Abstract
TCF7L2 transcription factor is a downstream effector of the canonical Wnt/β-catenin signaling, which controls cell fate and homeostasis. However, the complexity of TCF7L2 expression with numerous mRNA isoforms coding for proteins with distinct N- and C-termini allows variability in TCF7L2 functions and regulations. Here, we show that although TCF7L2 mRNA isoforms distinguish fetal, immortalized and adult differentiated endothelial cells (EC), they cannot explain the lack of significant β-catenin/TCF7 activities in ECs. Lithium, a Wnt-signaling activator, increases TCF7L2 mRNA levels and induces an RNA isoform switch favoring the expression of TCF7L2-short forms lacking the C-termini domains. Although the latter occurs in different cell types, its extent depends on the overall increase of TCF7L2 transcription, which correlates with cell-responsiveness to Wnt/β-catenin signaling. While GSK3β down-regulation increases TCF7L2 expression, there is no concomitant change in TCF7L2 mRNA isoforms, which demonstrate the dual effects of lithium on TCF7L2 expression via a GSK3β-dependent up-regulation and a GSK3β-independent modulation of RNA-splicing. TCF7L2E-long forms display a repressor activity on TCF7L2-promoter reporters and lithium induces a decrease of the endogenous TCF7L2 forms bound to native TCF7L2-promoter chromatin at two novel distal TCF7-binding sites. Altogether our data reveal a lithium-induced RNA switch favoring the expression of TCF7L2-short forms, which results in a transcriptional de-repression of lithium-target genes negatively regulated by TCF7L2-long forms, like TCF7L2, and thus to an amplification of Wnt-signaling in responsive cells.
Keywords: β-catenin, alternative-splicing, endothelial cells, immortalization
Introduction
Canonical Wnt/β-catenin signaling is involved in the control of cell fate during development and in stem cell renewal in the adult [1]. In this pathway, inhibition of β-catenin phosphorylation by GSK3β triggers β-catenin stabilization and its accumulation in the cytoplasm and subsequently in the nucleus where β-catenin acts as co-activator for the TCF7/LEF1 transcription factors. TCF7/LEF-1 factors belong to a small High-Mobility-Group (HMG) Box subfamily comprising 4 members: TCF7/TCF1, TCF7L1/TCF3, TCF7L2/TCF4 and LEF-1. Although they all bind the consensus A/TT/ACAAAG DNA sequence and share similar protein-protein interaction domains such as the β-catenin-binding domain and interacting domain for the co-repressors Groucho/TEL, they display differential activities regarding regulation of β-catenin/TCF7 target genes in different tissues and cell types [2].
Functional TCF7L2 diversity is also achieved by alternative RNA-splicing and exon-skipping that result in the expression of TCF7L2 forms with different N- and C-termini and thus variability in interacting partners and regulatory networks in a cell-context and target-gene dependent manner [3, 4]. The variable domains are, in the C-terminus, two binding sites for the co-repressor CtBP and a C-clamp involved in DNA-binding specificity and affinity, and in the N-terminus, the β-catenin binding domain and small intervening sequences between the β-catenin- and Groucho-binding domains, which may regulate these TCF7L2-interactions [2, 4–6].
While activation of the Wnt/β-catenin signaling, via mutations of β-catenin and several Wnt-signaling components, is highly associated with the development and progression of colon- and hepato-carcinoma [1], only rare mutations of TCF7L2 have been reported. Instead, specific TCF7L2-RNA isoforms were found enriched in colon carcinoma [3, 5]. Common polymorphisms distributed all along TCF7L2 gene have been associated with increased risks to develop Type-2 diabetes [7, 8] and its associated cardiovascular complications [9]. In this setting also, differential TCF7L2 mRNA levels and specific TCF7L2 mRNA isoforms are thought to depend upon TCF7L2 genotypes [10, 11].
