Abstract
Mutations in LRRK2 are thus far the most frequent known cause of autosomal dominant and idiopathic Parkinson’s disease (PD) with prevalent mutations being found within the GTPase (R1441C/G) and kinase (G2019S) domains. Previous in vitro studies have revealed that R1441C and G2019S mutations are associated with increased kinase activity. To better understand LRRK2-linked PD pathogenesis in vivo, we have generated transgenic C. elegans overexpressing human LRRK2 wild type, R1441C and G2019S in dopaminergic (DA) neurons. Overexpression of these LRRK2 proteins causes age-dependent DA neurodegeneration, behavioral deficits, and locomotor dysfunction that are accompanied by a reduction of dopamine levels in vivo. In comparison, R1441C and G2019S mutants cause more severe phenotypes than the wild type protein. Interestingly, treatment with exogenous dopamine rescues the LRRK2-induced behavioral and locomotor phenotypes. In contrast, expression of the GTP binding defective mutant, K1347A, or knockout of the C. elegans LRRK2 homolog, LRK-1, prevents the LRRK2-induced neurodegeneration and behavioral abnormalities. Hence, our transgenic LRRK2 C. elegans models recapitulate key features of PD including progressive neurodegeneration, impairment of dopamine-dependent behavior and locomotor function, and reduction in dopamine levels. Furthermore, our findings provide strong support for the critical role of GTPase/kinase activity in LRRK2-linked pathologies. These invertebrate models will be useful for studying pathogenesis of PD and for development of potential therapeutics for the disease.
Keywords: leucine-rich repeat kinase 2, Parkinson’s disease, Caenorhabditis elegans, dopamine, neurodegeneration
Introduction
Parkinson’s disease (PD) is a common neurodegenerative disorder characterized clinically by tremor, rigidity, bradykinesia, and postural instability, and pathologically by selective loss of dopaminergic (DA) neurons in affected brain regions. Mutations in leucine-rich repeat kinase 2 (LRRK2) represent the most frequent cause of autosomal dominant and idiopathic PD identified to date (Paisan-Ruiz et al., 2004; Zimprich et al., 2004). LRRK2 is a large multi-domain protein with both a GTPase and a kinase domain within the same molecule (Cookson et al., 2007; Gandhi et al., 2009; Guo et al., 2006; Mata et al., 2006). About thirty LRRK2 sequence variations have been identified (Brice, 2005; Farrer et al., 2005; Farrer, 2007; Nichols et al., 2007; Zabetian et al., 2005; Zimprich et al., 2004). Among them, the R1441C/G mutation within the GTPase domain, and the G2019S mutation within the kinase domain of LRRK2 occur more frequently and have been definitively linked to PD, highlighting an unprecedented role of GTPase and kinase signaling in the pathogenesis of the disease.
The normal function of LRKK2 or its closely related LRRK1 is not well understood. Null alleles of lrk-1 that encodes the sole LRRK2 homolog (LRK-1) in Caenorhabditis elegans (C. elegans) exhibit some defects in polarized sorting of synaptic vesicles (Sakaguchi-Nakashima et al., 2007). Knockdown of LRRK2 has been shown to promote neurite outgrowth, while overexpressing mutant LRRK2 has an opposing effect (MacLeod et al., 2006). However, it is unknown whether loss-of-function in LRK-1 and LRRK2 affects survival of DA neurons. Recent in vitro studies have suggested a possible gain-of-function mechanism for some PD-linked mutations. Biochemical investigations have shown that LRRK2 mutations such as R1441C and G2019S mutations lead to an increase in kinase activity of LRRK2 when overexpressed in cultured cells (Gloeckner et al., 2006; West et al., 2005). Further studies have demonstrated that LRRK2 is a GTP binding protein and a kinase that is activated intramolecularly by its own GTPase domain. Specifically, non-hydrolyzable analogs of GTP and the R1441C mutation have been shown to stimulate kinase activity while artificial mutations (K1347A and T1348N) that block GTP-binding to LRRK2 completely inhibit kinase activity (Guo et al., 2007; Ito et al., 2007; Li et al., 2007; Smith et al., 2006; West et al., 2007). Therefore, LRRK2 likely integrates GTPase and kinase signaling pathways relevant to the pathogenesis of PD. However, recent transgenic R1441C or R1441G mouse models have shown only some deficiency in DA neurotransmission without overt neurodegeneration (Li et al., 2009; Tong et al., 2009). In contrast, overexpression of LRRK2 wild type and G2019S in Drosophila and C. elegans has been reported to induce neurodegeneration and modulate mitochondrial function (Imai et al., 2008; Liu et al., 2008; Saha et al., 2009), demonstrating the power of an invertebrate model in studying the possible gain-of-function phenotype acquired by LRRK2 mutations. It remains to be definitively determined if GTPase or kinase activity is required for the pathogenesis of LRRK2-mediated neurodegeneration.
To further investigate the role of pathogenic R1441C and G2019S mutations within the respective GTPase and kinase domains and the requirement for GTPase/kinase activity in the survival and function of DA neurons, we have generated transgenic C. elegans overexpressing human LRRK2 wild type (WT), R1441C and G2019S mutants, and a LRRK2-inactive mutant K1347A in DA neurons. We have found that expression of active LRRK2 proteins including WT, R1441C and G2019S leads to age-dependent degeneration of DA neurons, behavioral deficit and locomotor dysfunction, and depletion of dopamine, whereas LRRK2-induced phenotypes are ameliorated by the GTP-binging defective K1347A mutation and by the loss of endogenous LRK-1.
Materials and methods
Nematode strains and maintenance
Several C. elegans strains were obtained from the Caenorhabditis Genetics Center or elsewhere as indicated. They include: WT reference strain N2 Bristol; lrk-1(km41) with a deletion of nearly all LRK-1 functional domains and lrk-1(km17) with a deletion of the LRK-1 kinase and WD40 domains as described [kind gifts from N. Hisamoto; (Sakaguchi-Nakashima et al., 2007)]; cat-2(e1112) defective in dopamine synthesis due to a loss-of-function mutation in the tyrosine hydroxylase homolog (Sawin et al., 2000); lin-15(765ts) with a temperature-sensitive loss-function mutation in lin-15 resulting in the multivulva (Muv) phenotype; and transgenic CAT-2 line (strain UA57, Pdat-1::CAT-2; Pdat-1::GFP).
All C. elegans strains were cultured on standard NGM (nematode growth medium) agar plates seeded with E. coli OP50 as a food source according to established protocols (Hope, 1999). NGM was made with Bacto agar and Bacto peptone from BD Biosciences and other chemicals from Sigma. Mixed stage animals were maintained as bulk culture on NGM agar at room temperature (22 °C). For most experiments, C. elegans strains were age-synchronized by egg-laying for 3 hours on NGM plates to obtain newly hatched animals that underwent development from larval stage L1 to L4, which was considered as the beginning of adulthood and counted as adult day 0. L4 worms were then picked and transferred to new NGM plates for growing to adults. For assays in worms with different adult stages, synchronized animals were maintained on NGM containing 0.02 mg/ml of 5′-fluorodeoxyuridine to suppress progeny growth.
