate these genes and thus Dictyostelium development [4, 5]. However, much remains to be understood about the proteins encoded by the genome. For instance, during development, Dictyostelium cells secrete a large number of different proteins [6], but the nature and function of most of the proteins are unknown. High-throughput identification of proteins in cells, or media conditioned by cells, can be done using mass spectrometry (MS), allowing MS-based proteomic techniques to generate proteome profiles that complement microarray data. Since many intracellular pathways are regulated by extracellular ligands, in this report we identify proteins that are secreted by developing Dictyostelium cells.
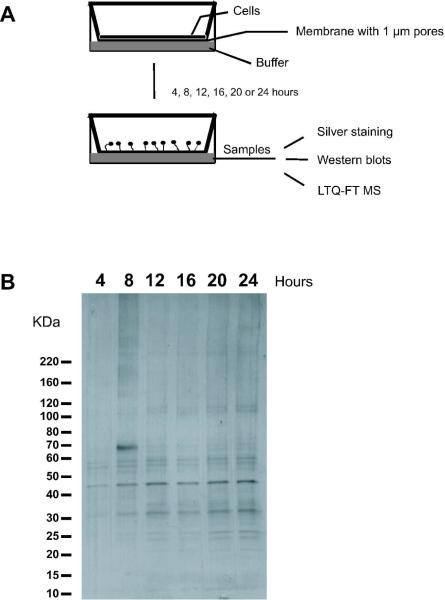
To collect secreted proteins, wild-type A×2 cells were grown in shaking culture as previously described [7]. Cells at mid-log phase (3 × 106 cells/ml) were collected by centrifugation at 1,500 × g for 3 minutes and washed twice by resuspending the pellets in PBM (20 mM KH2PO4, 10 μM CaCl2, 1 mM MgCl2, pH 6.1) and collecting cells by centrifugation. The final cell pellet was resuspended in PBM to 1 × 107cells/ml. Cells (107) were then pipetted on to a Type 353102 1 μm pore sized polyethylene terephthalate membrane six well format cell culture insert (Becton Dickinson, Franklin Lakes, NJ). After 10 minutes, the cells had settled on, and attached to, the membrane, and the buffer was gently removed from the insert. PBM was added into the well of a six well plate, and the insert with cells was then placed in the well (Figure 1A). The amount of buffer in the well was adjusted to just touch the membrane of the insert to keep the membrane and cells moist. The cells developed normally, with cells aggregating starting at 6 hours and forming fruiting bodies at 24 hours (data not shown). A different membrane and well was used for each time point. After the indicated hours (4, 8, 12, 16, 20, or 24) of development at 21°C, the conditioned starvation buffer in the well was collected and samples were stored at −80°C.
Figure 1. Proteins secreted by developing Dictyostelium cells.
(A) Experimental setup with wild-type cells on a porous membrane in contact with buffer. The developing cells aggregate and form fruiting bodies. During this period, molecules are secreted into the buffer. (B) Aliquots of the conditioned buffer were collected at the indicated times. Proteins in the conditioned buffer were separated by electrophoresis on a SDS-PAGE gel, which was then silver-stained.
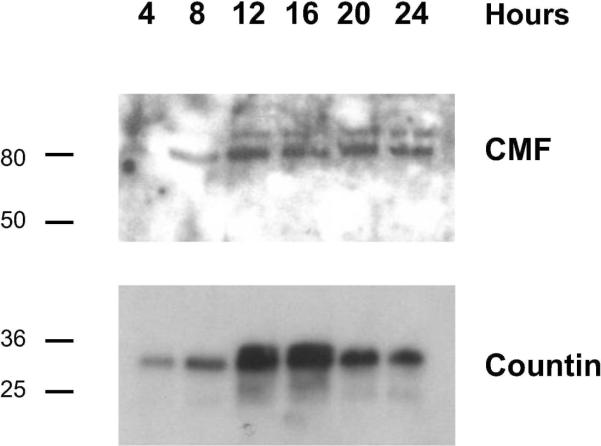
To verify that the samples contained proteins, 10 μl of conditioned buffer was boiled with 3 μl of 6× SDS sample buffer for 5 minutes, and proteins were separated by electrophoresis on 4–15 % Tris-Glycine gels (Biorad Laboratories, Hercules, CA). The gel was silver-stained as described previously [8]. Figure 1B shows a silver stained protein gel with a prominent band at 45 kDa and many other protein bands. We previously identified two proteins, conditioned medium factor (CMF) and countin, that are secreted by developing cells [9, 10]. To verify that these known secreted proteins were present in the conditioned medium samples, western blots were stained with anti-CMF or anti-countin antibodies. We found that CMF, a secreted protein that regulates aggregation [11] was present in our samples throughout development, with barely detectable levels at 4 hours and higher levels during later development (Figure 2). We found that countin, a secreted protein that determines aggregate size [10], was also present in our samples throughout development (Figure 2).