Lithium inhibits GSK3β and thereby activates β-catenin/TCF7 transcriptional activities in different immortalized and cancer cells [12]. However, we have previously shown that lithium does not induce β-catenin/TCF7 transcriptional activities in primary ECs, but induces rather a cell cycle arrest and activation of the tumor suppressor p53 leading subsequently to the establishment of cell senescence [13, 14]. Similarly, expression of stable β-catenin forms leads to development of features associated with cell senescence in vitro and tissue aging in vivo [15, 16]. Sustained activation of the Wnt3a/β-catenin signaling pathway in the skin has also been associated with premature tissue aging and increased TCF7-dependent activities were detected in aging skeletal muscles [17, 18]. Since TCF7L2 itself is a β-catenin target gene [19], we tested the hypothesis that specific expression of TCF7L2 variants in a cell context-dependent manner mirrors the responsiveness to and/or the outcome of Wnt/β-catenin signaling activation by comparing the expression of TCF7L2 forms and their Wnt-signaling-dependent regulations in ECs and in the responsive stem-like HEK293 cells.
Material and Methods
Materials
LiCl was purchased from Sigma-Aldrich and all the primary and secondary antibodies were from Cell Signaling. The oligonucleotide primers were synthesized by Integrated DNA Technologies and the sequences are reported in the Supplementary Table S1.
Mammalian expression constructs
The S37A-β-catenin-HA tagged construct was previously described [20]. The full-length cDNAs for the TCF7L2 variants expressed in ECs were amplified by RT-PCR from HUVEC RNAs, N-terminus HA-tagged and cloned into pCR3.1-TA vector (Invitrogen).
Cell culture and transfection
Primary BAEC (Cambrex) were maintained in DMEM containing 1g/L D-glucose, 5% FBS and 100U/ml penicillin-100μg/ml streptomycin (Gibco-Invitrogen). HEK293T (Invitrogen) and the retinal RF6A (ATCC) cells were maintained in DMEM containing 4.5g/L D-glucose, 10% FBS and antibiotics. DNA transfections were performed using the Exgen500 reagent (Fermentas).
Short-interfering RNA transfection
The human/bovine siRNA-GSK3β duplex (5’-ACACUAUAGUCGAGCCAAAuu-3’) was previously described [14]. The on-target-plus Non-targeting-siRNA#1 was used as control and the si-RNA transfections were achieved using Dharmafect-I reagent (Dharmacon Inc.).
RNA extraction, reverse transcription and real-time PCR
RNA extractions were performed using Trizol-reagent (Invitrogen). Total RNAs (1μg) were subjected to DNAse-I treatment and reverse-transcription prior to PCRs with an equivalent of 10ng RNA per reaction. Quantitative real-time PCRs were performed in duplicates using SyBr-Green core reagents and the ABI7000/ABI7900 apparatus (Applied-Biosystems) as previously described [21] and normalized with the levels of rpL30 mRNAs.
Reporter assays
Cells were transiently co-transfected in duplicates with 5ng pCMV-β-galactosidase for normalization, 125ng of the luciferase-reporters and 400ng of the S37A-β-catenin-HA or HA-TCF7L2 variants. The luciferase reporters used in this study are i) the TOP-/FOP-Flash and the OT/OF reporters that contain either wildtype (TOP/OT) or mutated (FOP/OF) TCF7-binding sites either directly in front of the luciferase-cDNA (OT/OF) or upstream of the minimal thymidine kinase-promoter (TOP/FOP) [22] and ii) two TCF7-dependent promoter-luciferase constructs, the −6150/+476-HuTCF7L2P [19, 23] and the 1G-2235/+60-MMP1P [14, 24]. The cells were lysed 36h post-transfection and the firefly luciferase and β-galactosidase activities were quantified using Luciferase assays (Promega) and Galacto Light Plus assays (Applied Biosystems) respectively and an LMax-II luminometer (Molecular Devices). The activities of firefly luciferase were normalized with those of β-galactosidase.
Whole cell extracts and Western Blot analysis
After cold PBS washes, the cells were lysed in 50mM HEPES pH7.4, 150mM NaCl, 1%Triton-X100 supplemented with protease and phosphatase inhibitors (Sigma-Aldrich). Equal amount of proteins were fractionated on SDS-polyacrylamide gels and transferred onto Immobilon-P membrane (Millipore). After incubation with primary and secondary HRP-conjugated antibodies, the immuno-reactive proteins were detected using SuperSignal® chemiluminescence (Pierce Chemical Co).
