Abstract
Dietary restriction (DR) has been used for decades to retard aging in rodents, but its mechanism of action remains an enigma. A principal roadblock has been that DR affects many different processes, making it difficult to distinguish cause and effect. To address this problem, we applied a quantitative genetics approach utilizing the ILSXISS series of mouse recombinant inbred strains. Across 42 strains, mean female lifespan ranged from 380 to 1070 days on DR (fed 60% of ad libitum [AL]) and from 490 to 1020 days on an AL diet. Longevity under DR and AL is under genetic control, showing 34% and 36% heritability, respectively. There was no correlation between lifespans on DR and AL; thus different genes modulate longevity under the two regimens. DR lifespans are significantly correlated with female fertility after return to an AL diet after various periods of DR (R = 0.44, P = 0.006). We assessed fuel efficiency (FE, ability to maintain growth and body weight independent of absolute food intake) using a multivariate approach and found it to be correlated with longevity and female fertility, suggesting possible causality. We found several quantitative trait loci responsible for these traits, mapping to chromosomes 7, 9, and 15. We present a metabolic model in which the anti-aging effects of DR are consistent with the ability to efficiently utilize dietary resources.
Keywords: aging, food restriction, lifespan, fertility, metabolic efficiency, quantitative trait loci, genetic mapping, physiology
Introduction
Dietary restriction (DR) extends lifespan and slows aging across a variety of taxa and also causes myriad other physiological changes (Weindruch and Walford, 1998; Mobbs et al., 2007). To ascertain which of these processes affected by DR might be causally responsible for DR’s benefits, we have utilized genetic variation underlying a variety of physiological traits in the ILSXISS (formerly the LXS) series of recombinant inbred (RI) strains of mice (Williams et al., 2004). The ILSXISS is one of the largest RI panels available in mice (65 strains extant), which makes it advantageous for applying genetic approaches, similar to those that have advanced our understanding of aging in invertebrate models (Longo and Finch, 2003; Tissenbaum and Johnson, 2008). These strains of mice are now available from the Jackson Laboratory.
Previous studies have identified several physiological responses to DR, such as lower body temperature and reduced body weight (BW), that exhibit genetic variation in the ILSXISS; heritability was 35% for body temperature and 42% for BW (Rikke et al., 2003; Rikke et al., 2004; Rikke et al., 2006; Rikke and Johnson, 2007). Here we suggest a role for metabolic efficiency in specifying longevity and other anti-aging actions of DR. This is consistent with observations of Weindruch et al. (1986), who found that individual, within-strain variation in BW during DR was positively correlated with lifespan in four out of four cohorts (peak R’s of 0.21, 0.26, 0.26, and 0.36; P’s < 0.01 to 0.1). This association was also noted by Harper et al. (2006) in their study of wild-derived mice. Since long-term DR does not reduce metabolic rate (Weindruch and Walford, 1988; Masoro et al., 1982; McCarter et al., 1985; Yu et al., 1985; McCarter and McGee, 1989), Weindruch et al. (1986) proposed that the association between BW during DR and longevity results from greater metabolic efficiency (ME) in the longer-lived and heavier mice. ME was defined as “the fraction of the energy content of ingested foods actually absorbed and trapped in a biologically useful form (ATP)” (Weindruch and Walford, 1998), but ME was not directly assessed except as BW. Weindruch and Walford (1988, p. 248–249) also defined ME in terms of energy usage as “the percentage of trapped energy used for essential physiological functions, versus that wasted as heat generation,… or in other non-essential ways.” They noted “growth” as being one of these essential functions.
Although ME has long been thought to increase survival during energy restriction (Lee and Lucia, 1961), Weindruch et al. (1986) were the first to suggest that variation in ME also underlies DR’s anti-aging effects. Ramsey et al. (2000) and López-Lluch et al. (2006) have since added to the molecular model, suggesting that the increased efficiency of ATP production stems from reduced mitochondrial proton leakage, which concurrently reduces the generation of reactive oxygen species (ROS), leading to a slower accumulation of cellular damage, thus retarding aging.
In this study, we used measures of weight maintenance and ability to maintain hair and tail growth as indices of efficiency of food utilization. To emphasize this aspect of ME and the fact that we have not performed metabolic measurements, we will refer to these measures as indices of “fuel efficiency” (FE). Here we demonstrate a positive genetic relationship between fuel efficiency under DR and DR’s coordinated effect on two aging-related traits: lifespan and litters per female after return to an AL diet. These findings are consistent with the predictions of the ME model.
Results
Variation in longevity
We assessed the lifespan of female mice from 42 ILSXISS strains fed either AL or at 60% of the AL consumption rate of that strain (DR) (Figure 1A; Supplementary Tables 1, 2). AL mean lifespans ranged 2-fold, from 490 to 1020 days (typically 10 mice per strain, 406 total). DR mean lifespans ranged almost 3-fold, from 380 to 1070 days (typically 12 mice per strain, 476 total). The lifespans under DR showed no significant relationship to the lifespans under AL (R = 0.14, P = 0.18, 1-tailed); this lack of correlation suggests that different genes modulate lifespan under each condition. The estimated heritability of lifespan was 34% for the AL and 36% for DR (95% confidence intervals 22% to 50% for both).
Figure 1. Variation of longevity under AL and DR conditions.
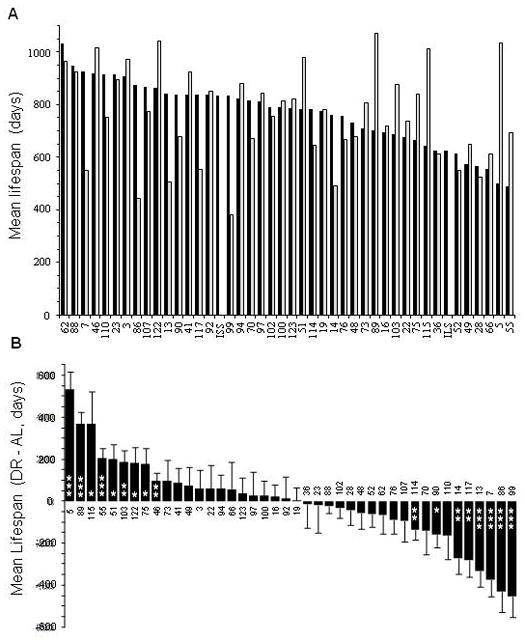
(A) Mean lifespans of 42 ILSXISS RI strains (all female, typical Ns per strain are 10 for AL, 12 for DR, housed 5 AL or 6 DR per cage). Also shown are female lifespans of the parental strains from which these RIs were generated: ILS (N = 28) and ISS (N = 31), measured in a previous cohort under AL conditions. (B) Difference in mean lifespan (DR – AL) for each ILSXISS strain arranged in order by positive effect of DR. * P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; 2-tailed t-tests, no Bonferroni corrections. Error bars, S.E.
For 22 strains, mean female lifespan under DR was numerically longer than it was under AL (Figure 1B). This included nine strains that reached statistical significance and three strains (ILSXISS 5, 55, 89) in which the difference was statistically significant even after the very conservative Bonferroni correction (P < 0.05/42 strains, 2-tailed t-tests, equal variances not assumed). This included one strain (ILSXISS 5) in which mean lifespan under DR was 106% longer (1.5 years) than under AL. Two other strains had DR means that were > 50% longer (≥ 1 year) than AL. These results show that DR’s longevity benefit is segregating among the strains.
Surprisingly, we also found that about half of the strains had mean female lifespans under DR that were shorter than AL (Figure 1B). Four of these differences were statistically significant experiment-wise (Strains 7, 13, 86, and 99). One DR strain had a mean that was 54% shorter than AL (1.25 years, SE = 0.27); five others had DR means that were shorter than AL by 36% to 49%. Overall average female lifespan under DR (762 days; SE = 28) was virtually identical to the AL grand mean lifespan of 769 days (SE = 19) (strains equally weighted).