Construction of transgenic C. elegans lines
Human cDNA encoding full-length C-terminally FLAG-tagged human LRRK2 WT and mutants (R1441C, G2019S, and K1347A) generated by site-directed mutagenesis were previously constructed in pCEP4 vector as described (Guo et al., 2007). The C. elegans promoter for dopamine transporter dat-1 (Pdat-1) was used to drive transgene expression in DA neurons (Nass et al., 2002). The Pdat-1 promoter sequence was PCR amplified from the upstream 719-bp region containing the initiation codon ATG using the corresponding Promoterome clone (Geneservice, UK) as the template. The plasmid pFXneEGFP used previously as a C. elegans expression vector (Kuwahara et al., 2006) was modified by switching the original BamHI site to the region 17-bp downstream of the NotI site. The PCR product of Pdat-1 was subsequently inserted into KpnI and HindIII sites immediately upstream of the enhanced green fluorescent protein (GFP) of the modified pFXneEGFP to create Pdat-1::GFP ( serving as a fluorescent reporter for DA neurons). The FLAG-tagged human LRRK2 cDNA was excised from NotI and BamHI sites of pCEP4 and subcloned into the same sites in the Pdat-1-containing pFXneEGFP, generating Pdat-1::LRRK2. Young adult C. elegans hermaphrodites with lin-15(765ts) allele (strain MT1898) were injected with a mixture of DNA consisted of Pdat-1::GFP (20 ng/μl) alone or together with Pdat-1::LRRK2 (20 ng/μl), as well as lin-15+ selection marker (50 ng/μl; pJM23, a gift from L. Avery) and an empty vector pSL1180 (20 ng/μl). The injected animals (about 20 animals for each genotype) were grown at 22 °C and their F1 progenies were first selected against the Muv phenotype and then were selected for the presence of GFP fluorescence in DA neurons in successive offspring. The resulting C. elegans transgenic animals with stable extrachromsomal arrays were generated, corresponding to those carrying Pdat-1::GFP only (GFP reporter lines) or those carrying both Pdat-1::GFP and Pdat-1::LRRK2 (LRRK2 WT, R1441C, G2019S, and K1347A). Multiple independent stable lines were obtained (5-7 lines for each genotype) and at least three of these lines were used initially for experimental analysis. Subsequent chromosomal integration was achieved by treatment with trimethylpsoralen and long-wave UV, followed by outcrossing with the N2 strain for at least three times. Additional strains were generated by crossing LRRK2 WT or G2019S integrated lines with LRK-1 deletion mutants carrying the lrk-1(km41) or lrk-1(km17) allele. Representative integrated lines from each strain were used in repeated experiments throughout this study, and their detailed genotype and strain designation is listed in Table 1.
Table 1.
Transgenic C. elegans strains used in this study
| Designation | Genotype | Strain |
|---|---|---|
| GFP | lin-15(n765ts) X; cwrIs730 [Pdat-1::GFP, lin-15(+)] | SGC730 |
| LRRK2+ | lin-15(n765ts) X; cwrIs722 [Pdat-1::GFP, Pdat-1::LRRK2(WT), lin-15(+)] | SGC722 |
| R1441C+ | lin-15(n765ts) X; cwrIs851 [Pdat-1::GFP, Pdat-1::LRRK2(R1441C), lin-15(+)] | SGC851 |
| G2019S+ | lin-15(n765ts) X; cwrIs856 [Pdat-1::GFP, Pdat-1::LRRK2(G2019S), lin-15(+)] | SGC856 |
| K1347A+ | lin-15(n765ts) X; cwrIs862 [Pdat-1::GFP, Pdat-1::LRRK2(K1347A), lin-15(+)] | SGC862 |
| LRRK2+; lrk-1(km17) | lrk-1(km17) I; lin-15(n765ts) X; cwrIs722 [Pdat-1::GFP, Pdat-1::LRRK2(WT), lin-15(+)] | SGC100 |
| LRRK2+; lrk-1(km41) | lrk-1(km41) I; lin-15(n765ts) X; cwrIs722 [Pdat-1::GFP, Pdat-1::LRRK2(WT), lin-15(+)] | SGC101 |
| G2019S+; lrk-1(km17) | lrk-1(km41) I; lin-15(n765ts) X; cwrIs722 [Pdat-1::GFP, Pdat-1::LRRK2(G2019S), lin-15(+)] | SGC102 |
Single worm PCR
Individual worms were lysed by treatment with proteinase K and heat, and resulting worm lysates were used for PCR to detect LRRK2 and lrk-1. For detection of LRRK2 transgene, a ~500-bp of LRRK2 open-reading frame was amplified by conventional PCR with the primers: forward 5′-CAGGCTGTTAAGACAAGAGC-3′ and reverse 5′-GGGCACAACCATATTCTTTA-3′. Duplex PCR was used to confirm the presence of deletions of lrk-1 in the lrk-1(km17) and lrk-1(km41) alleles using the following primers: km17 (forward 1: 5′-CCAAGATGGGATACCTGAAAAA-3′; forward 2: 5′-TTAACTGATCCGGGTGAATTGT-3′; and reverse: 5′-AGCTTCCAATAACCAGGTGAAA-3′) and km41 (forward 1: 5′-ATATACTCGGGAAGTTGGCTCA-3′; forward 2: 5′-GGTATTGCAGTTCTCTTTTTGGA-3′; and reverse: 5′-TAATCGAAATCGACACAGTTGG-3′).
Immunohistochemistry and Western blotting
Approximately 500 age-synchronized worms (adult day 2) were washed in M9 buffer (0.3% KH2PO4, 0.6% Na2HPO4, 0.5% NaCl, 1mM MgSO4) and were fixed with 4% paraformaldehyde for 30 min at room temperature. Fixed worms were placed on 1mg/ml poly-D-lysine coated slides, covered with glass coverslips, and left on dry ice for 10 min and transferred to liquid nitrogen for additional 20 min. After peeling off the coverslips, the glass slides were submerged in an acetone/methanol (50%/50%, v/v) bath pre-cooled to −80 °C for 5 min, and rehydrated through an ethanol:phosphate-buffered saline (PBS) series (9:1, 7:3, 1:1, 3:7, 1:9). The slides were blocked in PBST (0.1% Triton X-100 in PBS) for 6 h at 4 °C, incubated with 1:500 rabbit anti-LRRK2 antibody (catalog number 268, Novus Biologicals) overnight at room temperature, washed with PBS and then incubated with 1:1000 Alexa Fluor 568-conjugated secondary antibody (Invitrogen) for 6 h at room temperature. The immunostaining was detected using a confocol laser scanning microscope (Zeiss LSM 510 META).
Protein extracts were prepared from mixed stage worms grown to near confluence on five 100 mm NGM plates. Worms were washed with M9 buffer and collected by brief centrifugation at 3,000 × g for 1 min. The worm pellet was homogenized in ice-cold lysis buffer (1% Triton X-100 in PBS) supplemented with a cocktail of protease inhibitors (Roche), and the homogenate was clarified after centrifugation at 14,000×g for 10 min. The supernatant was collected and was immunoprecipitated with anti-FLAG M2 agarose (Sigma). Following washing with 0.5% Triton X-100 in PBS, the immunoprecipitates were collected for SDS-PAGE and Western blotting using anti-LRRK2 antibody 268 (1:3,000; Novus Biologicals) as descried previously (Guo et al., 2007). Anti-tubulin antibody E7 (1:8,000; Developmental Studies Hybridoma Bank, Iowa City, IA) was used for protein loading control.