Chromatin Immunoprecipitation (ChIp) assays
Cells were treated with either 10mM NaCl or 10mM LiCl for 36h prior to cell harvesting and native chromatin isolation. Chromatin-DNA fragments were obtained by microccocal-DNAse-I digestion (Enzymatic ChIp-assay, Cell Signaling) and 100μg of chromatin/DNA were incubated with non-immune-Ig (negative control), anti-Histone-H3 antibodies (positive control) or anti-TCF7L2 antibodies for 16h at 4ºC. The immune complexes were pulled down in presence of protein-G-magnetic beads and the associated DNA fragments were purified and identified by PCR using primer sets encompassing consensus TCF7-binding sites within the distal hu-TCF7L2 promoter (Supplementary Table S1).
Statistical analysis
Student’s t-tests were performed using GraphPad Prism version 4.0 (GraphPad Software) and results were considered significant at p<0.05.
Results and Discussion
Increased expression of endothelial-TCF7L2 variants cannot reverse the lack of significant β-catenin/TCF7 transcriptional activity in EC
Unlike HEK293 cells, ECs of different origins do not display significant β-catenin/TCF7 transcriptional activity in response to lithium [13] and stable forms of β-catenin (Fig. 1B). To determine whether differential TCF7L2 expression was responsible for this lack of β-catenin/TCF7 activity, we characterized TCF7L2 isoform expression in primary and immortalized ECs by performing RT-PCR with primer sets encompassing the alternative exons-4/6 and exons-13–16 (Fig. 1B) and by cloning the full-length TCF7L2 cDNAs expressed in HUVEC (Fig. 1A). The four major TCF7L2 splice variants expressed in HUVEC lack the exon-16, which is in agreement with its reported tissue-restricted expression [4, 10]. HUVEC express the long-TCF7L2E form containing the CtBP-binding sites and C-clamp, and the short-TCF7L2B form lacking both domains (Fig. 1A). HUVEC express also TCF7L2C and TCF7L2D variants that display the same C-termini coded by the association of Exon-14 with an A(10) stretch in Exon-17 [3], and thus contain the CtBP-binding sites but only an incomplete C-clamp [2, 4]. TCF7L2D contains also the Exon-4-coded domain that may regulate TCF7L2 interactions with β-catenin and/or Groucho.
Figure 1.
Expression and activity of TCF7L2 isoforms in ECs. (A) Schematic representation of human TCF7L2 exon organization [3, 4], of TCF7L2 domains and of the major TCF7L2 forms expressed in HUVEC. The primer sets used to analyze the expression of total TCF7L2 mRNA (T) and the alternative exons-4/6 (Ex3–9) and exons-13/16 (Ex12–17) are represented. (B) RT-PCR analysis of the TCF7L2 mRNA isoforms expressed in ECs with different tissue origins and immortalization status. The PCR products were resolved on 1% agarose gels. (C) Analysis of the transcriptional β-catenin/TCF7L2 isoforms activity in BAEC using co-transfection of S37A-β-catenin with TCF7L2 variants as indicated and TOP/FOP-Flash reporter assays (Mean±SEM, n=4–5). A representative western-blot analysis of the endogenous and exogenous TCF7L2 isoforms is shown.