We also analyzed DR’s effect on maximum lifespan, considered an essential aspect of DR’s ability to retard the “fundamental process of aging” (Wang et al., 2004; Allison et al., 2001). For the DRs, these maxima (longest-lived individual of each strain and diet) ranged from 711 to 1312 days; the AL controls ranged from 721 to 1244 days (Supplementary Table 2). The DR and AL maxima correlated highly with their respective means (DR: R = 0.86, P = 2 × 10−13 AL: R = 0.81, P = 6 × 10−11). Although these are obviously not independent data sets, the correlations indicate that DR had essentially the same effect on maximum lifespan as it had on mean lifespan; i.e. the effect of DR on mean lifespan is not disproportionately affected by early deaths (the correlation between earliest death and mean lifespan was 0.72 for ALs and 0.67 for DRs). Maximum lifespan under DR showed a significant correlation with AL maximum lifespan (R = 0.43; P = 0.0025; for a correlation matrix of all longevity constructs, see Supplementary Table 3).
Effects on female fertility
Although the effects of DR on lifespan are important, any “anti-aging” effects of DR are most likely the result of natural selection acting to improve female fertility during times of limited food availability (Allison et al, 2001; Holliday, 1989). Previous studies have in fact shown that DR females returned to AL will produce more litters and more pups per litter than females fed AL continuously (Weindruch and Walford, 1988; Merry and Holehan, 1991; Johnston et al., 2006). Consequently, we asked whether DR could prolong the period of female fertility and whether the same genes modulating longevity under DR also modulate this extension of fertility in females.
We assessed female fertility across 51 ILSXISS strains (2 strains tested twice). Because females typically stop estrus cycles during DR (Nelson et al., 1985; Merry and Holehan, 1991), fertility was measured after we returned animals to an AL diet after 7 or 11 months of DR (see Methods: Fertility, Statistical Methods-Regressions). We used litters per female as our measure for fertility, which allowed us to quantitate fertility even in cases where no litters were produced. We found that females that had been on DR produced significantly more litters than their AL controls: averaging 2.8 litters per female vs 1.4 (P = 10−6; 1-tailed, paired-sample t-test, n = 51 strains). This increase was associated with more pups per litter (averaging 4.4 pups per litter for the DR vs 3.3 for the AL, P = 0.0002; 1-tailed, paired-sample t-test; n = 20), assessable only in a subset of 20 strains that produced a countable number of pups (ie, not cannibalized for both DR and AL). In these DR studies, litters per female correlated significantly with age-at-last-litter; however, age-at-last-litter did not differ between AL and DR. Since these data were derived from three successive cohorts, we removed cohort effects by regressing litters per female after DR on litters per female maintained on AL, which also corrected for the cohort differences in the length of DR (see Methods for details). These fertility studies were conducted on singly-housed mice fed 60% AL under SPF conditions for the entire study. DR was not started until 2 months of age, when females are mature enough to produce their first litter.
In contrast to lifespan, we found few strains in which DR caused a reduction in female fertility compared with controls fed AL continuously. In 41 of the 47 strains producing one or more litters, the mice returned to AL feeding after DR produced more litters per female than did their AL controls. This improved litter production after DR showed a highly significant correlation with mean lifespan under DR (R = 0.44, P = 0.006, n = 33 strains, 1-tailed; Figure 2), indicating that a genetic component co-specifies DR longevity and female fertility.
Figure 2. Scatterplot showing genetic correlation between longevity and female fertility responses to DR.
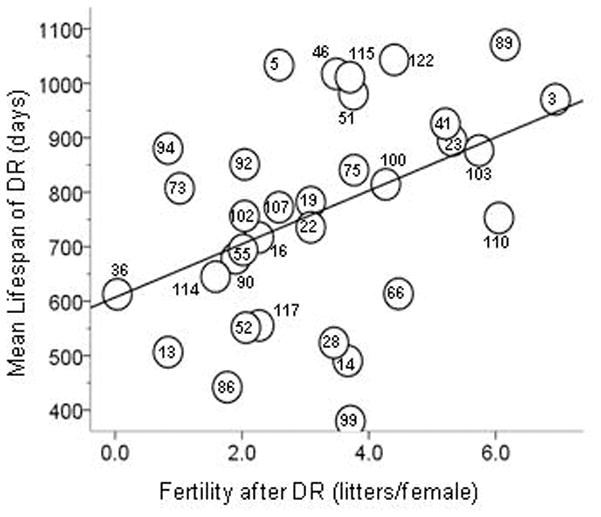
Fertility and longevity were measured in separate cohorts. The data are from the 33 ILSXISS strains common to both studies. The numbers within or next to the data points are the strain names. R = 0.44, P = 0.006, 1-tailed.
Fuel efficiency (FE) under DR
We observed a positive correlation between BW and the rate of hair growth on DR (R= 0.40, P = 0.004, N = 51 strains. UCB; 0.59, P = 0.00002, N = 44 strains, UTHSCSA; Supplementary Figure 1) (both regressed on comparable scores for mice fed AL, which removes the effects observed in the AL condition). This correlation means that, under DR conditions, similar genes act to maintain body weight and hair growth. This suggests that the mice are segregating differences in a metabolic component specifying these two traits. We have called this component “fuel efficiency” (FE), to refer to how well weight and growth are maintained per gram of food consumed. Because the BW and growth measures defining FE under DR are regressed on AL, FE under DR is independent of the baseline FE under AL.
Thus we defined FE operationally as a coordinated effect on BW and growth parameters (tail growth rate and hair regrowth rate) under DR. We made the effect on BW independent of differences in food intake by simultaneous regression on BW and food intake under the AL condition; we will refer to this new parameter as BW efficiency. We applied a PCA (Principal Components Analysis, see Methods) to define FE as a single factor representing the covariation among the three variables—BW efficiency, rate of hair regrowth, and rate of tail growth (“Statistical Methods, principal components analysis”). We found that FE was a highly significant predictor of fertility after DR (R = 0.34, P = 0.007, 1-tailed, n = 51 strains; Figure 3). FE also showed the same correlation (R = 0.34, P = 0.026, 1-tailed, n = 33 strains) with Aging Measure 1, defined below as the coordinated effect on fertility, mean lifespan, and maximum lifespan.
Figure 3. Fuel efficiency (FE) predicts fertility after DR.
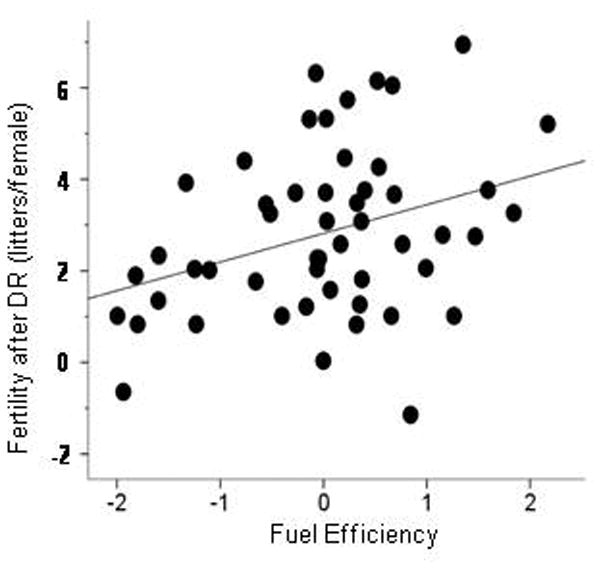
The same mice from 51 ILSXISS strains were measured for both traits. FE was defined as the principal components factor representing the positive covariation among DR BW efficiency, DR hair growth, and DR tail growth. R = 0.34, P = 0.007, 1-tailed.
Quantitative trait loci (QTLs)
We then asked if we could detect QTLs for any of these traits: DR longevity, extended fertility in response to DR, Aging Factor 1, and FE. A provisional QTL for AL longevity was identified on chromosome 19 (Figure 4). Genome scans revealed a potential QTL on chromosome 7 for both longevity and female fertility on DR that just missed the cut-off for genome-wide significance (Figure 5). The ILS allele at this chromosome 7 locus was associated with a positive effect on both longevity and fertility, consistent with the positive correlation between these traits. To increase our statistical power for identifying QTLs affecting both fertility and longevity, we applied a PCA that used litters per female after DR, DR mean lifespan, and DR maximum lifespan to develop a single factor (Aging Measure 1), representing the covariation among the three variables (see Supplementary Table 3 for correlation matrix). This procedure was suggested by Allison et al., (2001) in that the inclusion of maximum lifespan helps remove extraneous variance and gives more weight to the effect of DR on mortality later in life. In addition, we expected DR’s anti-aging mechanism would most likely manifest itself as a coordinated effect on lifespan and fertility. Using Aging Measure 1, we found that the chromosome 7 QTL achieved genome-wide statistical significance (P = 0.029). This QTL, designated Lfdr1 for “longevity and fertility response to dietary restriction, QTL 1,” explained ~20% of the genetic variance in Aging Measure 1; the 95% confidence interval covered a region of ~8 cM centered about the microsatellite D7Mit351.