Neurodegeneration Assay
Degeneration of DA neurons in live C. elegans was analyzed essentially as described (Berkowitz et al., 2008). Briefly, age-synchronized worms were paralyzed by treatment with 3 mM levamisole and mounted on glass slides. The DA neurons in the head regions [four cephalic (CEP) neurons and two anterior dierdic (ADE) neurons] were visualized for GFP fluorescence under a Zeiss Axiovert 200M microscope, and monitored at different ages (adult day 0 (L4) through day 9). The numbers of CEP neurons, which were larger than ADE neurons and were free of interference from endogenous autofluorescence even in old animals, were scored for the presence of intact neurons. CEP neurons missing the cell body or with broken neurites were counted as degenerating. For each strain about 30 worms were analyzed in at least three independent experiments, with a total of approximately 90-120 animals scored in the same manner. The percent of intact CEP neurons remaining was calculated against the numbers of CEPs expected if no degeneration occurred (four in each animal). Fluorescent images of DA neurons in the head region of worms were taken with a Zeiss Axiovert 200M microscope using 1 s exposure time at 20X magnification.
Behavioral assay
Well-fed worms with intact DA neural circuitry move slower in the presence of bacterial food than in its absence. This basal slowing response (or food-sensing response) was assayed as described (Sawin et al., 2000). Briefly, food-containing assay plates were prepared by spreading the bacterial food, E.coli (OP50), in a ring on NGM agar, and together with non-coated assay plates, were incubated at 37°C overnight and cooled to room temperature prior to assay. Age-synchronized worms (about 10 worms each strain) were transferred from their food-containing culture plate to a non-food plate and washed in S basal buffer (100 mM NaCl, 10 μg/ml cholesterol, 50 mM potassium phosphate, pH 6.0) twice to get rid of the remaining food in their bodies. After that, worms were transferred to the food-containing or non-food assay plates prepared above, settled for 5 min, and their body bends in 20 s intervals were recorded for each of the worms. For DA treatment, worms grown to adult day 4 were pre-incubated for 6 h at room temperature on NGM plates containing bacterial food doped with 2 mM dopamine hydrochloride prior to the assay for basal slowing response as described above.
Locomotion assay
The locomotor speed of worms was monitored using an automated worm tracking system. This system consisted of a stereomicroscope (Zeiss Stemi 2000C) mounted with a digital camera (Cohu 7800), and a digital motion system (Parker Automation) controlled by a computer software as previously described (Feng et al., 2004). The temperature (20-22 °C) and humidity (around 40%) were strictly controlled during each assay. Worms were synchronized at embryo stage, and their locomotor speed was analyzed at the different ages of adult day 0, 2, 4, 6 and 8. Each worm was transferred to a freshly made NGM plate, and was immediately tracked for 10 min by the worm tracking system. The stereomicroscope was fixed at 20X magnification during automated operation, and the imaging density was 170 pixels/mm. The locomotion velocity of the animal at each time point, computed as centroid displacement of pixels per second, was recorded in real time during automated tracking. The speed (mm/min) for each 1-min duration was converted using the equation locomotor speed=Pixel velocity × 60 ÷ 170. The average locomotor speed was calculated by averaging all data points collected during the entire 10 min period from about 20 worms for each strain.
Dopamine measurement
Age-synchronized worms were collected from 100 mm NGM plates using M9 buffer, washed once and incubated in M9 for 30 min at room temperature to clear off remaining bacteria, and collected by brief centrifugation. Worm pellets were kept frozen at −80 °C until analysis. Samples were resuspended in 200 μl of 0.05 N perchloric acid, sonicated on ice three times for a duration of 15 s each time, and centrifuged at 16,000xg for 15 min. The supernatant was used for measurement of DA by reverse phase HPLC with electrochemical detection, essentially as described previously (Pehek et al., 2006). Samples (20 μl) were injected onto a 2×100 mm Phenomenex (Torrance, CA) column (Ultracarb™, 3 mm particle size, ODS 20) maintained at 37 °C.. The mobile phase consisted of 32 mM anhydrous citric acid, 54 mM sodium acetate trihydrate, 0.074mM EDTA, 0.215 mM octylsulfonic acid, and 3 % methanol (vol/vol), pH 4.2. Dopamine was detected with a BAS LC-4C electrochemical detector and a glassy carbon electrode maintained at a potential of +0.60V relative to an Ag/AgCl reference electrode. The dopamine levels of different worm strains were normalized to total amount of protein present in each sample. Protein was determined by the method of Bradford using the protein assay dye reagent (Bio-Rad).
Statistical analysis
Data were presented as the means±SEM. Statistical significance of differences between results was evaluated using one-way ANOVA followed by the Tukey-Kramer post hoc test for multiple comparisons.
Results
Generation of transgenic C. elegans lines overexpressing LRRK2 WT, R1441C and G2019S in DA neurons
We targeted the LRRK2 transgene to DA neurons to better mimic a PD-relevant cellular environment. In C. elegans hermaphrodites, there are only eight DA neurons and their anatomic locations can be visualized easily with the use of green fluorescent protein (GFP) coupled with a DA-neuron-specific promoter for the dopamine transporter dat-1 (Pdat-1). We generated transgenic lines overexpressing either the GFP reporter alone (Pdat-1::GFP) or together with the full-length LRRK2 proteins (Pdat-1::GFP; Pdat-1::LRRK2s) corresponding to LRRK2 WT, PD-linked mutants R1441C and G2019S, and a LRRK2-inactive mutant K1347A. These transgenic worms were designated as GFP, LRRK2+, R1441C+, G2019S+, and K1347A+, respectively. A total of 3-7 independent transgenic lines per genotype were established, and representative strains carrying chromosomally integrated transgene were used throughout this study (Table 1).
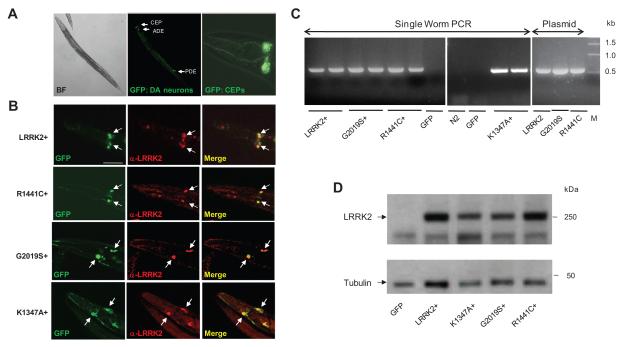
We performed a series of experiments to verify the expression of LRRK2 transgene in these transgenic lines. As reported previously by others (Cao et al., 2005; Kuwahara et al., 2006; Nass et al., 2002), we confirmed that the Pdat-1 promoter directed transgene expression exclusively in DA neurons of all LRRK2 transgenic worms, as revealed by the location of fluorescent GFP reporter (Fig. 1A). Immunohistochemistry showed that LRRK2 immunoreactivity co-localized with the GFP reporter in DA neurons of the LRRK2 transgenic lines (LRRK2+, R1441C+, G2019S+, and K1347A+) (Fig. 1B). The presence of LRRK2 transgene was confirmed using single-worm PCR for a fragment of human LRRK2 cDNA, which was detected only in the transgenic lines transformed with the LRRK2 gene, but not in the GFP reporter line or nontransgenic N2 worms (Fig. 1C). Western blotting confirmed that human LRRK2 proteins were expressed at similar levels with an expected Mr of about 280-kDa in the LRRK2+, R1441C+, G2019S+ and K1347A+ lines (Fig. 1D). Thus we successfully established C. elegans transgenic lines overexpressing human LRRK2 proteins (WT, R1441C, G2019S, and K1347A) in DA neurons.
Fig. 1.