This distribution of TCF7L2 variants in HUVEC can also be followed by PCRs using primer sets encompassing Exons-3–9 and Exons-12–17 (Fig. 1B), where the relative abundances of TCF7L2 mRNA isoforms containing Exon-4 (low) and Exon-14 (high) are consistent with the cloning data. Exon-15 and Exon-16, which have the same length in most species, cannot be distinguished on agarose and acrylamide gels (Figs. 1B–2B), but based on our cloning data the bands labeled Exon15/16 contain in majority Exon-15. The expression of Exons-4/6 and Exon-14 vary among the ECs studied. RF6A and HUVEC, which have embryonic origins, present the lowest relative levels of Exon-4-containg forms like the embryonic stem-like HEK293 cells (Fig. 1B). ECs derived from adult tissues display significant expression of the Exon-4 though it is more prominent in the primary BAEC than in the immortalized HMEC1 [25] and Br-MVEC1 [26]. The immortalization processes induced by viral T-antigens include senescence by-pass and acquisition of stem-like features [27], and thus the decreased expression of TCF7L2-Exon-4 in the immortalized adult ECs seems to reflect those phenotypic changes rather than differences between macro- and micro-vascular ECs. In contrast, the expression of Exons-13,14 RNA forms seems to differentiate macro- and micro-vascular ECs, since the lowest relative levels are found in BAEC, which instead express higher levels of the Exons-13,15 and Exons-13,14,15 RNA forms (Fig. 1B). As expected, these differential expressions of TCF7L2 RNA isoforms translate into differential expressions of TCF7L2-long and -short forms between BAEC and RF6A that can be visualized by western blotting (see Fig. 2C).
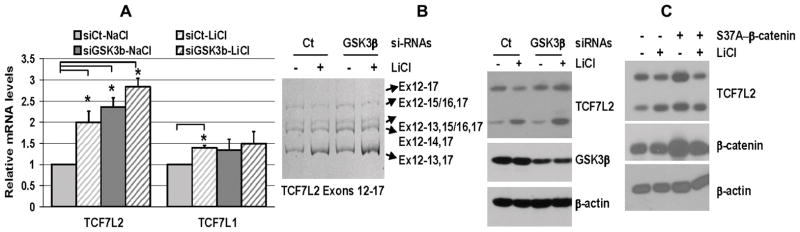
Figure 2.
Lithium increases TCL7L2 mRNA expression and favors the expression of TCF7L2 RNA isoforms coding for TCF7L2-short forms. Cells were treated with either 10mM NaCl or 10mM LiCl for 36h prior to RNA isolation and analysis of total TCF7L2 RNA levels by real-time PCR (Mean±SEM, n=4–5, *p<0.05) (A), of TCF7L2 isoforms (Exons-3–9, Exons-12–17) by standard PCR and 8%-acrylamide gel electrophoresis (B), and prior to whole-cell lysis and analysis of TCF7L2 forms by western blotting (C).
Although all the major TCF7L2 variants expressed in ECs contain the N-terminal β-catenin-binding domain, they failed to increase β-catenin/TCF7 activity in BAEC when co-expressed with the stable S37A-β-catenin form (Fig. 1C). The short-TCF7L2B forms increased the activity of both FOP- and TOP-Flash reporters indicating general effects on transcription rather than TCF7-dependent effects (Fig. 1C). In contrast, the expression of the TCF7L2E variant decreased the activities of both FOP- and TOP-Flash reporters resulting in a 40% decreased TOP/FOP ratio, while TCF7L2D had no effect (Fig 1C). A similar TCF7L2E-mediated decrease of the basal TOP/FOP ratio was also observed in HEK293 (see Fig. 3A) and in RF6A (not shown). These data are consistent with the increased DNA binding affinity and transcriptional activity conferred by the C-clamp [4, 6] and pinpoint a repressor role for TCF7L2E-long forms on this TOP/FOP system. Nevertheless, the expression of specific TCF7L2 isoforms is insufficient to explain the poor β-catenin/TCF7 activity in BAEC and RF6A since they express predominantly the transcriptional active TCF7L2-long forms and the “permissive” TCF7L2-short forms respectively.
Figure 3.
Down-regulation of GSK3β and expression of stable β-catenin forms affect TCF7L2 levels but not the balance of TCF7L2 variants. (A) Analysis of TCF7L2 RNA levels (Mean±SEM, n=3, *p<0.05), RNA isoform patterns and protein variants in HEK293 cells transfected with either si-control#1 (Ct) or si-GSK3β duplexes and treated with either 10mM NaCl or 10 mM LiCl. (B) Analysis of TCF7L2 isoform expression by western blotting in HEK293 cells transfected with either PCR3-control or S37A-β-catenin constructs and treated 16h later with either 10mM NaCl or 10mM LiCl for an additional 24h period.