Figure 4. QTL map for longevity under AL.
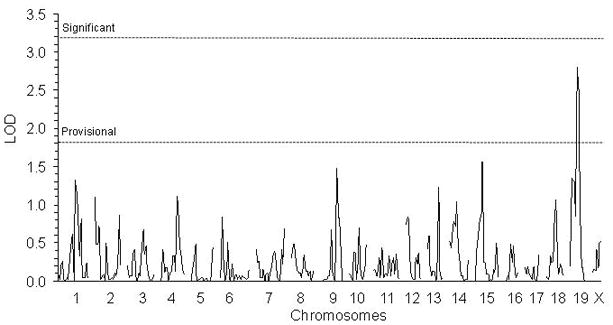
The threshold for statistical significance as determined by permutation testing was LOD 3.2. The cut-off for a provisional QTL was LOD 1.8.
Figure 5. QTL map for longevity under DR and fertility after DR.
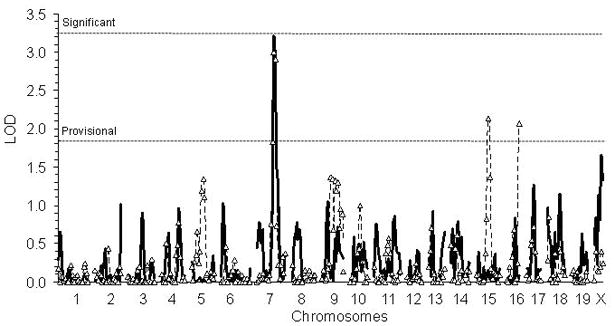
The thick solid line is the genome scan for DR mean lifespan. The dashed line with triangles at each marker is the scan for litters/female. The horizontal dashed lines indicate thresholds for significant (LOD 3.3) and provisional QTLs (LOD 1.8) as determined by permutation testing.
We also conducted a genome-wide scan for QTLs affecting FE, using BW efficiency, hair regrowth rate, and tail growth rates collected both at UCB and at UTHSCSA. We performed whole genome scans for each of these measures. Because the strains in the UCB and UTHSCSA cohorts only partially overlapped and the traits had low between-site correlations, we integrated the results by applying a rank-based genome-scan meta-analysis strategy (GSMA) to the linkage statistics from the five genome scans (Levinson et al, 2003). This strategy, originally developed for human linkage studies, rank orders the highest LOD values taken from each of 158 genomic regions covering the entire genome and calculates the average rank across all scans and tests for average rank scores higher than expected by chance. The results revealed a genome-wide significant QTL on chromosome 9, which explains about 20% of the genetic variance and will be referred to as Fedr1 for “Fuel efficiency response to dietary restriction, QTL 1”. We also detected a provisional QTL on chromosome 15 (explaining roughly 15% of the genetic variance; Figures 6A, B, C). The ILS alleles at both loci were associated with a positive effect on all traits at both study sites. This result is consistent with our model that these loci specify FE in response to DR.
Figure 6. Mapping on fuel efficiency.
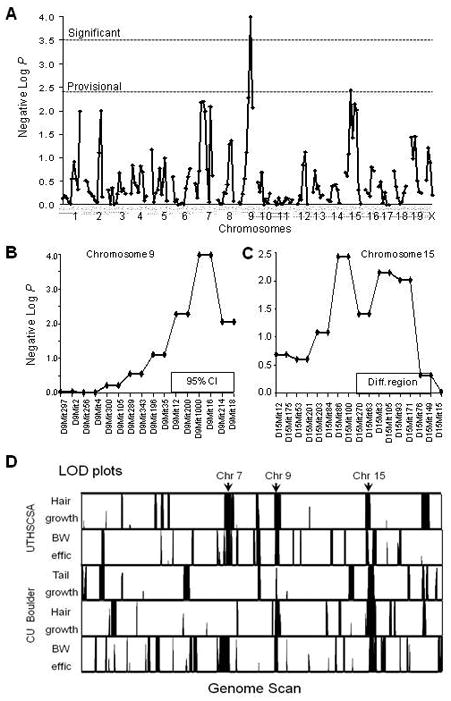
(A) Negative logarithm (base 10) of the P values from a genome scan meta-analysis of the BW efficiency and growth-related responses to DR. Dashed lines indicate thresholds for significant and provisional QTLs. (B) Expanded chromosome 9 scan. Box shows 95% confidence interval. (C) Expanded chromosome 15 scan. Box shows location of ILS differential locus in ISS congenic ISS.ILS-Lore5 (5LA). (D) Genome-scan bar codes showing potential sites of overlap for the same five traits. Each scan is truncated below 0.44 LOD to remove noise and above 0.6 to visualize overlaps. BW effic = body weight efficiency as described in the text. LODs are displayed for loci positively correlated with BW efficiency at UCB. Arrows indicate loci discussed in the text. Chromosome lengths are proportional to the number of markers.
We also developed a novel “bar code” approach (see Methods) for displaying the results from all five scans (Figure 6D). This method indicated an overlap in all five scans at three markers: D9Mit1000 on chromosome 9 and two adjacent markers on chromosome 15, D15Mit86 and D15Mit100; both coincided with loci found in the GSMA scan. The ILS alleles at the Fedr1 locus on chromosome 9 were associated with a weak but positive effect on fertility and lifespan (Figure 4); the single-marker P was 0.01 for fertility after DR, 0.07 for mean lifespan, and 0.01 for DR maximum lifespan.
Congenic with increased fuel efficiency under DR
Previously, Bennett et al. (2002) developed a congenic strain to capture a QTL affecting ethanol sensitivity—ISS.ILS-Lore5 (abbreviated 5LA)—that happened to partially overlap with the chromosome 15 region (95% CI) for the FE QTL (Figure 6C). If the congenic captured the FE QTL, the 5LA congenic strain should weigh 0.3 g more during DR than its ISS control, after weight is stabilized. This estimate is based on the fact that the QTL explains about 15% of the genetic variance. We tested this prediction by monitoring the BWs of 5LA congenics and ISS controls; the strain means were carefully weight-matched to within 0.1 g (both females and males), when we initiated DR at 50 days of age (Figures 7A, B). Both strains were fed DR rations of 1.8 g per day per mouse (~60% AL), and both strains lost weight precipitously during the first three weeks (Figures 7A, B). When BW began to stabilize at Week 4 and beyond, the 5LA congenic consistently maintained a higher mean BW than ISS, and this was true for both sexes. From Weeks 13 through 18, the congenics had a mean BW that averaged 0.6 g greater than ISS for the females and 0.3 g greater for the males (P = 0.04 for main effect of strain, 1-tailed, no sex by strain interaction, repeated measures ANOVA), suggesting that the congenics carry a locus from the ILS strain that confers increased ability to maintain BW under DR, which we will call Fedr2.
Figure 7. BWs of chromosome-15 congenic (5LA) and ISS control under DR.