Figure 2. The conditioned starvation buffer contains known secreted proteins.
Proteins in the conditioned buffer were separated by electrophoresis on SDS-PAGE gels, and transferred to membranes. The membranes were then stained with affinity purified anti-CMF or anti-countin antibodies. The position of molecular mass markers (in kDa) is indicated at left.
To identify secreted proteins, the conditioned starvation buffer samples were then sent to the University of Utah mass spectrometry and proteomics core facility. This facility did a trypsin digest of the proteins in the conditioned medium. LC/MS/MS analyses on these samples were done as described previously described [12, 13], with the following modifications. The tryptic digests were reconstituted in 10 μL, and 5 μL of this was loaded on a 100 μm × 75 mm NanoLC column. To elute the peptides, a gradient of 6 to 86% solvent B was used at a flow rate of 350 nL/min for 78 minutes. The tandem MS/MS allowed peptides to be identified based on both the peptide mass and the mass of subfragments. All peptides used to identify a protein had MASCOT scores >19.
For each time point, 76 to 235 different proteins were identified giving a total of 349 identified proteins (Table 1 and supplementary information). A listing of the 349 proteins, along with the spectra and identified peptides, is also available in the PRIDE database (http://www.ebi.ac.uk/pride) with accession number 9943. In addition, lists of the peptide assignments are in the supplementary information. To identify functional classes of proteins, Cytoscape (http://cytoscape.org/index.php) was used as a software platform for GO analysis using the BiNGO plugin (http://www.psb.ugent.be/cbd/papers/BiNGO/). Statistical analysis for over-represented individual categories was done using hypergeometric tests with the Benjamini & Hochberg False Discovery Rate (FDR) correction at a significance level of p < 0.05. Protein identifications from the MS data file were converted to Dictybase identifiers to facilitate Gene Ontology (GO) analysis. The gene association file was downloaded from www.dictybase.org, which contains GO terms for all genes in the Dictyostelium genome.
Table 1.
Biological process of the identified secreted proteins
| Category | 4h | 8h | 12h | 16h | 20h | 24h | Total secreted protein |
|---|---|---|---|---|---|---|---|
| Protein metabolic process | 24 | 42 | 43 | 44 | 57 | 0 | 67 |
| Proteolysis | 12 | 16 | 22 | 21 | 24 | 12 | 29 |
| Carbohydrate metabolic process | 9 | 14 | 20 | 19 | 17 | 10 | 25 |
| Cell adhesion | 0 | 0 | 0 | 7 | 8 | 0 | 9 |
| Negative regulation of proliferation | 0 | 0 | 2 | 2 | 2 | 2 | 2 |
| Regulation of aggregate size | 0 | 0 | 3 | 0 | 0 | 0 | 3 |
| Others | 12 | 40 | 50 | 65 | 70 | 73 | 110 |
|
| |||||||
| unknown function | 19 | 33 | 50 | 65 | 57 | 30 | 104 |
|
| |||||||
| Total proteins identified | 76 | 145 | 190 | 223 | 235 | 127 | 349 |
The GO annotation categories of the identified secreted proteins that are significantly over-represented (based on BiNGO analysis) are listed at left. For each time point, the number of secreted proteins in that category is indicated. The total secreted protein column shows the total number of proteins in a given GO annotation category for all the time points; since many proteins are found at multiple time points, this number is not simply the sum of the numbers in the different time points. The dictybase number and protein description for each identified protein is given in the supporting information.