Lithium increases TCF7L2 mRNA expression and affects TCF7L2 mRNA isoform patterns to favor the expression of TCF7L2-short forms
Since increased β-catenin/TCF7 activity can occur in part through TCF7L2 up-regulation, such as in response to vitamin-D in Wnt/β-catenin responsive cells [23], we investigated whether TCF7L2 expression was differentially regulated upon cell responsiveness to Wnt/β-catenin signaling by assessing the mRNA and protein levels of TCF7L2 isoforms after lithium treatment in BAEC and RF6A as well as in the Wnt/β-catenin responsive HEK293 cells. As shown in Fig. 2A, lithium treatment resulted in a significant 2.4 fold increase of total TCF7L2 mRNAs in HEK293 cells, while there was a trend for an increase in BAEC (1.4 fold) and a slight decrease in RF6A cells (Fig. 2A). TCF7L1 mRNA expression was similarly regulated by lithium though to a lesser extent in BAEC and HEK293 cells (Fig. 2A). In addition to increase TCF7L2 mRNA expression in HEK293 cells, lithium induced also drastic changes in the patterns of TCF7L2 mRNA isoforms containing the alternative Exons-13–16. An overall 50% decrease of the mRNA forms containing Exons-13,14,15/16, Exons-13,15/16 and Exons-13,14 was observed, while the mRNA forms containing only Exon-13 increased ~ 1.5 fold (Fig. 2B). This led to the concomitant decrease of the TCF7L2-long forms and increase of the TCF7L2-short forms as detected by western blotting (Fig. 2C), and thus to an inversion of the ratio TCF7L2-long/-short forms (3 to 0.25). Although lithium treatment induced similar TCF7L2 RNA isoform switch in BAEC (Fig. 2B) its limited extent, which follows the smaller increase of total TCF7L2 mRNA levels (Fig. 2A), failed to reverse the TCF7L2-long/-short form ratio (3 to 1.8) (Fig. 2C). A slight increase of the TCF7L2-short forms was also detected in lithium-treated RF6A though without significant effect on TCF7L2 form balance since it was already in favor of TCF7L2-short forms (0.3 to 0.25) (Figs. 2B–C). In all the cells, acute lithium treatments had no significant effect on the expression of TCF7L2-RNA isoforms containing the Exons-4/6 (Fig. 2B). The concomitant decrease of the longer TCF7L2-RNA forms (Exons-13,14,15; Exons-13,15; Exons-13,14) and increase of the shorter TCF7L2-Exons-13 RNA form argue for an effect of lithium on TCF7L2-RNA splicing and more precisely on exon-skipping rather than on the stability of specific TCF7L2-RNA isoforms. Thus, lithium affects TCF7L2 mRNA isoform pattern in a rather specific fashion and its extent in different cells seems to correlate with the strength of β-catenin/TCF7 activities.
GSK3β down-regulation and β-catenin-stabilization increase TCF7L2 mRNA expression without affecting TCF7L2-RNA isoform patterns
To determine the involvement of lithium-induced GSK3β inhibition on TCF7L2-RNA expression and patterns, GSK3β down-regulation by specific si-RNA duplexes was undertaken as previously described [14]. Down-regulation of GSK3β levels by 60–50% in HEK293 cells resulted in 2.2 and 1.4 fold increase of TCF7L2 and TCF7L1 mRNA levels respectively (Fig. 3A). Lithium treatment did not significantly increase further both TCF7L2 and TCF7L1 mRNA levels (Fig. 3A), indicating that lithium-induced TCF7L2 up-regulation is GSK3β-dependent. However, GSK3β down-regulation did not affect the distribution of TCF7L2 mRNA isoforms containing the alternative Exons-13–16 as well as the lithium-induced TCF7L2-RNA isoform switch favoring TCF7L2-short forms (Fig. 3A). Similarly, the ectopic expression of the stable S37A-β-catenin form increases the levels of both TCF7L2-long and -short forms but had no significant effect on the balance between TCF7L2 variants (Fig. 3B). These data are thus consistent with two inter-related but independent lithium-induced events: a GSK3β-dependent increase of TCF7L2 transcription and a GSK3β-independent effect on TCF7L2-RNA splicing. The former event is in agreement with TCF7L2 being itself a Wnt/β-catenin target gene [19]. Since TCF7L2 variants have clearly differential transcriptional activities (Fig. 1C, [4, 5]), the latter represents a novel lithium-dependent modulation of Wnt-signaling which distinguishes Wnt/β-catenin signaling responsive and unresponsive cells.