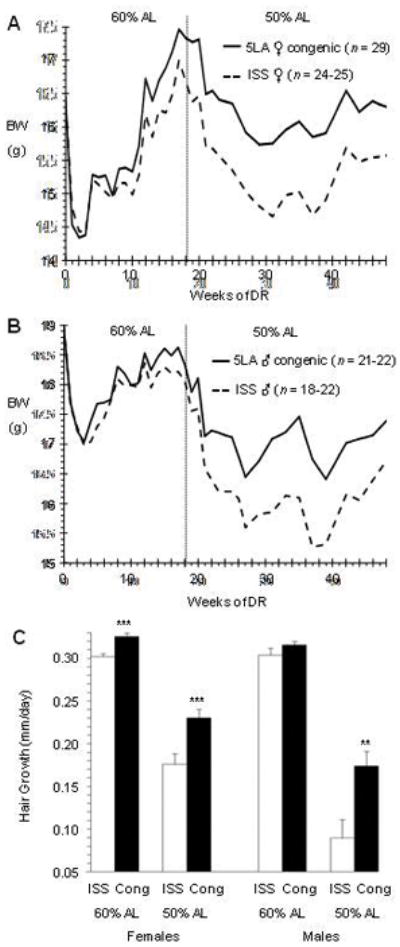
Congenic (solid line) is identical to ISS (dashed line) except for a differential region on chromosome 15 introgressed from ILS by ten generations of backcrossing (Bennett et al., 2002). For females (A) and males (B), the initial strain means for BW were identical (within 0.1 g) and both strains were fed identical rations per mouse (1.8 g/mouse/day = 60% AL, 1.5 g/mouse/day = 50% AL). BWs were measured just before feeding once each week through Week 23 and every other week thereafter. The rations were switched to 50% AL at Week 19. (C) Hair growth rate was measured under 60% AL between Weeks 14 and 17 of DR (23 days) and under 50% AL between Weeks 28 and 31 (23 days). For females, n = 24–25 for ISS and 29 for 5LA. For males, n = 21–22 for ISS and 22 for 5LA. ** P < 0.005, *** P < 0.001, 1-tailed t-tests. Cong = congenic strain.
We then asked whether switching the mice to a more severe level of food restriction would generate a greater difference in BW in response to DR. Therefore, starting at Week 19, the rations were reduced from 1.8 to 1.5 g/mouse/day (~50% AL), which produced another large drop in BW for both strains that continued for many weeks (Figures 7A, B). For Weeks 19 through 48, the average difference in BW between the congenic and ISS increased 2-to-3-fold for both females and males (P < 10−4 for main effect of strain, 1-tailed, no sex by strain interaction, repeated measures ANOVA), with the absolute difference being ~1 g for both sexes. These results suggested that the differential region in these congenics had captured Fedr2 for “fuel efficiency in response to dietary restriction, QTL 2.”
We also tested whether the improved maintenance of BW by the congenics was associated with better growth by measuring the rate of hair growth during Weeks 14 to 17 of DR (~60% AL, 23 days of growth, Figure 7C). We found that hair growth was 8% faster for the female congenics than it was for the ISS females, a small but highly significant difference (P = 0.0002, 1-tailed t-test). Hair growth was also faster in the congenic males (4%) but not significantly so (P = 0.11, 1-tailed t-test). ANOVA indicated a highly significant main effect of strain (P = 0.001), no effect of sex (P = 0.43), and no strain by sex interaction (P = 0.26). When the mice were switched to 50% AL, the difference in hair growth between the congenics and controls increased markedly (Figure 7C). For the males, hair growth was 94% faster for the congenics compared to ISS, which was highly significant (P = 0.002, 1-tailed). For females, hairs of the congenic mice grew 31% faster, also highly significant (P = 0.0006, 1-tailed). These results validated the presence of a gene in the differential region affecting FE.
Discussion
We report the outcomes of a quantitative genetic study on aging and longevity in the mouse. We studied an extant series of recombinant inbred strains (ILSXISS) that have been used both in DR aging studies as well as to study alcohol sensitivity (Williams et al., 2004). The major advantage of using RI strains is that their genotypes have been extensively assessed, thus minimizing costs, and detailed characterizations of numerous traits have already been conducted. Subsequent testing for significant relationships among traits in a stable set of RIs is another key advantage of using a large genetic panel for studying complex mechanisms (Chesler et al., 2008). These strains were recently imported by the Jackson Laboratory and 65 strains should eventually be available.
This study indicated a large amount of genetic variation for mouse longevity; heritability was 34% for AL and 36% for DR (60% of AL food intake). There was no significant correlation between mean longevity under these two conditions, although maximum lifespans of the AL and DR mice were significantly correlated. Similar observations were made at the UTHSCSA on the ILSXISS RI mice (Liao et al., 2010a, b; Mattson 2010), where they also observed similar heritability (28% AL males, 36% AL females, 55% DR males, 53% DR females). These two sets of observations were generated under different conditions (e.g., SPF status, age at which DR was initiated, one or both sexes tested, additional tests performed at each site, etc).
A very unexpected observation is that in about half of the strains, longevity was shortened by DR compared with the longevity under AL feeding. Similar results were also obtained at the UTHSCSA (Liao et al. 2010a). They also saw differential effects of DR on longevity in which some strains showed a positive response but about one-third showed a negative response and about one-third showed no significant response at all. Thus the observations of differential effects are replicable, providing a critical validation of this unexpected result. These findings are not completely novel but no previous study has examined more than two or three genotypes simultaneously. Forster et al. (2003) and Turturro et al. (1999) previously suggested that DBA mice get less life extension under DR than C57BL/6 mice, but no studies have found anything close to the range of variation observed here and no genetic studies have been carried out.
The lifespan shortening under DR was unexpected based on the wide efficacy of DR in many species. This and other issues have led some to question the relevance of these studies and suggest several possibilities that could make this observation of less interest. Nutrient deficiency might explain some of the life-shortening, but there were no obvious signs of malnutrition. First, all strains were able to establish a stable BW on the DR diet. Second, few animals died in the first three months and those that died early did not appear to be slowly wasting away, but usually died without being noticeably moribund or cachectic; the diet composition provided multiples of all micronutrients except selenium and choline (Liao et al., 2010). Third, the shortened lifespans did not cluster as might be expected if there were one overriding cause of death. Because most of the mice in our lifespan study were cannibalized before they were found, we did not conduct pathology studies, nor did we have sufficient funds to perform detailed autopsies.
It’s also important to note that our lifespan data correlated significantly with female fertility, post DR (R = 0.44, P = 0.006, N = 33 strains). This observation suggests genetic segregation of a common anti-aging component, which we called Aging Measure 1. Several previous studies of female reproductive capabilities under DR (Weindruch and Walford, 1988; Merry and Holehan, 1991; Johnston et al., 2006) found that females returned to an AL diet, after as long as 16 months on the DR diet, were, in general, able to regain fertility. This fertility data can also be examined to ask if there were negative effects of DR on female fertility, as might be expected if it were harmful to the mice: We found few negative effects on fertility caused by DR; early deaths and SPF status were not an issue. In addition, we collected fertility data on three of the four strains with significantly shorter DR lifespan and found that fertility was improved for two and only slightly decreased in the third (Supplementary Table 4). This also argues against a malnutrition effect of the DR regimen.
A related question is whether the observed lifespan shortening was genetically distinct from the observed lifespan extension. Under that model we would expect very different correlations with female fertility—which was almost uniformly affected positively by DR. Of the 33 strains tested for both fertility and lifespan, 19 strains had life extension under DR, and the correlation between DR fertility and DR mean and maximum lifespan was 0.36 and 0.24 (P = 0.06, 0.16, respectively, 1-tailed). In the 14 remaining strains, which showed lifespan shortening under DR, the correlation between DR fertility and mean and maximum lifespan was essentially the same: 0.32 and 0.47 (P = 0.13 and 0.04, respectively, 1-tailed). The significant QTL on chromosome 7 modulating both fertility and longevity also suggests common uniform action. The simplest interpretation is that lifespan shortening and lengthening under DR were determined by common genes.
In our analysis of metabolic efficiency, we found significant differences in how much weight the mice could maintain on DR that correlated with maintenance of tail growth rate and the rate of hair regrowth. BW and hair growth rate were correlated at each study site, which led us to define a new parameter that is a composite of BW and growth, a trait which we have called fuel efficiency (FE), which was extracted using Principal Components Analysis. This FE factor was found to be genetically specified and significantly correlated with Aging Measure 1 (R = 0.34, P = 0.026). As noted, this factor was defined independent of FE under AL; consequently, whether FE under AL predicts longevity under DR as suggested by Ferguson et al. (2007) is a separate issue from the analyses conducted in this study (the AL efficiency model will be tested in future studies).