Generation of transgenic C. elegans overexpressing LRRK2 in DA neurons. (A) Representative images of adult worms from LRRK2 transgenic lines visualized by bright-field light microscopy (BF, left) and epifluorescence microscopy (GFP) of whole body (middle) or enlarged head region (right). The positions of DA neurons (2 pairs of cephalic neurons CEPs, 1 pair of anterior deirid neurons ADEs, and 1 pair of posterior deirid neurons PDEs) are indicated by arrows. (B) Immunostaining of human LRRK2 in transgenic C. elegans overexpressing LRRK2 WT, R1441C, G2019S and K1347A (designated as LRRK2+, R1441C+, G2019S and K1347A+, respectively). Shown are head regions containing DA neurons CEPs visualized by GFP fluorescence (left) or anti-LRRK2 immunostaining (middle), followed by the merged images (right). (C) Detection of LRRK2 transgene. Single-worm PCR for a ~500 bp fragment of LRRK2 cDNA. LRRK2 transgene was detected in the LRRK2+, R1441C+, G2019S+, and K1347A+ lines, but not in the GFP reporter line or non-transgenic strain N2. Plasmid constructs used for transformation served as positive controls. M, DNA size marker in kb. (D) Detection on Western blots of LRRK2 proteins in C. elegans overexpressing LRRK2 WT, K1347A, G2019S, and R1441C, but not in the GFP reporter line. Worm lysate was affinity purified with anti-FLAG agarose, followed by Western blotting with an antibody against LRRK2. Scale bar = 5 μm.
LRRK2 expression causes age-dependent degeneration of DA neurons
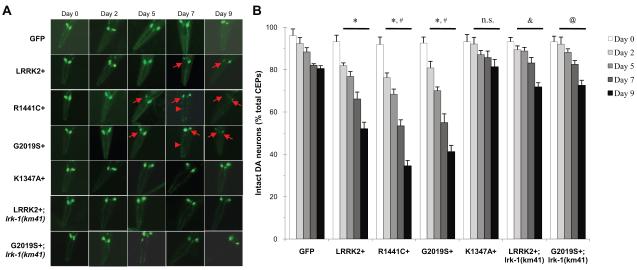
We investigated whether overexpression of LRRK2 WT and PD-linked mutants R1441C and G2019S induces neurodegeneration in C. elegans DA neurons. Previous studies have established that DA neurodegeneration can be reliably assayed in live worms by monitoring the morphological changes of DA neurons co-expressing GFP reporter (Berkowitz et al., 2008; Nass et al., 2002). We scored transgenic worms throughout their life span for the survival of each of the four cephalic neurons (CEPs), as these DA neurons are relatively large in size and are free from interference by autofluorescence that can develop in old animals. We observed no overt morphological changes within the DA neurons in any of the LRRK2 transgenic lines before the worms reached adulthood (up to the last larval stage L4, referred to hereafter as adult day 0). However, we found that transgenic worms overexpressing LRRK2 WT, R1441C or G2019S in DA neurons manifested adult-onset and age-dependent neurodegeneration characterized by the loss of DA neuron soma and breakdown of neurites as compared to the GFP control line (Fig. 2A). LRRK2+, R1441C+ and G2019S+ worm lines exhibited significant neuronal loss that began in young adults (at about adult day 2) and proceeded with progressive worsening in older animals in middle to old age (adult day 5-9) relative to the GFP reporter line (Fig. 2B).
Fig. 2.
LRRK2 expression leads to DA neurodegeneration. (A) C. elegans expressing GFP marker contained intact DA neurons (CEP neurons) and smooth neurites, whereas the C. elegans LRRK2 overexpression lines (LRRK2+, R1441C+ and G2019S+) displayed prominent loss of DA neurons (arrows) and broken neuritis (arrowheads) during adulthood. Days in adulthood (number of days past larval stage L4) is indicated on top. (B) The age-dependent DA neurodegeneration was quantified by the loss of CEP neurons in transgenic animals with different ages. Synchronized worms at adult day 0 (L4 larval stage), 2, 5, 7, and 9 were mounted on agar-containing slides for live imaging and scoring for the presence of each of the four CEP neurons visualized by GFP fluorescence. About 20-30 animals per strain were used in each of the three to four independent experiments. The total number of CEP neurons expected from all animals (4 per animal) is regarded as 100%, and numbers of CEP neurons detected in each experiment are used to calculate the percent intact DA neurons remaining. LRRK2+ and G2019S+ worms were crossed with the lrk-1(km41) mutant worms to generate the LRRK2+; lrk-1(km41) and G2019S+; lrk-1(km41) strains, respectively. Error bars indicate SEM. One-way ANOVA: * p<0.01 versus GFP control. # p<0.05 versus LRRK2+. & p<0.01 versus LRRK2+. @ p<0.01 versus G2019S. n.s., not significant compared to GFP reporter line.
Moreover, R1441C+ and G2019S+ mutant lines showed a more severe loss of DA neurons than the WT LRRK2+ line. There were 34% loss of DA neurons on adult day 7 and 48% loss on day 9 in the LRRK2+ worms, 47% loss on day 7 and 65% loss on day 9 in the R1441C+ worms, and 45% loss on day 7 and 60% loss on day 9 in the G2019S+ worms (Fig. 2B). In contrast, less than 20% loss of DA neurons was attributable to natural aging as observed in GFP worms that lived for 9 days in adulthood. These results are consistent with the observations that R1441C and G2019S are both activating mutations with an enhanced GTP-bound state and increased kinase activity in vitro, respectively (Guo et al., 2007; Ito et al., 2007; Lewis et al., 2007; Li et al., 2007; West et al., 2005; West et al., 2007). To further define the role of GTPase/kinase activity of LRRK2 in LRRK2-induced DA degeneration, we examined a mutant line overexpressing an engineered K1347A mutation in DA neurons. Previous studies have shown that the K1347A mutation impairs GTP binding to the GTPase domain and subsequently inactivates kinase activity of LRRK2 in vitro (Guo et al., 2007; Ito et al., 2007; Li et al., 2007; Smith et al., 2006; West et al., 2007). Transgenic worms overexpressing the LRRK2-inactive mutant K1347A (K1347A+) did not exhibit a neurodegeneration phenotype as seen in animals overexpressing LRRK2 WT or its activating mutants R1441C and G2019S (Fig. 2A & B). Taken together, these findings suggest that overexpression of LRRK2 WT and PD-linked LRRK2 mutants R1441C and G2019S are sufficient to acquire a toxic gain-of-function leading to DA neurodegeneration, which is likely mediated by the GTPase/kinase activities of LRRK2.