Lithium-induced TCF7L2 isoform switch is accompanied by a decrease of distal TCF7L2-promoter occupancy by TCF7L2 in HEK293 cells
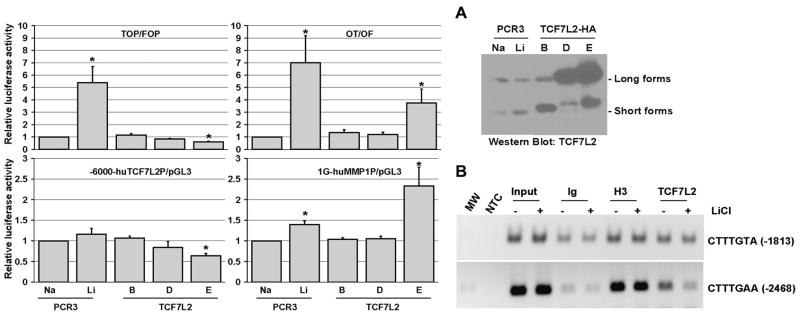
To determine the consequences of lithium-induced TCF7L2 isoform switch, we assessed the transcriptional activities of TCF7L2 variants on different TCF7-dependent reporters: the synthetic TOP/FOP and OT/OF reporter systems that differ by the absence of minimal promoter sequences in the latter and two promoter-luciferase reporters, -6150/+476-huTCF7L2P-pGL3 and 1G-2235/+60-MMP1P-pGL3 [14, 23], corresponding to two lithium-target genes, TCF7L2 (Fig. 2A) and MMP-1 [14], containing consensus TCF7-binding sites in their proximal promoters [19, 24]. In HEK293 cells, the TCF7L2E variant acted as a repressor on the TOP/FOP system (50% decrease) and as an activator on the OT/OF system (3 fold increase) (Fig. 4A), which further confirms the dual transcriptional activities of TCF7L2E depending on the neighboring promoter sequences [2, 4]. TCF7L2D displayed a weaker repressor activity (20% decrease) but no activator role, while the short-TCF7L2B variant had no activity (Fig. 4A). In contrast to its expected positive effects on MMP1-promoter reporter [24], TCF7L2E repressed unexpectedly TCF7L2-promoter activity by 50% (Fig. 4A). Since all the activities were tested in the absence of β-catenin overexpression, they likely represent the basal TCF7L2 activities on the corresponding promoters in HEK293 cells. Compared to the increase of TCF7L2 (2.4 fold) and MMP1 (6 fold) RNA levels observed in response to lithium (Fig. 2A and [14]), there were only weak increases of the corresponding promoter activities: 1.2 and 1.7 respectively (Fig. 4A). Nonetheless, lithium had the opposite effects of TCF7L2E on the -6150-huTCF7L2-promoter reporter as well as on the -2068-muTcfl2-promoter reporter [23] (not shown), suggesting that, instead of a TCF7L2-dependent activation, a decrease of TCF7L2E-repressor activity on TCF7L2-promoter may occur in response to lithium via its effect on the balance between active and permissive TCF7L2 variants.
Figure 4.