Other studies have also reported that individual mice that maintained the highest BW were likely to be the longest-lived individuals among cohorts of genetically identical mice (Weindruch et al., 1986; Harper et al., 2006). This is consistent with a model in which life extension by DR is due, at least in part, to benefits that are a by-product of FE. This might be expected if FE is itself a surrogate measure of metabolic efficiency, which is a survival response in which animals under DR adopt a new metabolic strategy limiting wastage of energy. In brief (Figure 8), the model suggests that reduced food availability during DR is a signal to reallocate energy by decreasing proton leakage across the mitochondrial membrane (Ramsey et al., 2000; Lopez-Lluch et al., 2006). This leads to decreased heat production, increased ATP production, and presumably a concomitant decrease in the production of senescence-promoting reactive oxygen species (ROS) (Ramsey et al., 2000; Lopez-Lluch et al., 2006). The higher ATP levels are generally available and help preserve BW, growth, and many other processes, including maintenance and repair functions that might combat aging (Weindruch and Walford, 1988; Weindruch et al., 1986). Additional studies will be needed to confirm the molecular details of the ME model.
Figure 8. Metabolic Efficiency Model.
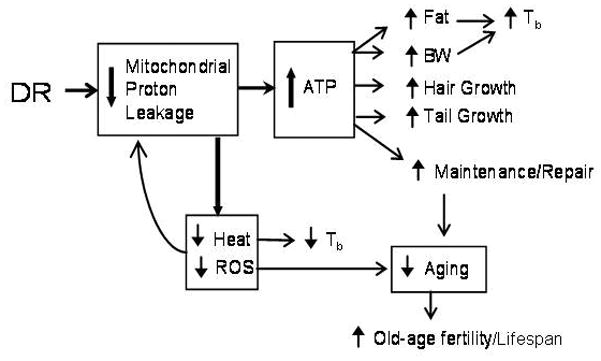
Abbreviations: ATP, adenosine triphosphate; BW, body weight; ROS, reactive oxygen species; Tb = body temperature. Up arrows indicate increased or enhanced; down arrows indicate decreased or slowed. Although reduced heat production is central to the model, the net effect on Tb is ambiguous because increased fat and BW reduce heat loss (Rikke and Johnson, 2007).
We found three significant QTLs (genetic regions harboring genes controlling these various aging traits, Supplementary Table 5). On chromosome 7, we found a QTL affecting lifespan and fertility after DR that we have named Lfdr1 for “longevity and fertility response to dietary restriction, QTL 1; this QTL also has suggestive effects on FE (Fig. 5D). Two QTLs having significant effects on FE were identified on chromosomes 9 and 15. These we have named Fedr1 and Fedr2, respectively, for “fuel efficiency response to dietary restriction” QTLs 1 and 2. The QTL on chromosome 9 is also suggestive for an effect on lifespan and fertility in the direction expected (ie, ILS allele associated with increased lifespan and fertility). The QTL on chromosome 15 was not suggestive for an effect on lifespan and fertility, but there is not sufficient statistical power to rule out a QTL of the expected size (only 15% of the genetic variance).
The ME model also aids in predicting what genes are strong candidates for the QTLs identified in this study. Potential candidates include Ppara (peroxisome proliferator-activated receptor alpha) on chromosome 9 (Figure 6b), which has previously been proposed to modulate not only DR’s health benefits (Corton et al., 2004; Masternak and Bartke, 2007) but also ME during DR (Lopez-Lluch et al., 2006). Candidates on chromosomes 7 and 15 include Cck (cholecystokinin) and Cckbr (cholecystokinin receptor B), respectively; the orthologs of these genes have recently been shown to be necessary for DR-induced life extension in C. elegans (Park and Johnson, 2009). However, the haplotypes of Ppara, Cck, and Cckbr, along with their promoter regions, are not polymorphic between ILS and ISS, illustrating how strong candidate genes can also be immediately ruled out.
Among those genes that are in polymorphic haplotype regions, we also identified several candidates that were particularly interesting. These included the genes Ucp-2 and Ucp-3 encoding mitochondrial uncoupling proteins, which are obvious candidates for effects on metabolic efficiency; the Ucp-2 gene exhibits a missense polymorphism between ISS and ILS. Another notable candidate gene is Lass5, an ortholog of the longevity assurance gene lag-1 in yeast (Jazwinski 2002). Lass5 has an alanine-proline polymorphism between ISS and ILS. We also identified a number of other candidate genes with potential to modulate ATP, NAD, and thyroid hormone signalling consistent with the ME model (Supplementary Table 5).
We also looked for polymorphic candidate genes among those identified by Swindell (2009) as being significantly up or down regulated in a meta-analysis of murine gene expression during DR. One such candidate in the chromosome 7 QTL region was Serpinh1, serine (or cysteine) peptidase inhibitor, clade H, member 1. This gene is consistently down regulated in variety of tissues (heart, liver, hypothalamus, hippocampus, striatum). According to the UniProt database (http://www.uniprot.org/uniprot/P19324), the Serpinh1 protein is inducible by heat shock and functions as a possible chaperone that binds specifically to collagen.
For decades, DR has been a prominent and useful tool for retarding aging in rodents. This study demonstrates that applying a systems genetics approach to the analysis of DR (the combination of genetics and detailed physiological analyses) has great potential for elucidating the mechanisms behind DR’s many health and aging benefits.
Materials and Methods
Ethics Statement
All animal procedures were conducted in accordance with NIH guidelines and approved by the UCB and the UTHSCSA Institutional Animal Care and Use Committees.
Longevity
The longevity study was conducted at the Institute for Behavioral Genetics at UCB on mice that were inbred for ~30 generations. We used only females to maximize our ability to detect a correlation with female fertility in response to DR. Females are also less likely to fight under multiple-housing conditions (used for the longevity studies but not for the FE and fertility studies during DR). Even so, eight strains were removed from the study because of fighting.
The two longevity cohorts were 3 months different in birth date but were otherwise studied concurrently. The older cohort comprised 24 strains that were age-matched to within 10 days of each other (March 27 to April 5, 2005), weaned at 22 to 25 days of age, and started on DR at 30 to 40 days of age (May 5, 2005). The second cohort comprised 18 strains that were also age-matched to within 10 days of each other (June 28 to July 7, 2005), weaned at 22 to 25 days of age, and started on DR at 28 to 38 days of age (August 4, 2005). The ALs were housed 5 per cage, the DRs 6 per cage because of their smaller size; two cages were studied per strain and diet. One or two days prior to starting DR, the mean BWs of the DR and AL mice of each strain were weight-matched to within 0.3 g of each other and litters were split up as much as possible.
All of the mice were housed under the same conditions in the same colony room, which had a 12 hour light:dark cycle (7 am:7 pm), temperature 21 to 24 °C, and relative humidity 15% to 60%. The AL and DR cages were maintained side by side in alternating columns of AL and DR to minimize the potential for shelf-level variation in AL food intake (Greenman et al., 1983).
Rice-grain-size transponders (~0.1 g) (Bio Medic Data Systems, Seaford, DE) for reading identification were used on all but two mice per cage. The transponders were injected by syringe under a loose fold of skin posterior to the scruff of the neck. The mice without transponders were given a left-ear or right-ear punch for identification. An ANOVA of lifespan using diet, strain, and transponder as between group factors indicated no main effect of the transponders (P = 0.22) and no interaction with diet or strain or both (Ps > 0.8).
The diet was Harlan-Teklad 7012 (Madison, WI), which we have used in all of our studies (see Liao et al., 2010 for nutritional content). The DR rations of 60% AL were calculated each week separately for each strain using a 3-week moving average of the AL food intake (measured each week). The DR rations were also adjusted by the differential amount of food wasted by the AL and DR mice as measured every 10 to 12 weeks. Initially, the DR mice were fed double rations on Mondays and Wednesdays and a triple ration on Fridays as done in our previous studies of singly-housed mice (Rikke et al., 2003; Rikke et al., 2004; Rikke et al., 2006; Rikke and Johnson, 2007). However, to mitigate the potential for fighting, we switched to feeding every day starting on May 4, 2006 (~1 year after starting DR). All rations were weighed to the nearest 0.1 g and manually delivered to the bottom of the cage rather than the food hopper because gnawed pellets in the hopper occasionally became too difficult for the mice to grasp. Because AL mice exhibit a substantial decline in their food intake with age, the DR rations were kept constant after Weeks 73–74 of DR (1.4 years old).