A data file listing predicted proteins (see supporting information) was generated from two independent experiments for each time point. Of the 349 identified proteins, 8% are associated with proteolysis, 19% with protein metabolism, 7% with carbohydrate metabolic processes, 36% are associated with other functions, and 30% are of unknown function (Table 1). At 4 hours, we detected several enzymes associated with proteolysis, in agreement with the observation that lyzosomal enzymes are secreted during early development [14, 15]. We found CMF and countin in the 8, 12, 16, 20, and 24 hour samples, indicating that known secreted proteins were detected by the MS. We also found adenylyl cyclase in the 16 hour samples; this may be due to the secretion of adenylyl cyclase-rich multivesicular bodies that act as chemoattractant-releasing exosomes providing a trail for migrating cells during aggregation [16]. The process of secreting multivesicular bodies is dependent on actin and protein synthesis [16]. We detected actin and ribosomal proteins among the secreted proteins (supplementary information); one possibility is that these proteins are associated with multivesicular bodies.
In summary, we identified 349 proteins secreted by developing Dictyostelium cells. We detected known proteins that are secreted as individual glycoproteins (CMF), secreted proteins that form a complex in the extracellular environment (countin), and proteins that are secreted in multivesicular bodies (adenylyl cyclase). Other known extracellular signaling proteins that were identified in this study include the proliferation-inhibiting proteins AprA and CfaD [7, 17], and the quorum sensing proteins DicA and B [18]. The proteins that we identified probably do not include proteins that accumulate to a very low level, or secreted proteins that immediately bind to cells, the substratum, or plastic. The absence of glycolytic enzymes suggests that very few of the proteins we detected were in the extracellular medium due to cell lysis. Thus at least 349 different proteins, and thus at least 2.6% of the 13600 proteins predicted to be encoded by the genome, appear to be secreted during Dictyostelium development.
The proteins we identified will be of interest for further research. We found secreted proteins with similarity to proteins with known enzymatic properties and which are parts of signal pathways [19–26]. These include dipeptidyl peptidase, cathepsin D, alpha-mannosidase, cysteine protease, phospholipase-D, protein phosphatase 2A and acid ceramidase. Some secreted proteins are inhibitors of these enzymes, and are also worth exploring. For instance, one of the secreted proteins we found is cystatin. Cystatins are potent inhibitors of cysteine proteases, and decreased expression of cystatins is associated with some tumors [27]. In the process of identifying proteins secreted during development, we found known secreted proteins as well as previously hypothetical proteins. The identification of proteins which accumulate in the extracellular medium during development will be valuable information for the Dictyostelium community, and in future this method could be used, for instance, to screen mutants with defects in secretory pathways.
Supplementary Material
Acknowledgments
This research was supported by NIH R01 GM074990. The authors thank Dr. Siddhartha Basu and Dictybase for help with converting the gene list to dictybase ID's. We also thank Dr. Krishna Parsawar for valuable discussions regarding MS analysis.
Footnotes
The authors have declared no conflict of interest.