TCF7L2E variant represses TCF7L2 transcription and lithium decreases TCF7L2 occupancy onto TCF7L2-promoter native chromatin. (A) Analysis of the transcriptional activity of TCF7L2 variants in HEK293 cells using co-transfection with either synthetic TCF7-dependent reporter systems (TOP/FOP and OT/OF) or TCF7-target gene promoter-reporters containing consensus TCF7-binding sites (−6150-huTCF7L2P-pGL3 and 1G-2235-huMMP1P-pGL3). Results are expressed as relative luciferase activity with control cells equal to 1 (mean±SEM of 3–5 independent experiments performed in duplicates, *p<0.05) and a representative western blot analysis of the endogenous and exogenous expression of TCF7L2 forms is shown. (C) ChIp analysis of endogenous TCF7L2-bound onto the distal TCF7L2-promoter regions in HEK293 cells treated with either 10mM NaCl or 10mM LiCl for 24h. The PCR products, corresponding to the DNA-fragments containing the indicated consensus TCF7-binding sites at position −1813 and −2468 relative to the transcription start and immunoprecipitated with either the negative (non-immune-Ig) or positive (anti-Histone-H3) antibody controls or with anti-TCF7L2 antibodies, were resolved on agarose gels.
Next, we analyzed the changes in the huTC7L2-promoter occupancy by endogenous TCF7L2 in response to lithium using ChIp assays. First, in addition to the proximal +80 site previously described [19], two putative TCF7-binding sites located at -1813 and -2468 were found highly conserved between species. Both -1813 and -2468 distal sites showed specific binding of endogenous TCF7L2 by ChIp assays (Fig. 4B) and thus represent novel functional TCF7-binding sites within TCF7L2-promoter. After lithium treatment, a significant decrease of TCF7L2-bound to the more distal -2468 site was observed as well as to a lesser extent to the -1813 site (Fig. 4B), indicating that lithium affects negatively TCF7L2-promoter occupancy by TCF7L2 forms. Altogether these data, based on the drastic differences in transcriptional activity of TCF7L2 variants, are thus consistent with a basal repression of TCF7L2-promoter activity through binding of the TCF7L2E variants to the distal -2468 and -1813 TCF7-binding sites, which is released by the lithium-induced switch to the permissive TCF7L2-short forms. Beside an intrinsic decreased DNA-binding activity, it is likely that changes in interactions with neighboring partners participate also in the differential binding of TCF7L2-short form onto TCF7L2-promoter.
In summary this study reports novel effects of lithium on Wnt/β-catenin signaling via dual modulations of TCF7L2 expression. Lithium increases TCF7L2 mRNA levels in a GSK3β-dependent manner and induces a GSK3β-independent RNA-isoform switch favoring the expression of TCF7L2-short forms. The extent of the latter mirrors the ability of lithium to increase β-catenin/TCF7 activities. Our data confirm the differential transcriptional activities of TCF7L2 variants and emphasize the ability of TCF7L2E to act either as an activator or repressor depending on the promoter-context [2, 4]. Furthermore, we identified the repression of TCF7L2-promoter activity by TCF7L2E and showed that lithium-induced increase of TCF7L2-short forms is accompanied with a decrease of TCF7L2-bound onto TCF7L2-promoter native chromatin indicative of a de-repression mechanism of TCF7L2 transcription rather than TCF7-dependent activation per se. Such de-repression of TCF7-target genes may participate in an amplification of Wnt/β-catenin signaling, which further distinguish responsive and unresponsive cells.