Serology samples during all of our studies were collected from sentinel mice every three months and sent to Charles River Laboratories for testing on a standard battery of ten pathogens used to define SPF colonies. At the same time, veterinary services tested for pin worms and fur mites. The colony was SPF until Mouse Hepatitis Virus (MHV) and Mouse Norovirus (MNV) were detected in our facility in December 2005 (Cohort 1 was ~8 months old, Cohort 2 was ~5 months), and the entire facility remained infected thereafter (although the reason for the SPF break was not discovered, the break was associated with the construction of a new facility above the SPF). Although the potential for causing lifespan variation in our study is unknown and a valid concern, MHV and MNV have not been shown to affect mortality except in mice engineered to be immunodeficient. Typically, infected mice develop immunity within three weeks (Committee on Infectious Diseases of Mice and Rats, 1991; Liberati, 2009). The mice in our study did not exhibit diarrhea or otherwise appear to be adversely affected. All other cohorts used for our studies of BW, hair growth, tail growth, and fertility were conducted under SPF conditions for their entire duration. Moreover, there was no break of the SPF barrier at UTHSCSA.
The number of unnatural deaths and whether they were censored is detailed in Supplementary Table 1 for both cohorts. When half of the mice from the same diet and strain died, the remaining mice were combined in a single cage.
Fertility
The study of fertility after DR was conducted at UCB. This study comprised three cohorts born November 1999, May 2000, and February 2003; these cohorts preceded the cohorts studied for longevity, with no overlap. The ages, inbreeding, and Ns associated with these cohorts are detailed in our reports on their BW and body temperature responses to DR (Rikke et al., 2006; Rikke and Johnson, 2007). In brief, all mice were female, singly housed, and closely age-matched within cohorts. DR was started at 50 to 60 days of age. Typically, four mice were assessed per strain and diet. Fifty-one strains were studied in total and two strains were studied twice. DR was maintained at 60% of AL as calculated separately for each strain based on weekly measures of AL food intake. The rations were adjusted for food wastage measured every 10 weeks. The DR mice were given their rations on a Monday-Wednesday-Friday feeding schedule.
Fertility was measured after taking the mice off DR after 7, 11.5, or 11 months of DR in Cohorts 1 (11 strains), 2 (9 strains), and 3 (33 strains), respectively. The DRs were allowed to eat ad libitum for one week before all mice were set up in continuous harem matings with young (2 to 4 months old) C57BL/6J (B6) males (IBG substrain). The DR mice were allowed to continue eating ad libitum while they were mated but are still referred to as the DRs. In Cohorts 1 and 2, we combined two females from the same diet but different strains. In Cohort 3, we combined two females from the same diet and strain. Sixteen males were randomly selected near the end of the Cohort-2 study for fertility testing with 1or 2 young B6 females; these 14-month-old males all sired litters within 30 days. Male fertility was also tested in Cohort 2 by swapping out the males that didn’t produce pregnancies within 2 months (n = 20); these males were also set up with 1or 2 young B6 females and all sired litters within 30 days. In Cohort 3, the males were rotated among the harem cages every 1 to 2 months to minimize the potential effects of variation in male fertility. The females in all cohorts were checked each week for pregnancy and daily when pregnancy was suspected. Whether the females were pregnant was assessed visually in Cohort 1. In Cohorts 2 and 3, BW was also monitored, and a 4-g or more gain in BW, followed by a similar drop in BW within two weeks, was found to be a reliable indicator of pregnancy and delivery with subsequent cannibalisation. Pregnant females were counted as having produced a litter even if no pups were observed under the assumption that the litter was cannibalized (a frequent occurrence in Cohort 3, probably due to rotating the males). All pups were removed 2 to 7 days after birth. In Cohorts 1 and 2, the females were typically kept in the study > 100 days after their last litter, which was 4.5 standard deviations longer than the average time between litters. In Cohort 3, the females were monitored at least 120 days after their last litter. Females that died while mated were treated as if they had stopped producing litters. Mice that died prior to mating were given a zero score for litter production.
Body weight and body weight efficiency
The studies of BW and BW efficiency were conducted at UCB and at UTHSCSA. The BW data from the UCB mice was the same as previously reported for Weeks 17 to 30 of DR (6 to 9 months of age) in Rikke et al. (2006).
The BW and BW efficiency data from the UTHSCSA mice were collected on 42 strains (Liao et al., 2010). Typically for each strain, two cages each of males and females were housed five per cage. The DR mice were fed 60% of AL as determined by weekly measurements of the average AL food intake per gram BW for each strain prior to the start of DR. For each strain, one cage of each sex was randomly assigned to the DR group at 4 to 5 months of age and fed the same DR rations throughout the study (same chow as in our UCB studies). Because average BW of the DR and AL groups differed prior to DR, food given to the DR group was corrected for any difference in mean BW. The DR rations were placed in the cage food hopper daily, 1 hour before lights off. BW data were collected ~12 months after starting DR, when the mice were ~17 months of age.
Growth
The study of hair growth was conducted at UCB and UTHSCSA. Hair growth rates were measured by plucking a small bald patch (approximately 1 cm2) on the back of each mouse with tweezers. After the hair had partially regrown, a sample was plucked for length measurements and the number of days of growth was recorded. In the UCB cohorts, the AL hairs were typically plucked after 14 to 15 days of growth in Cohorts 1 and 2, and 20 days in Cohort 3. Because the DR hairs generally grew slower, these hairs were typically plucked after 22 to 29 days in Cohorts 1 and 2, and 25 to 32 days in Cohort 3. In the UTHSCSA cohort, the AL hairs were plucked after 17 days and the DR hairs after 24 days of growth. Eleven representative hairs from each sample were measured under a microscope using an ocular micrometer. To avoid errors due to inadvertently measuring broken or full-length hairs, we automatically excluded the longest and shortest measurement from each sample. The remaining nine measurements were averaged, divided by the days of growth, and converted to units of mm/week. The 5LA congenic and ISS hairs regrown under 60% AL were measured blind by B.A.R. and under 50% AL by a research assistant with no knowledge of the expected result.
Tail growth, studied at UCB, was measured each week for approximately 5 months immediately after initiating DR. The tails were measured by holding the mice by the scruff of the neck and laying the tails alongside a ruler.
Statistical Methods
Statistical significance was defined as P < 0.05 unless noted otherwise, and all analyses were conducted using SPSS for Windows 16.0. All correlation coefficients (Rs) are Pearson coefficients. The QTL mapping was conducted using MapManager QTXb20 for Windows and 310 microsatellite markers. Statistical significance was determined empirically by permutation testing (10,000 trials).
Regressions
Unless noted otherwise, all references to a DR response refer to values that have been corrected for baseline by regressing on their respective AL values. This procedure was done so that the variation under AL would not be confounded with the new variation imposed by DR (Allison et al., 2001). For example, if there were strain variation in fertility under AL conditions and DR had no effect on this fertility, failure to regress on AL would lead to an erroneous conclusion that DR affects fertility; however, if fertility on DR is first regressed on fertility under AL we obtain the correct conclusion: that there is no variation due to DR. Regressing on AL was also sufficient to control for the cohort differences in litter production rates due to the different ages at which the matings were initiated. When we regressed the UTHSCSA data on AL, we also adjusted for sex differences at the same time by specifying sex as a fixed factor in a univariate, general linear model. As we have shown empirically for BW under DR (Rikke et al., 2006), regression is more effective at removing the AL correlation than straight division (however, the residuals have been converted to % AL values to be physiologically meaningful as noted in Methods of Rikke et al., 2006), and mathematically, regression guarantees removal of a correlation without requiring any assumptions as to which correction method is more appropriate (Kaiser, 1989). In the case of DR mean lifespan, there was no correlation with AL mean lifespan and the effect of regression on our analyses was negligible; therefore, we have not used any correction.