LRRK2 expression causes DA-specific behavioral dysfunction
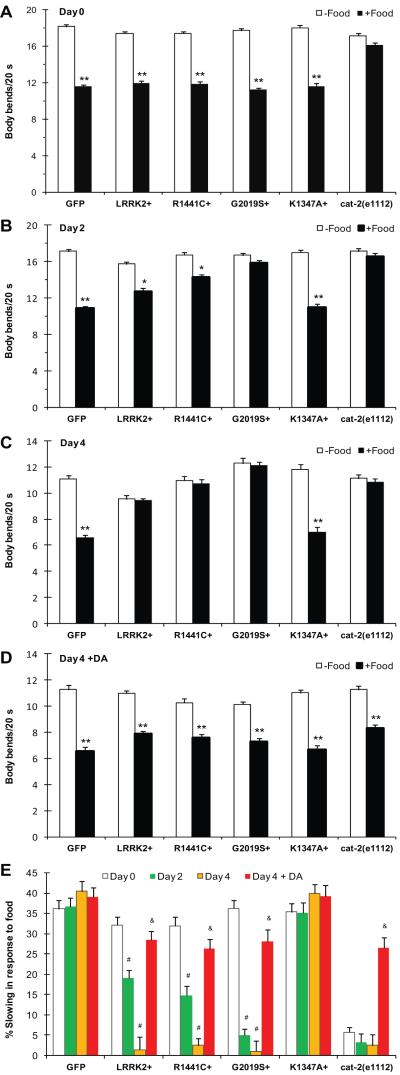
To evaluate if LRRK2 overexpression disrupts dopamine-dependent function and behavior, we performed a well-established dopamine-specific behavior assay in C. elegans. Well-fed worms slow down their body bending in the presence of bacterial food relative to the absence of bacteria. This behavior, termed basal slowing response or food-sensing, is mediated exclusively through a dopamine-mediated neural circuit, and the behavior is lost in the cat-2(e1112) mutant strain defective for tyrosine hydroxylase (Sawin et al., 2000). As expected, GFP animals reduced their body bending in response to bacterial food throughout adult day 0 to day 4, whereas the cat-2(e1112) worms failed to slow their bending rate (Fig. 3A-C). Transgenic C. elegans lines overexpressing LRRK2 WT, R1441C and G2019S showed normal behavior at the beginning of the adulthood (adult day 0), but they manifested a progressive loss of the basal slowing response from adult day 2 to day 4 (Fig. 3A-C). In contrast, the LRRK2-inactive mutant K1347A transgenic worms showed no behavior abnormality. The severely impaired basal slowing behavior in LRRK2 transgenic animals and cat 2(e1112) worms on adult day 4 could be rescued following feeding with exogenous dopamine (Fig. 3D). The behavioral deficits induced by LRRK2 overexpression were age-dependent. LRRK2+, R1441C+, and G2019S+ worms exhibited intermediate behavioral impairment on day 2, but by day 4 they had completely lost the basal slowing response similar to cat-2(e1112) worms (Fig. 3E). It was noteworthy that the G2019S+ worms exhibited more rapid progression of this behavioral deficit than R1441C+ and LRRK2 WT+ worms. Nevertheless, treatment with exogenous dopamine led to a symptomatic relief to a similar extent in all transgenic worms overexpressing LRRK2 WT, R1441C and G2019S (Fig. 3E). We concluded that expression of LRRK2 WT and mutant R1441C and G2019S in DA neurons leads to age-dependent behavioral impairment that is responsive to treatment with exogenous dopamine.
Fig. 3.
LRRK2 overexpression impairs dopamine-specific basal slowing behavior in C. elegans. (A-C) Animals were monitored for body bending in the absence (-Food) or presence of bacterial food (+Food) on adult day 0, 2, and 4. They include transgenic C. elegans lines overexpressing GFP reporter alone or together with LRRK2 WT, R1441C, G2019S, and K1347A in DA neurons. The cat-2(e1112) worms deficient for dopamine synthesis were used as a control. (D) Rescue of impaired basal slowing response at adult day 4 following treatment with exogenous dopamine. Animals on adult day 4 were treated for 6 h with 2 mM dopamine hydrochloride applied to bacterial food. Their body bends were then recorded in the absence (-Food) or presence (+Food) of bacterial food. (E) Data from A-D were presented as percentage of slowing in body bending in the presence of food relative to that in the absence of food on adult day 0, 2, and 4. About 10 animals per strain were used for behavioral assays under each condition. Error bars indicate SEM. *p<0.05 and **p<0.01 versus no food. # p<0.01 versus adult day 0. & p<0.01 versus untreated.
LRRK2 overexpression induces locomotor dysfunction
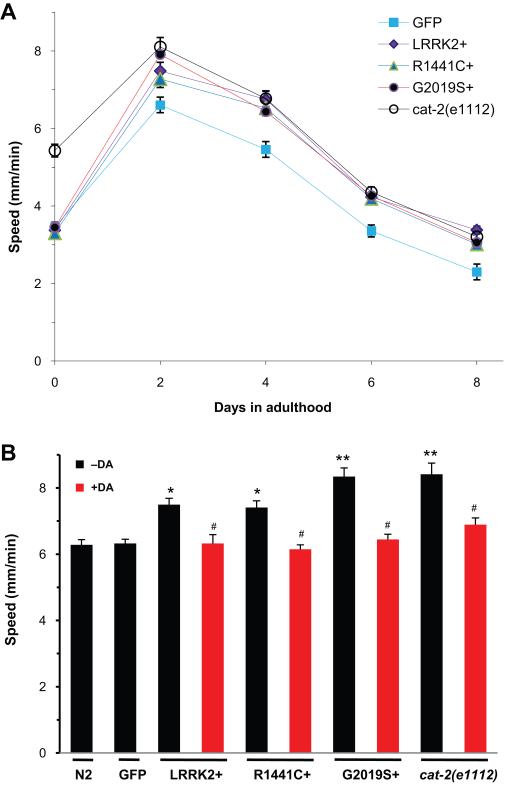
To identify possible locomotor dysfunction in transgenic lines overexpressing LRRK2 WT, R1441C and G2019S, we utilized a machine vision based worm tracking system to monitor worm movement. The spontaneous movement of undisturbed and freely moving worms was automatically detected and continuously analyzed by a computer-controlled motion system and a high-speed digital camera (Feng et al., 2004). Quantitative analysis of locomotor speed revealed that LRRK2 overexpression led to an adult-onset dysfunction in locomotion. As shown in Fig. 4A, GFP reporter worms or LRRK2+, R1441C+ and G2019S+ worms showed similar motility at the beginning of adulthood (adult day 0). However, all three LRRK2 expressing lines manifested abnormally hyperactive locomotion from adult day 2 (young) through day 8 (old) as compared to the GFP reporter line (Fig. 4A). The G2019S+ worms exhibited slightly more severe locomotor phenotype than the LRRK2+ or R1441C+ on adult day 2, the age during which worms were most active in locomotion (Fig. 4B). We explored the possibility that LRRK2 induced hyperactive locomotion might be related to the dysfunction of DA neurotransmission especially on adult day 2, even though a majority of DA neurons are still intact in the LRRK2 expressing lines at this age (Fig. 2). Indeed, the LRRK2-induced locomotor dysfunction mimicked that displayed in the cat-2(e1112) mutant line deficient for dopamine biosynthesis except that cat-2(e1112) worms already exhibited the hyperactive locomotion on adult day 0 (Fig. 4A). Moreover, treatment with exogenous dopamine rescued the locomotor dysfunction in LRRK2+, R1441C+ and G2019S+ lines as well as cat-2(e1112) worms on adult day 2 (Fig. 4B). These results demonstrate that expression of LRRK2 WT and PD-linked mutants in DA neurons leads to locomotion dysfunction that can be rescued by treatment with dopamine.
Fig. 4.
LRRK2 overexpression causes locomotor dysfunction in C. elegans. (A) Transgenic C. elegans lines overexpressing GFP reporter alone or together with LRRK2 WT, R1441C and G2019S were analyzed for locomotion on adult day 0 through 8. An automated behavior tracking system was used for tracking and recording of real-time locomotor speed of C. elegans freely moving on bacterial lawn for 10 min. The average locomotor speed of 10-20 worms per each strain was plotted a function of animal age. (B) Average locomotor speed was analyzed from various C. elegans strains that were either untreated or treated for 6 h with 2 mM dopamine hydrochloride applied to bacterial food. Non-transgenic strain N2 and cat-2(e1112) mutant strain deficient for dopamine synthesis were used as controls. Error bars indicate SEM. *p<0.05 and **p<0.01 compared to the GFP control.