Supplementary Material
Acknowledgments
We thank Drs. Clevers, Vogelstein, Toborek, and Andres for kindly sharing DNA constructs and Brain-MVEC mRNAs. This work was supported by NIH-AG031999 and College of Health Sciences-Pilot Research Funds, University of Kentucky (CDM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 2.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 3.Duval A, Rolland S, Tubacher E, Bui H, Thomas G, Hamelin R. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872–3879. [PubMed] [Google Scholar]
- 4.Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wöhrle S, Hecht A. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/{beta}-catenin targets. Nucleic Acids Res. 2009;38:1964–1981. doi: 10.1093/nar/gkp1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuilliere-Dartigues P, El-Bchiri J, Krimi A, Buhard O, Fontanges P, Fléjou JF, Hamelin R, Duval A. TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene. 2006;25:4441–4448. doi: 10.1038/sj.onc.1209471. [DOI] [PubMed] [Google Scholar]
- 6.Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 8.Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjögren M, Florez JC, Almgren P, Isomaa B, Orho-Melander M, Lindblad U, Daly MJ, Tuomi T, Hirschhorn JN, Ardlie KG, Groop LC, Altshuler D. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890–2895. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 9.Sousa AG, Marquezine GF, Lemos PA, Martinez E, Lopes N, Hueb WA, Krieger JE, Pereira AC. TCF7L2 polymorphism rs7903146 is associated with coronary artery disease severity and mortality. PLoS One. 2009;4:e7697. doi: 10.1371/journal.pone.0007697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott LJ, Usher N, Tong M, Sprau A, Swift A, Bonnycastle LL, Erdos MR, He Z, Saxena R, Harmon B, Kotova O, Hoffman EP, Altshuler D, Groop L, Boehnke M, Collins FS, Hall JL. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet. 2009;18:3795–3804. doi: 10.1093/hmg/ddp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, Berney T, Montanya E, Mohlke KL, Lieb JD, Ferrer J. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao CD, Hoang P, DiCorleto PE. Lithium inhibits cell cycle progression and induces stabilization of p53 in bovine aortic endothelial cells. J Biol Chem. 2001;276:26180–26188. doi: 10.1074/jbc.M101188200. [DOI] [PubMed] [Google Scholar]
- 14.Struewing IT, Durham SN, Barnett CD, Mao CD. Enhanced endothelial cell senescence by lithium-induced matrix metalloproteinase-1 expression. J Biol Chem. 2009;284:17595–17606. doi: 10.1074/jbc.M109.001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damalas A, Kahan S, Shtutman M, Ben-Ze'ev A, Oren M. Deregulated beta-catenin induces a p53- and ARF-dependent growth arrest and cooperates with Ras in transformation. EMBO J. 2001;20:4912–4922. doi: 10.1093/emboj/20.17.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M, Yu Q, Subrahmanyam R, Difilippantonio MJ, Ried T, Sen JM. Beta-catenin expression results in p53-independent DNA damage and oncogene-induced senescence in prelymphomagenic thymocytes in vivo. Mol Cell Biol. 2008;28:1713–1723. doi: 10.1128/MCB.01360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo CJ, Gutkind JS, Hwang PM, Finkel T. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 18.Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 19.Saegusa M, Hashimura M, Kuwata T, Hamano M, Okayasu I. Upregulation of TCF4 expression as a transcriptional target of beta-catenin/p300 complexes during trans-differentiation of endometrial carcinoma cells. Lab Invest. 2005;85:768–779. doi: 10.1038/labinvest.3700273. [DOI] [PubMed] [Google Scholar]
- 20.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 21.Struewing IT, Barnett CD, Tang T, Mao CD. Lithium increases PGC-1alpha expression and mitochondrial biogenesis in primary bovine aortic endothelial cells. FEBS J. 2007;274:2749–2765. doi: 10.1111/j.1742-4658.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 22.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 23.Beildeck ME, Islam M, Shah S, Welsh J, Byers SW. Control of TCF-4 expression by VDR and vitamin D in the mouse mammary gland and colorectal cancer cell lines. PLoS One. 2009;4:e7872. doi: 10.1371/journal.pone.0007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jean C, Blanc A, Prade-Houdellier N, Ysebaert L, Hernandez-Pigeon H, Al Saati T, Haure MJ, Coluccia AM, Charveron M, Delabesse E, Laurent G. Epidermal growth factor receptor/beta-catenin/T-cell factor 4/matrix metalloproteinase 1: a new pathway for regulating keratinocyte invasiveness after UVA irradiation. Cancer Res. 2009;69:3291–3299. doi: 10.1158/0008-5472.CAN-08-1909. [DOI] [PubMed] [Google Scholar]
- 25.Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, Lawley TJ. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- 26.Weksler BB, Subileau EA, Perrière N, Charneau P, Holloway K, Leveque M, Tricoire-Leignel H, Nicotra A, Bourdoulous S, Turowski P, Male DK, Roux F, Greenwood J, Romero IA, Couraud PO. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005;13:1872–1874. doi: 10.1096/fj.04-3458fje. [DOI] [PubMed] [Google Scholar]
- 27.Ali SH, deCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11:15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.