Subtracting AL mean lifespan from the DR mean
Although we used a difference score to highlight the negative effects that DR had on the lifespans of many strains, difference scores were not appropriate for the statistical analyses conducted in this study. If the AL means represented a baseline to be subtracted, then, minimally, the DR and AL lifespans would need to be correlated (Kaiser 1989). The absence of correlation means that subtracting only adds to the variance; it is no different than subtracting quantities chosen from a table of random numbers with the same mean and SD. The end result is less statistical power for detecting correlations with other traits and similarly less power for conducting the QTL mapping (Vickers 2001). Even worse, subtracting in our study would have created an artifactual negative correlation with the AL mean lifespans (R = 0.49, P = 4.6 × 10−6)—instead of removing a baseline effect, it falsely creates one (Kaiser et al., 1989).
Principal components analysis
DR mean lifespan, DR maximum lifespan (no regression on AL), DR litters/female, AL mean lifespan, and AL maximum lifespan were used as the input variables. Only Eigen values greater than 1 were considered. Rotation was done using the varimax method of SPSS. Two factors were obtained: one specific for the three DR variables (Rs = 0.7–0.9, R ≤ 0.2 for the AL variables), the other specific for the AL variables. The DR-specific factor scores were used for genetic mapping and the correlation analyses. For the nine strains measured for longevity but not fertility, we used the factor scores from repeating the PCA without including the fertility variable DR litters/female (the correlation between factor scores calculated with fertility as a variable versus those calculated without fertility was 0.97; Supplementary Table 3). In this manner, we were able to utilize our data from all 42 strains for the genetic mapping.
Fuel efficiency in our statistical analysis was defined as the common factor responsible for the covariation among BW efficiency, hair growth, and tail growth as measured in the 51 strains studied for fertility after DR. This factor was extracted using the same PCA procedure described above. The input variables were DR BW efficiency, DR hair growth, and DR tail growth. Body temperature was not included in our physiological assessment of metabolic efficiency because the strain differences in body temperature reflect variation in the rate of heat loss, not heat production (Rikke et al., 2007).
QTL mapping
QTLs were mapped using MapManager as we have previously described (Rikke et al. 2006). The permutation testing to assess statistical significance was conducted using 10,000 permutations. A provisional threshold permits a single false-positive per genome scan (genome-wide P < 0.63, single-marker P typically less than 0.01); QTLs exceeding this threshold require confirmation (Lander and Kruglyak, 1995).
The QTL on chromosome 7 affecting the principal components factor for DR longevity and fertility after DR explained 32% of the genetic variance according to MapManager’s output. This effect size was then adjusted for low statistical power as previously described and referenced in Rikke et al. (2004). In brief, using Figure 2 of Belknap et al. (1996) and adjusting for N = 42 strains, we estimated that the power available for detecting this QTL was 0.45. The overestimate of effect size is (1/power)0.6, suggesting that the QTL was overestimated by a factor of 1.615, yielding a corrected size of ~20% (32%/1.615).
The 95% confidence interval was estimated as previously described and referenced in Rikke et al. (2004). Given the heritability of DR mean lifespan of 36% (h2RI) and an average sample size of 11 DR mice per strain (476/42), we used Figure 1 of Belknap (1998) to estimate that the heritability of the strain means was ~85% (h2RImean). Using the equation NF2 = 2(1-0.5h2RI)(h2RImean/h2RI)Nstrain, we then calculated that testing 476 mice from 42 strains (Nstrain) was equivalent to testing 163 F2 mice [2*(1-0.5(0.36))*(0.85/0.36)*42]. The 95% confidence interval was then estimated from the formula 530/(NF2*VQTL), where VQTL equals half of the RI effect size of 20%. The result is 530/(163*(0.20/2)), which equals 32.5 cM. This result was then divided by 4 because an RI recombination map is expanded four-fold relative to an F2 map; the resulting 95% confidence interval was thus ~8 cM.
Genome scan meta-analysis strategy
We followed the methods of Levinson et al. (2003) and specific details, including power calculations, for GSMA can be found there. We used the GSMA software package (http://mmg.umds.ac.uk/GSMA) to execute the analysis. Briefly, the genome was divided into the recommended 158 bins: bin width was designed to ensure that there were at least two bins on the smallest chromosome and at least one marker in each bin (Levinson et al., 2003). Within each bin, the most statistically significant linkage result was selected. Each bin was then ranked within each genome scan. We then weighted each scan according to the square root of the number of strains, and multiplied each ranking by this weight. The average rank at each bin was then calculated across all scans, and the chi-square statistical significance of this score was empirically assessed by permutation (10,000 replicates). Scores that exceeded a significance threshold of 0.00032 (Bonferroni correction for 158 bins) were considered genome-wide significant, and those exceeding 0.004 were considered provisional (i.e. one false-positive expected per genome scan = genome-wide P < 0.63/158 bins) (Lander and Kruglyak, 1995).
Bar code analysis
We truncated the bottom and top portion of each scan to create a “bar code” for visualizing where all five traits had positively correlated, overlapping peaks. The bottom was truncated where the empirical probability (determined by permutation testing) that all five genome scans would display an overlap at even one marker by chance was < 0.05.
Candidate gene search
Using Positional Medline (http://omicspace.riken.jp/PosMed) with the key words “aging” and “caloric restriction”, we identified genes in the 95% confidence intervals of the QTL regions (Supplementary Table 4) associated in the literature with pathways previously implicated in DR’s mechanism of action. We then used the SNP browser of GeneNetwork (http://www.genenetwork.org/webqtl/snpBrowser.py) to identify those genes also belonging to different haplotype blocks when comparing ILS and ISS.
Supplementary Material
Acknowledgments
We thank Professional Research Associates Matt Battaglia, Christine Martin, Colin Larson, Galen Miller, Kristina Williams, and John Yerg for data collection and animal husbandry. We thank Phyllis Carosone-Link for managing the database and assisting with the statistical analyses. We also thank the dozens of undergraduate students who assisted with data collection and animal care over the years.
Funding was provided by the Ellison Medical Foundation and the National Institute on Aging. The Ellison Medical Foundation also provided special funding so that the ILSXISS, ILS, ISS, and the 5LA congenic are available for purchase from the Jackson Laboratory, Bar Harbor, ME.