LRRK2 overexpression leads to dopamine deficiency
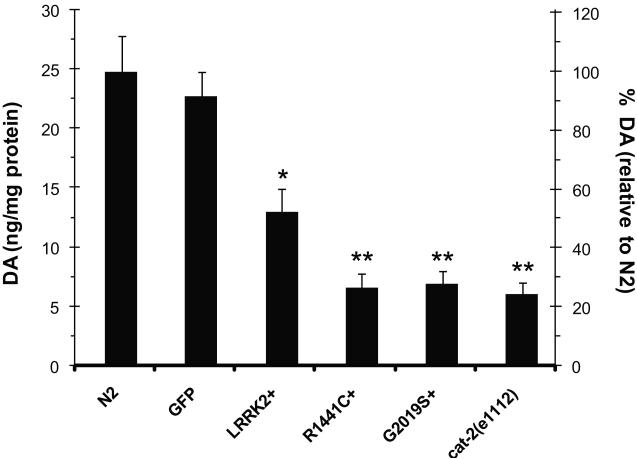
Since both the behavioral deficit and locomotor dysfunction caused by LRRK2 WT, R1441C and G2019S in transgenic worms could be rescued by dopamine treatment (Fig. 3 and Fig 4), we explored the possibility that these LRRK2 transgenic worms contain reduced levels of dopamine. We measured the whole animal levels of dopamine in age-synchronized LRRK2 transgenic worms and control animals on adult day 4. LRRK2 overexpression caused a significant decrease in dopamine levels, with ~50% dopamine loss in LRRK2+ worms, and ~72% dopamine loss in R1441C+ and G2019S+ worms relative to the native strain N2 (Fig. 5). In comparison, the GFP reporter line showed no DA deficiency and the DA-deficient reference strain cat-2(e1112) exhibited ~75% reduction of dopamine levels as compared to N2. The LRRK2-induced dopamine deficiency was age-dependent since no apparent changes in dopamine levels were observed in LRRK2+ and G2019S+ worms as compared to N2 or GFP reporter strains (data not shown).
Fig. 5.
LRRK2 overexpression leads to dopamine deficiency. Dopamine levels in transgenic worms expressing GFP reporter alone or together with LRRK2 WT, R1441C and G2019S were measured on adult day 4. Non-transgenic N2 worms and cat-2(e1112) mutant worms deficient in dopamine synthesis were used as controls. The dopamine contents were normalized with total proteins present in each strain (left axis) and percent dopamine was shown relative to N2 (right axis). Synchronized worms grown to adult day 4 were collected for dopamine measurements using reverse phase HPLC with electrochemical detection. Error bars indicate SEM. *p<0.05 and **p<0.01 compared to N2.
Loss of LRK-1 attenuates LRRK2-induced neurodegenerative and behavioral phenotype
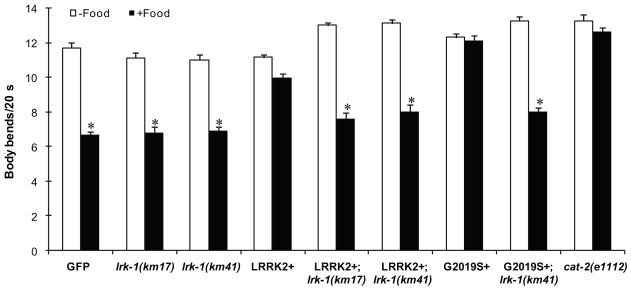
C. elegans contains a single homolog LRK-1 encoded by lrk-1 with a similar domain structure as that of human LRRK2. A previous study generated two deletion mutations of lrk-1, termed km41 and km17, that likely represent null alleles of lrk-1 due to the elimination of all functional domains (km41) or the C-terminal region containing the kinase domain (km17) (Sakaguchi-Nakashima et al., 2007). We crossed the LRRK2 overexpressing lines into the lrk-1 null background and examined the phenotype in these cross lines. We found that loss of endogenous LRK-1 via lrk-1(km41) attenuated DA neurodegeneration induced by overexpression of either LRRK2 WT or G2019S through the adulthood (Fig. 2). Moreover, both lrk-1(km41) and lrk-1(km17) null alleles rescued the behavioral deficit in the basal slowing response caused by overexpression of LRRK2 WT and G2019S in DA neurons on adult day 4 (Fig. 6). These results suggest that LRK-1 is functionally similar to LRRK2 and loss of LRK-1 can antagonize the LRRK2-induced neurodegenerative and behavioral phenotype.
Fig. 6.
Loss of endogenous LRK-1 attenuates LRRK2-induced behavioral deficits. Basal slowing response was examined on adult day 4 in transgenic C. elegans lines overexpressing LRRK2 WT and G2019S mutant (LRRK2+ and G2019S+), lrk-1 deletion lines km17 and km41, and their cross progeny [LRRK2+; lrk-1(km17), LRRK2+; lrk-1(km41), and G2019S+; lrk-1(km41)]. Worms carrying lrk-1(km17) and lrk-1(km41) null alleles had good basal slowing response. In contrast, LRRK2+ and G2019S+ worms exhibited prominent loss of basal slowing response as compared to the GFP reporter line. This behavioral impairment was rescued in the cross progeny with the deletion of endogenous LRK-1 as shown by the recovery of basal slowing response in the LRRK2+; lrk-1(km17), LRRK2+; lrk-1(km41), and G2019S+; lrk-1(km41) worms. Age-synchronized worms were grown to adult day 4 on NGM plates. About 10 worms were used for behavioral assay in each strain. The cat-2(e1112) worms defective in dopamine synthesis served as a control. Error bars indicate SEM. *p<0.01 versus no food.
We also examined whether loss of LRK-1 protects DA neurons from other causes of stresses. We took advantage of another C. elegans neurodegeneration model in which CAT-2 (tyrosine hydroxylase homolog) is overexpressed in DA neurons using the Pdat-1 promoter (stain UA57, Pdat-1::CAT-2; Pdat-1::GFP). Previous studies have shown that overexpression of CAT-2 leads to toxic levels of DA, a known source of oxidative stress, and consequent DA neurodegeneration (Cao et al., 2005). Consistent with the previous report (Cao et al., 2005), the CAT-2 overexpression line (designated as CAT-2+) displayed marked DA neurodegeneration on adult day 4 and progressive loss of basal slowing response on adult day 2 and 4, but not on adult day 0 (Supplemental Fig. 1). We crossed CAT-2+ line with the lrk-1(km41) deletion line to produce the cross progeny (CAT-2+; km41) and assessed DA neurodegeneration and behavioral phenotype. The CAT-2+; km41 line displayed typical DA neuron morphology and basal slowing behavior during the adulthood, indicating that the lrk-1(km41) loss-of-function allele rescued neurodegeneration and behavioral deficits induced by oxidative stress following CAT-2 overexpression (Supplemental Fig. 1). Taken together, our results suggest that loss of LRK-1 may protect DA neurons from different neurodegenerative stressors.
