Abstract
The proline-rich γ-carboxyglutamic acid (Gla) proteins (PRGPs) 1 and 2 are the founding members of a family of vitamin K-dependent single-pass integral membrane proteins characterized by an extracellular amino terminal domain of approximately 45 amino acids that is rich in Gla. The intracellular carboxyl terminal region of these two proteins contains one or two copies of the sequence PPXY, a motif present in a variety of proteins involved in such diverse cellular functions as signal transduction, cell cycle progression, and protein turnover. In this report, we describe the cloning of the cDNAs for two additional human transmembrane Gla proteins (TMG) of 20–24 kDa named TMG3 and TMG4. These two proteins possess extracellular Gla domains with 13 or 9 potential Gla residues, respectively, followed by membrane-spanning hydrophobic regions and cytoplasmic carboxyl terminal regions that contain PPXY motifs. This emerging family of integral membrane Gla proteins includes proline-rich Gla protein (PRGP) 1, PRGP2, TMG3, and TMG4, all of which are characterized by broad and variable distribution in both fetal and adult tissues. Members of this family can be grouped into two subclasses on the basis of their gene organization and amino acid sequence. These observations suggest novel physiological functions for vitamin K beyond its known role in the biosynthesis of proteins involved in blood coagulation and bone development. The identification and characterization of these proteins may allow a more complete understanding of the teratogenic consequences of exposure in utero to vitamin K antagonists, such as warfarin-based anticoagulants.
Vitamin K is an essential cofactor in the posttranslational conversion of glutamate residues to γ-carboxyglutamate (Gla) residues during the biosynthesis of vitamin K-dependent proteins (1, 2). This reaction is catalyzed by γ-glutamyl carboxylase (3) and takes place within the lumen of the endoplasmic reticulum (4). Carboxylation of glutamate residues within the amino terminal regions of the vitamin K-dependent proteins is directed by a propeptide of approximately 20 amino acids first observed in factor IX (5). The propeptide contains the carboxylase recognition site (6, 7), which is subsequently cleaved to yield the mature protein. In vertebrates, proteins modified by γ-glutamyl carboxylation can be divided into three groups. The first group is characterized by the presence of an amino terminal Gla domain, a protein module of approximately 45 amino acid residues containing 9–12 Gla residues. The Gla domains of these proteins mediate calcium-dependent association with anionic phospholipid-containing membranes, notably at sites of vascular injury and on the surface of activated platelets (8–10). Members of this class of Gla proteins include the vitamin K-dependent blood coagulation factors (prothrombin, factor VII, factor IX, and factor X) and coregulators of blood coagulation (protein C, protein S, and protein Z). A more recent addition to this group is Gas6, the ligand for the receptor tyrosine kinases Axl and Sky (11, 12). Gas6 has been characterized as a cell survival factor (13), a growth factor (14), and a cofactor in the uptake of apoptotic cells by macrophages (15). A second group of Gla-containing proteins includes osteocalcin and matrix Gla protein (MGP), which contain three or five Gla residues, respectively (16, 17). These proteins are required for regulation of bone growth and extraosseous calcification (18, 19). Lastly, the presence of Gla residues within the γ-glutamyl carboxylase itself has recently been reported (20).
The identification and functional characterization of Gas6 underscores the possibility that vitamin K plays a much broader physiological role than previously understood. Indeed, several lines of evidence support the hypothesis that vitamin K is involved in the development of the central nervous system (CNS). A dietary deficiency of vitamin K or administration of warfarin, a vitamin K antagonist, to young mice has been shown to decrease the biosynthesis of sulfatide, an important component of myelin, and this phenomenon was corrected by administration of vitamin K (21, 22).
Additional support for the hypothesis that vitamin K is required for CNS development is provided by clinical studies of the fetopathic consequences of warfarin exposure in utero. Warfarin embryopathy presents clinically as a constellation of developmental abnormalities, including bone malformation, stippled epiphyses, and nasal hypoplasia (23). CNS malformations are observed less consistently and appear to depend on the dosage, timing, and duration of maternal warfarin prophylaxis. These malformations include ventriculomegaly, hydrocephaly, agenesis of the corpus callosum, optic atrophy, congenital cataracts, and mild to severe mental retardation (24, 25).