Abbreviations
- 5LA
ISS.ILS-Lore5 congenic strain
- AL
ad libitum
- B6
C57BL/6J
- BW
body weight
- cM
centiMorgan
- DR
dietary restriction
- FE
fuel efficiency, a metric of the ability to maintain body weight and growth rates (e.g., hair) under DR conditions
- GSMA
genome scan meta-analysis
- h2
heritability
- IBG
Institute for Behavioral Genetics
- ILS
Inbred Long Sleep
- ISS
Inbred Short Sleep
- LOD
logarithm of the odds of linkage
- ME
metabolic efficiency
- PCA
principal components analysis
- QTL
quantitative trait locus
- RI
recombinant inbred
- ROS
reactive oxygen species
- SNP
single nucleotide polymorphism
- SPF
specific pathogen free
- SPSS
Statistical Package for the Social Sciences
- Tb
body temperature
- UCB
University of Colorado at Boulder
- UTHSCSA
University of Texas Health Science Center at San Antonio
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison DB, Miller RA, Austad SN, Bouchard C, Leibel R, Klebanov S, Johnson TE, Harrison DE. Genetic variability in responses to caloric restriction in animals and in regulation of metabolism and obesity in humans. J Gerontol A Biol Sci Med Sci. 2001;56:55–65. doi: 10.1093/gerona/56.suppl_1.55. [DOI] [PubMed] [Google Scholar]
- Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet. 1998;28:29–38. doi: 10.1023/a:1021404714631. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Mitchell SR, O’Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- Bennett B, Beeson M, Gordon L, Carosone-Link P, Johnson TE. Genetic dissection of quantitative trait loci specifying sedative/hypnotic sensitivity to ethanol: mapping with interval-specific congenic recombinant lines. Alcohol Clin Exp Res. 2002;26:1615–1624. doi: 10.1097/01.ALC.0000037136.49550.B3. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Infectious Diseases of Mice and Rats, Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council. Infectious Diseases of Mice and Rats. Washington, DC: National Academy Press, Washington, D.C; 1991. pp. 102–111. http://www.nap.edu/openbook.php?isbn=0309063329. [Google Scholar]
- Corton JC, Apte U, Anderson SP, Limaye P, Yoon L. Mimetics of caloric restriction include agonists of lipid-activated nuclear receptors. J Biol Chem. 2004;279:46204–46212. doi: 10.1074/jbc.M406739200. [DOI] [PubMed] [Google Scholar]
- Ferguson M, Sohal BH, Forster MJ, Sohal RS. Effect of long-term caloric restriction on oxygen consumption and body temperature in two different strains of mice. Mech Ageing Dev. 2007;128:539–545. doi: 10.1016/j.mad.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster MJ, Morris P, Sohal RS. Genotype and age influence the effect of caloric intake on mortality in mice. FASEB J. 2003;17:690–692. doi: 10.1096/fj.02-0533fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman DL, Bryant P, Kodell RL, Sheldon W. Relationship of mouse body weight and food consumption/wastage to cage shelf level. Lab Anim Sci. 1983;33:555–558. [PubMed] [Google Scholar]
- Harper JM, Leathers CS, Austad SN. Does caloric restriction extend life in wild mice? Aging Cell. 2006;5:441–449. doi: 10.1111/j.1474-9726.2006.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Food, reproduction and longevity: is the extended lifespan of calorie-restricted animals an evolutionary adaptation? Bioessays. 1989;10:125–127. doi: 10.1002/bies.950100408. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Growing old: metabolic control and yeast aging. Annu Rev Microbiol. 2002;56:769–792. doi: 10.1146/annurev.micro.56.012302.160830. [DOI] [PubMed] [Google Scholar]
- Johnston SL, Grune T, Bell LM, Murray SJ, Souter DM, Erwin SS, Yearsley JM, Gordon IJ, Illius AW, Kyriazakis I, Speakman JR. Having it all: historical energy intakes do not generate the anticipated trade-offs in fecundity. Proc Biol Sci. 2006;273:1369–1374. doi: 10.1098/rspb.2005.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser L. Adjusting for baseline: change or percentage change? Statistics in Medicine. 1989;8:1183–1190. doi: 10.1002/sim.4780081002. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lee M, Lucia SP. Some relationships between caloric restriction and body weight in the rat. I Body composition, liver lipids and organ weights. J Nutr. 1961;74:243–248. [Google Scholar]
- Levinson DF, Levinson MD, Segurado R, Lewis CM. Genome scan meta-analysis of schizophrenia and bipolar disorder, part I: Methods and power analysis. Am J Hum Genet. 2003;73:17–33. doi: 10.1086/376548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 2010a;9:92–95. doi: 10.1111/j.1474-9726.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. No evidence that competition for food underlies lifespan shortening by dietary restriction in multiply housed mice: response to commentary. Aging Cell. 2010b in press. [Google Scholar]
- Liberati TA. Murine Norovirus. Taconic, Animal Health Matters. 2009 website. http://www.taconic.com/wmspage.cfm?parm1=286.
- Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- López-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, de Cabo R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ, Yu BP, Bertrand HA. Action of food restriction in delaying the aging process. Proc Natl Acad Sci U S A. 1982;79:4239–4241. doi: 10.1073/pnas.79.13.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson M. Genes and behavior interact to determine mortality in mice when good is scarce and competition fierce. Aging Cell. 2010 doi: 10.1111/j.1474-9726.2010.00561.x. in press. [DOI] [PubMed] [Google Scholar]
- Masternak MM, Bartke A. PPARs in calorie restricted and genetically long-lived mice. PPAR Res. 2007:28436. doi: 10.1155/2007/28436. doi:10.1155 2007, 28436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter R, Masoro EJ, Yu BP. Does food restriction retard aging by reducing the metabolic rate? Am J Physiol. 1985;248:E488–E490. doi: 10.1152/ajpendo.1985.248.4.E488. [DOI] [PubMed] [Google Scholar]
- McCarter RJ, McGee JR. Transient reduction of metabolic rate by food restriction. Am J Physiol. 1989;257:E175–E179. doi: 10.1152/ajpendo.1989.257.2.E175. [DOI] [PubMed] [Google Scholar]
- Merry BJ. Dietary manipulation of ageing: an animal model. In: Bittle AH, Collins KJ, editors. The Biology of Human Ageing. Cambridge University Press; 1986. pp. 233–242. [Google Scholar]
- Merry BJ, Holehan AM. The effect of dietary restriction on the endocrine control of reproduction. In: Fishbein L, editor. Biological Effects of Dietary Restriction. Berlin: Springer-Verlag, Berlin; 1991. pp. 140–146. [Google Scholar]
- Miller RA, Nadon NL. Principles of animal use for gerontological research. J Gerontol A Biol Sci Med Sci. 2000;55:B117–B123. doi: 10.1093/gerona/55.3.B117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobbs CV, Yen K, Hof PR, editors. Mechanisms of dietary restriction in aging and disease. Karger; New York: 2007. [Google Scholar]
- Nelson JF, Gosden RG, Felicio LS. Effect of dietary restriction on estrous cyclicity and follicular reserves in aging C57BL/6J mice. Biol Reprod. 1985;32:515–522. doi: 10.1095/biolreprod32.3.515. [DOI] [PubMed] [Google Scholar]
- Park S-K, Johnson TE. Lifespan extension by dietary restriction is mediated by nlp-7 signaling and coelomocyte endocytosis in C. elegans. FASEB J. 2009 doi: 10.1096/fj.09-142984. published online 9/25/2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JJ, Harper ME, Weindruch R. Restriction of energy intake, energy expenditure, and aging. Free Radic Biol Med. 2000;29:946–968. doi: 10.1016/s0891-5849(00)00417-2. [DOI] [PubMed] [Google Scholar]
- Rikke BA, Battaglia ME, Allison DB, Johnson TE. Murine weight loss exhibits significant genetic variation during dietary restriction. Physiol Genomics. 2006;27:122–130. doi: 10.1152/physiolgenomics.00068.2006. [DOI] [PubMed] [Google Scholar]
- Rikke BA, Johnson TE. Physiological genetics of dietary restriction: uncoupling the body temperature and body weight responses. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1522–R1527. doi: 10.1152/ajpregu.00215.2007. [DOI] [PubMed] [Google Scholar]
- Rikke BA, Yerg JE, 3rd, Battaglia ME, Nagy TR, Allison DB, Johnson TE. Strain variation in the response of body temperature to dietary restriction. Mech Ageing Dev. 2003;124:663–678. doi: 10.1016/s0047-6374(03)00003-4. [DOI] [PubMed] [Google Scholar]
- Rikke BA, Yerg JE, 3rd, Battaglia ME, Nagy TR, Allison DB, Johnson TE. Quantitative trait loci specifying the response of body temperature to dietary restriction. J Gerontol A Biol Sci Med Sci. 2004;59:B118–125. doi: 10.1093/gerona/59.2.b118. [DOI] [PubMed] [Google Scholar]
- Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585–600. doi: 10.1186/1471-2164-10-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Johnson TE. Aging Processes in Caenorhabditis elegans. In: Guarente L, Partridge L, Wallace DC, editors. Molecular Biology of Aging. Cold Spring Harbor Press; Cold Spring Harbor, NY Press: 2008. pp. 153–183. [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- Vickers AJ. The use of percentage change from baseline as an outcome in a controlled trial is statistically inefficient: a simulation study. BMC Med Res Methodol. 2001;1:6. doi: 10.1186/1471-2288-1-6. Epub 2001 Jun 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Q, Redden DT, Weindruch R, Allison DB. Statistical methods for testing effects on “maximum lifespan”. Mech Ageing Dev. 2004;125:629–632. doi: 10.1016/j.mad.2004.07.003. 2006. Erratum in Mech Ageing Dev 127, 652. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford RL. The retardation of aging and disease by dietary restriction. Charles C. Thomas; Springfield, IL: 1988. [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Williams RW, Bennett B, Lu L, Gu J, DeFries JC, Rikke BA, Belknap JK, Johnson TE. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mamm Genome. 2004;15:637–647. doi: 10.1007/s00335-004-2380-6. [DOI] [PubMed] [Google Scholar]
- Yu BP, Masoro EJ, McMahan CA. Nutritional influences on aging of Fischer 344 rats: I. Physical, metabolic, and longevity characteristics. J Gerontol. 1985;40:657–670. doi: 10.1093/geronj/40.6.657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