Discussion
In the present study, we have generated transgenic C. elegans expressing human LRRK2 WT and two PD-linked mutants R1441C and G2019S in DA neurons. These transgenic worms manifest pathological phenotypes that recapitulate several important aspects of PD including adult-onset progressive neurodegeneration, impairment of dopamine-specific behavior, and locomotor dysfunction accompanied by a loss of dopamine neurotransmitter. The abnormal behavioral and locomotor phenotypes can be rescued by treatment with exogenous dopamine. Therefore, these LRRK2 transgenic worms represent useful in vivo LRRK2-linked PD models and hold great promise for deciphering PD pathogenesis. Our findings have provided several novel insights into the pathogenesis of LRRK2-linked pathogenesis. First, overexpression of all active variants of LRRK2 including LRRK2 WT, R1441C and G2019S, but not the inactive form K1347A is sufficient to induce neurodegeneration accompanied by behavioral deficits and depletion of dopamine. The pathologic phenotype is more severe in LRRK2 carrying PD-linked mutations R1441C and G2019S than that of LRRK2 WT. These results provide evidence in support of the notion that PD-linked mutations R1441C and G2019S initiate a harmful gain-of-function leading to neuronal degeneration and dysfunction, consistent with the autosomal dominant effect of these mutations in humans. Second, the LRRK2-inactive K1347A mutation appears to thwart LRRK2-induced DA neurodegeneration and behavioral deficits in C. elegans. Since K1347A mutation has been shown to directly impair GTP binding to the GTPase domain and subsequent inactivation of kinase activity of LRRK2 in vitro (Guo et al., 2007; Ito et al., 2007; Li et al., 2007; Smith et al., 2006; West et al., 2007), our findings suggest that either the reduced GTP binding or decreased kinase activity of LRRK2 underlie the normal neuronal phenotype in worms expressing K1347A. However, a second possibility that the K1347A mutation may reduce the stability of LRRK2 has also been suggested (Lewis et al., 2007). We have shown that the K1347A mutant worms express similar steady levels of LRRK2 compared to LRRK2 WT and other mutants as determined by immunohistochemistry and Western blotting (Fig 1). Therefore, we conclude that K1347A acts as a stable LRRK2-inactive variant, at least when expressed in C. elegans, and that our data are consistent with the hypothesis that GTP-binding or kinase activity is required for LRRK2-induced pathological phenotype. While R1441C/G and G2019S are more prevalent mutations and several studies support their role in the overall enhancement of kinase activity of LRRK2, there are other PD-linked mutations such as Y1699C and I2020T that do not show an consistent kinase-activating effect in vitro raising the possibility that these mutations may act differently (Greggio and Cookson, 2009). We envision that the scientific approach used in our present C. elegans models may serve as a platform to study the effect and pathogenic mechanisms of these mutations in vivo. Finally, our results have revealed that loss of endogenous C. elegans LRK-1 protects against the LRRK2-induced phenotype as well as neurotoxicity induced by CAT-2 overexpression in DA neurons. Because LRK-1 is expressed broadly in both neuronal and non-neuronal cells, it remains to be established whether LRK-1 has a selective role in the survival of DA neurons. Nevertheless, it is surprising that neurodegeneration (Fig. 2) and behavioral deficits (Fig. 6) induced by overexpression of LRRK2 WT or G2019S in DA neurons can be completely rescued by the LRK-1 null alleles that cause deletion of either all functional domains (km41) or the C-terminal region containing kinase domain (km17). These findings suggest that either LRK-1 contributes to neurotoxicity or that endogenous LRK-1 is required for transducing the noxious signals from hyperactive LRRK2 to cause neurodegeneration. Since LRRK2 is capable of forming both a homodimer and heterodimer with LRRK1 and dimerization is required for LRRK2 kinase activity (Greggio et al., 2008; Klein et al., 2009; Sen et al., 2009), it is conceivable that LRRK2/LRK-1 may together regulate neuronal signaling and survival in C. elegans expressing LRRK2 in the LRK-1 WT background. Further characterization of the role of the endogenous LRRK2 homolog such as C. elegans LRK-1 and mammalian LRRK1 in neuronal function will be required. Unlike humans with two structurally similar proteins LRRK1 and LRRK2, invertebrate animals such as Drosophila and C. elegans have only a single homolog of LRRK2 and multiple null-alleles are available for loss-of-function studies.
Only a few in vivo models of LRRK2 overexpression have been reported to date. It has been shown that pan-neuronal overexpression of LRRK2 or deletion of endogenous LRK-1, regulate mitochondrial function, stress response and neurite outgrowth in C. elegans (Saha et al., 2009; Sakaguchi-Nakashima et al., 2007; Samann et al., 2009). Overexpression of LRRK2 WT and G2019S in Drosophila have also been reported to induce neurodegeneration (Liu et al., 2008). Unfortunately, the mutant LRRK2-induced phenotype recently reported in transgenic mouse models is much more subtle. Transgenic mice carrying the R1441C mutation within the mouse genome are normal in gross behavior and showed no neurodegeneration, even though their evoked dopamine neurotransmission is compromised (Tong et al., 2009). Humanized transgenic mice expressing the R1441G mutation have been reported to cause some axonal abnormality without DA neuronal loss (Li et al., 2009). It is possible that these mild abnormalities manifested by R1441C/G mutation in mouse models represent pathogenic events preceding dopaminergic degeneration. However, these initial studies have neither systematically compared the pathogenic roles between LRRK2 WT and different PD-linked mutations (e.g. R1441C and G2019S), nor have they provided adequate insights into the critical role of GTPase/kinase activities in LRRK2-induced neurodegeneration and behavioral deficits. Hence, our LRRK2 C. elegans transgenic lines as described in this study will be particularly useful for exploring the mechanisms underlying LRRK2-mediated pathogenesis. In addition, as invertebrate models of human diseases have been increasingly utilized for early stage drug development (Artal-Sanz et al., 2006), these transgenic C. elegans models of LRRK2-linked PD may serve as versatile tools to aid rapid discovery and development of LRRK2-based therapeutics for PD prior to preclinical studies in mammalian models.
Supplementary Material
Supplemental Fig. 1. Loss of endogenous LRK-1 protects against neurodegenerative stress induced by CAT-2 overexpression in DA neurons. (A) C. elegans overexpressing CAT-2 and GFP marker in DA neurons [(Pdat-1::CAT-2; Pdat-1::GFP), designated as CAT-2+] or the cross progeny between CAT-2+ and lrk-1(km41) [designated as CAT-2+; lrk-1(km41)] were monitored for morphology of CEP neurons using bright field (BF) or fluorescence (GFP) microscopy. CAT-2+ worms displayed age-dependent neurodegeneration as indicated by missing or degenerating CEP neurons on adult day 4 (arrow) but not on adult day 0. In contrast, the CAT-2+; lrk-1(km41) worms showed no apparent neurodegeneration on adult day 4. Synchronized worms grown to adult day 0 and 4 were mounted on agar-containing slides for live imaging. (B) Basal slowing response was examined on adult day 0, 2, and 4 in the CAT-2+ and CAT-2+; lrk-1(km41) worms. The CAT-2+ worms manifested age-dependent progressive loss of basal slowing response on day 2 and 4 as compared to day 0. This behavioral deficit was rescued in the CAT-2+; lrk-1(km41) worms. Synchronized worms were grown to adult day 0, 2 and 4 on NGM plates, and were assayed for body bending in the absence (−Food) or presence (+Food) of bacterial food. The native strain N2 and mutant cat-2(e1112) worms defective in dopamine synthesis served as controls. About 10 worms were used for behavioral assay in each strain. Error bars indicate SEM. *p<0.01 versus no food.