References
- [1].Kessin RH. Dictyostelium - Evolution, cell biology, and the development of multicellularity. Cambridge Univ. Press; Cambridge, UK: 2001. [Google Scholar]
- [2].Eichinger L, Pachebat JA, Glockner G, Rajandream MA, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fey P, Gaudet P, Curk T, Zupan B, et al. dictyBase--a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Iranfar N, Fuller D, Loomis WF. Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Eukaryot Cell. 2003;2:664–670. doi: 10.1128/EC.2.4.664-670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Van Driessche N, Shaw C, Katoh M, Morio T, et al. A transcriptional profile of multicellular development in Dictyostelium discoideum. Development. 2002;129:1543–1552. doi: 10.1242/dev.129.7.1543. [DOI] [PubMed] [Google Scholar]
- [6].West CM, Erdos GW. The expression of glycoproteins in the extracellular matrix of the cellular slime mold Dictyostelium discoideum. Cell Differ. 1988;23:1–16. doi: 10.1016/0045-6039(88)90032-2. [DOI] [PubMed] [Google Scholar]
- [7].Bakthavatsalam D, Brock DA, Nikravan NN, Houston KD, et al. The secreted Dictyostelium protein CfaD is a chalone. J Cell Sci. 2008;121:2473–2480. doi: 10.1242/jcs.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Joachim Sasse SRG. John Wiley & Sons, Inc.; 2003. pp. 10.16.15–10.16.16. [Google Scholar]
- [9].Gomer RH, Yuen IS, Firtel RA. A secreted 80×10(3)Mr protein mediates sensing of cell density and the onset of development in Dictyostelium. Development. 1991;112:269–278. doi: 10.1242/dev.112.1.269. [DOI] [PubMed] [Google Scholar]
- [10].Brock DA, Gomer RH. A cell-counting factor regulating structure size in Dictyostelium. Genes Devel. 1999;13:1960–1969. doi: 10.1101/gad.13.15.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jain R, Yuen IS, Taphouse CR, Gomer RH. A density-sensing factor controls development in Dictyostelium. Genes Devel. 1992;6:390–400. doi: 10.1101/gad.6.3.390. [DOI] [PubMed] [Google Scholar]
- [12].Atkins JF, Wills NM, Loughran G, Wu CY, et al. A case for “StopGo”: reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go) RNA. 2007;13:803–810. doi: 10.1261/rna.487907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cassidy PB, Edes K, Nelson CC, Parsawar K, et al. Thioredoxin reductase is required for the inactivation of tumor suppressor p53 and for apoptosis induced by endogenous electrophiles. Carcinogenesis. 2006;27:2538–2549. doi: 10.1093/carcin/bgl111. [DOI] [PubMed] [Google Scholar]
- [14].Rossomando EF, Maldonado B, Crean EV, Kollar EJ. Protease secretion during onset of development in Dictyostelium discoideum. J. Cell Sci. 1978;30:305–318. doi: 10.1242/jcs.30.1.305. [DOI] [PubMed] [Google Scholar]
- [15].Dimond RL, Burns RA, Jordan KB. Secretion of Lysosomal enzymes in the cellular slime mold, Dictyostelium discoideum. J Biol Chem. 1981;256:6565–6572. [PubMed] [Google Scholar]
- [16].Kriebel PW, Barr VA, Rericha EC, Zhang G, Parent CA. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J Cell Biol. 2008;183:949–961. doi: 10.1083/jcb.200808105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Brock DA, Gomer RH. A secreted factor represses cell proliferation in Dictyostelium. Development. 2005;132:4553–4562. doi: 10.1242/dev.02032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kolbinger A, Gao T, Brock D, Ammann R, et al. A cysteine-rich extracellular protein containing a PA14 domain mediates quorum sensing in Dictyostelium discoideum. Euk. Cell. 2005;4:991–998. doi: 10.1128/EC.4.6.991-998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ehrmann M, Clausen T. Proteolysis as a regulatory mechanism. Annu Rev Genet. 2004;38:709–724. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- [20].Carl-McGrath S, Lendeckel U, Ebert M, Rocken C. Ectopeptidases in tumour biology: a review. Histol Histopathol. 2006;21:1339–1353. doi: 10.14670/HH-21.1339. [DOI] [PubMed] [Google Scholar]
- [21].Johnson MD, Torri JA, Lippman ME, Dickson RB. The role of cathepsin D in the invasiveness of human breast cancer cells. Cancer Res. 1993;53:873–877. [PubMed] [Google Scholar]
- [22].Calvo A, Catena R, Noble MS, Carbott D, et al. Identification of VEGF-regulated genes associated with increased lung metastatic potential: functional involvement of tenascin-C in tumor growth and lung metastasis. Oncogene. 2008;27:5373–5384. doi: 10.1038/onc.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- [24].Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Mol Cancer Res. 2003;1:789–800. [PubMed] [Google Scholar]
- [25].Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Park JH, Schuchman EH. Acid ceramidase and human disease. Biochim Biophys Acta. 2006;1758:2133–2138. doi: 10.1016/j.bbamem.2006.08.019. [DOI] [PubMed] [Google Scholar]
- [27].Rivenbark AG, Coleman WB. Epigenetic regulation of cystatins in cancer. Front Biosci. 2009;14:453–462. doi: 10.2741/3254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.