Acknowledgements
This work was supported in part by grants from National Institutes of Health, National Parkinson Foundation, American Parkinson Disease Association, Michael J. Fox Foundation for Parkinson’s Research, Johnson & Johnson Corporate Office of Science and Technology, and the Department of Veterans Affairs. We thank O. Kourennaia for technical assistance, S. Mitani (Tokyo Women’s Hospital, Japan) for providing the pFxneEGFP vector, L. Avery (University of Texas Southwestern Medical Center) for the pJM23 (lin-15+) plasmid, Z. Feng (Case Western Reserve University) for setting up and use of the worm behavioral tracking system, N. Hisamoto (Nagoya University, Japan) for the lrk-1(km 17) and lrk-1(km 41) strains, and J. Duerr (Ohio University) for advice on worm immunohistochemistry. Some C. elegans strains were obtained from the Caenorhabditis Genetics Center, which is supported by National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artal-Sanz M, de Jong L, Tavernarakis N. Caenorhabditis elegans: a versatile platform for drug discovery. Biotechnol J. 2006;1:1405–1418. doi: 10.1002/biot.200600176. [DOI] [PubMed] [Google Scholar]
- Berkowitz LA, Hamamichi S, Knight AL, Harrington AJ, Caldwell GA, Caldwell KA. Application of a C. elegans dopamine neuron degeneration assay for the validation of potential Parkinson’s disease genes. J Vis Exp. 2008 doi: 10.3791/835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice A. Genetics of Parkinson’s disease: LRRK2 on the rise. Brain. 2005;128:2760–2762. doi: 10.1093/brain/awh676. [DOI] [PubMed] [Google Scholar]
- Cao S, Gelwix CC, Caldwell KA, Caldwell GA. Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J. Neurosci. 2005;25:3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR, Dauer W, Dawson T, Fon EA, Guo M, Shen J. The roles of kinases in familial Parkinson’s disease. J. Neurosci. 2007;27:11865–11868. doi: 10.1523/JNEUROSCI.3695-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer M, Stone J, Mata IF, Lincoln S, Kachergus J, Hulihan M, Strain KJ, Maraganore DM. LRRK2 mutations in Parkinson disease. Neurology. 2005 doi: 10.1212/01.wnl.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- Farrer MJ. Lrrk2 in the limelight! Neurology. 2007;69:1732–1733. doi: 10.1212/01.wnl.0000279588.20391.88. [DOI] [PubMed] [Google Scholar]
- Feng Z, Cronin CJ, Wittig JH, Jr., Sternberg PW, Schafer WR. An imaging system for standardized quantitative analysis of C. elegans behavior. BMC Bioinformatics. 2004;5:115. doi: 10.1186/1471-2105-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi PN, Chen SG, Wilson-Delfosse AL. Leucine-rich repeat kinase 2 (LRRK2): a key player in the pathogenesis of Parkinson’s disease. J. Neurosci. Res. 2009;87:1283–1295. doi: 10.1002/jnr.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O’Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro. 2009:1. doi: 10.1042/AN20090007. art:e00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, Thomas KJ, Baekelandt V, Cookson MR. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp. Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Wang W, Chen SG. Leucine-rich repeat kinase 2: Relevance to Parkinson’s disease. Int. J. Biochem. Cell Biol. 2006;38:1469–1475. doi: 10.1016/j.biocel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Hope IA. C. elegans: a practical approach. Oxford University Press; New York: 1999. [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, Iwatsubo T. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson’s disease. Biochemistry (Mosc) 2007;46:1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- Klein CL, Rovelli G, Springer W, Schall C, Gasser T, Kahle PJ. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J. Neurochem. 2009;111:703–715. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- Kuwahara T, Koyama A, Gengyo-Ando K, Masuda M, Kowa H, Tsunoda M, Mitani S, Iwatsubo T. Familial Parkinson mutant alpha-synuclein causes dopamine neuron dysfunction in transgenic Caenorhabditis elegans. J. Biol. Chem. 2006;281:334–340. doi: 10.1074/jbc.M504860200. [DOI] [PubMed] [Google Scholar]
- Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 2007;357:668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. J. Neurochem. 2007;103:238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu W, Oo TF, Wang L, Tang Y, Jackson-Lewis V, Zhou C, Geghman K, Bogdanov M, Przedborski S, Beal MF, Burke RE, Li C. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson’s disease. Nat. Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, Ren Q, Jiao Y, Sawa A, Moran T, Ross CA, Montell C, Smith WW. A Drosophila model for LRRK2-linked parkinsonism. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2693–2698. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Nass R, Hall DH, Miller DM, 3rd, Blakely RD. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3264–3269. doi: 10.1073/pnas.042497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WC, Elsaesser VE, Pankratz N, Pauciulo MW, Marek DK, Halter CA, Rudolph A, Shults CW, Foroud T. LRRK2 mutation analysis in Parkinson disease families with evidence of linkage to PARK8. Neurology. 2007;69:1737–1744. doi: 10.1212/01.wnl.0000278115.50741.4e. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, de Munain A. Lopez, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Pehek EA, Nocjar C, Roth BL, Byrd TA, Mabrouk OS. Evidence for the preferential involvement of 5-HT2A serotonin receptors in stress- and drug-induced dopamine release in the rat medial prefrontal cortex. Neuropsychopharmacology. 2006;31:265–277. doi: 10.1038/sj.npp.1300819. [DOI] [PubMed] [Google Scholar]
- Saha S, Guillily MD, Ferree A, Lanceta J, Chan D, Ghosh J, Hsu CH, Segal L, Raghavan K, Matsumoto K, Hisamoto N, Kuwahara T, Iwatsubo T, Moore L, Goldstein L, Cookson M, Wolozin B. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J. Neurosci. 2009;29:9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi-Nakashima A, Meir JY, Jin Y, Matsumoto K, Hisamoto N. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Curr. Biol. 2007;17:592–598. doi: 10.1016/j.cub.2007.01.074. [DOI] [PubMed] [Google Scholar]
- Samann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J. Biol. Chem. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Sen S, Webber PJ, West AB. Leucine-Rich Repeat Kinase 2 (LRRK2) Kinase Activity: Dependence on Dimerization. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.025437. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Tong Y, Pisani A, Martella G, Karouani M, Yamaguchi H, Pothos EN, Shen J. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Zabetian CP, Samii A, Mosley AD, Roberts JW, Leis BC, Yearout D, Raskind WH, Griffith A. A clinic-based study of the LRRK2 gene in Parkinson disease yields new mutations. Neurology. 2005 doi: 10.1212/01.wnl.0000172630.22804.73. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Loss of endogenous LRK-1 protects against neurodegenerative stress induced by CAT-2 overexpression in DA neurons. (A) C. elegans overexpressing CAT-2 and GFP marker in DA neurons [(Pdat-1::CAT-2; Pdat-1::GFP), designated as CAT-2+] or the cross progeny between CAT-2+ and lrk-1(km41) [designated as CAT-2+; lrk-1(km41)] were monitored for morphology of CEP neurons using bright field (BF) or fluorescence (GFP) microscopy. CAT-2+ worms displayed age-dependent neurodegeneration as indicated by missing or degenerating CEP neurons on adult day 4 (arrow) but not on adult day 0. In contrast, the CAT-2+; lrk-1(km41) worms showed no apparent neurodegeneration on adult day 4. Synchronized worms grown to adult day 0 and 4 were mounted on agar-containing slides for live imaging. (B) Basal slowing response was examined on adult day 0, 2, and 4 in the CAT-2+ and CAT-2+; lrk-1(km41) worms. The CAT-2+ worms manifested age-dependent progressive loss of basal slowing response on day 2 and 4 as compared to day 0. This behavioral deficit was rescued in the CAT-2+; lrk-1(km41) worms. Synchronized worms were grown to adult day 0, 2 and 4 on NGM plates, and were assayed for body bending in the absence (−Food) or presence (+Food) of bacterial food. The native strain N2 and mutant cat-2(e1112) worms defective in dopamine synthesis served as controls. About 10 worms were used for behavioral assay in each strain. Error bars indicate SEM. *p<0.01 versus no food.