The mechanism(s) underlying the ontogeny of CNS malformation associated with warfarin embryopathy remains unclear. One possible hypothesis is that CNS lesions are secondary to a vascular incident during late gestation arising from undercarboxylation of vitamin K-dependent coagulation factors (24). However, this model is contradicted by a case in which the CNS sequellae of warfarin embryopathy were observed after maternal warfarin administration between gestational weeks 8 and 12, a period that precedes the onset of fetal coagulation factor expression (26). Further evidence that vitamin K plays a specific physiological role in the brain was provided by experiments in which administration of vitamin K was shown to up-regulate Src kinase activity and increase phosphorylation of the receptor tyrosine kinase Eyk, focal adhesion kinase, and paxillin, in the chick brain during embryogenesis (27). This effect was abrogated by warfarin.
It remains unclear whether these observations can be ascribed to an undiscovered function of vitamin K itself, or to the inadequate carboxylation of as-yet unidentified protein substrates. γ-Glutamyl carboxylase activity and uncharacterized substrates have been identified in a wide variety of tissues (28). Carboxylase expression has been shown to be developmentally regulated in rat embryogenesis, with prominent expression observed in neuroepithelia at a developmental stage preceding the onset of expression in the hepatic primordium (29).
The functional characterization of Gas6 represented the first exception to the notion that vitamin K is required solely for blood coagulation and the regulation of bone growth and soft tissue calcification. The subsequent identification of the integral membrane Gla proteins proline-rich Gla protein (PRGP) 1 and 2 and demonstration of their broad tissue distribution (30) further support this view. The influence of vitamin K and its antagonists on sulfatide metabolism and tyrosine phosphorylation, the demonstration of extrahepatic γ-glutamyl carboxylase expression during fetal development, and the embryopathology associated with in utero warfarin exposure further suggest that our understanding of vitamin K function is incomplete.
In this manuscript, we report the identification of two cDNAs encoding Gla-domain-containing proteins that are single-pass transmembrane proteins. These two proteins have been named transmembrane Gla proteins 3 and 4 (TMG3 and TMG4). Analysis of their deduced amino acid sequences reveals that, like the two PRGPs, they contain copies of the sequence PPXY, a motif known to mediate interactions with WW domain-containing proteins in a variety of cellular processes, including protein turnover, cell-cycle progression, and signal transduction (31). Members of this family of Gla proteins thus possess hallmarks of cell-surface receptors, namely, a transmembrane topology and potential cytoplasmic signaling motifs. In addition, the widespread expression of these proteins in the fetus raises the possibility that impairment of their function(s) by inadequate γ-glutamyl carboxylation may be partly responsible for the teratogenic effects of warfarin exposure in utero.
Materials and Methods
Isolation of the TMG3 cDNA.
An expressed sequence tag (EST) derived from a human ovarian carcinoma (GenBank no. AI266282) and predicted to encode a Gla domain, was identified in the EST database (dbEST) maintained by the National Center for Biotechnology Information (NCBI) by using the tblastn search algorithm (32). The query sequence was derived from a potential Gla domain-encoding sequence previously identified in an alternate reading frame of the 5′ end of the rabbit aortic cyclic nucleotide gated channel cDNA (ref. 33; GenBank no. X59668). Oligonucleotide primers based on this EST sequence were used to isolate the 5′ and 3′ ends of the TMG3 cDNA from a human adult spinal cord cDNA library by the PCR by using rapid amplification of cDNA ends (RACE; ref. 34).
Isolation of the TMG4 cDNA.
The amino acid sequence of the PRGP1 Gla domain was used to query the completed human genome sequence database at the NCBI using tblastn. This search identified a Gla domain-encoding sequence in a contiguous sequence (GenBank no. AL035400) derived from chromosome 11, band p13. Query of dbEST with this genomic sequence identified an EST from a human choriocarcinoma library that encoded a novel Gla domain-containing protein (GenBank no. BE732028). Oligonucleotide primers based on this EST sequence were used to isolate the 3′ end of the TMG4 cDNA from a human T47D breast carcinoma cell line cDNA library and the 5′ end of the TMG4 cDNA from a human MCF7 breast cancer cell line cDNA library. Full-length cDNAs were subsequently isolated from both T47D and MCF7 cDNA libraries.
Determination and Analysis of cDNA Sequences.
Sequencing reactions were performed by using a BigDye Primer Cycle Sequencing Kit (Applied Biosystems) and were analyzed on an Applied Biosystems PRISM 377 DNA Sequencer. In all cases, sequencing reactions were performed directly on PCR products, as well as on a minimum of five independent plasmid clones to ensure the absence of PCR-generated mutations. The cDNA sequences of PRGP1 and -2, and TMG3 and -4 were used to query the Human Genome Sequence Datatase and the unfinished High-throughput Genomic Sequence Database at NCBI by using the blastn program. These alignments allowed determination of the positions of intron/exon boundaries within these genes. The genomic sequences, identified by GenBank accession number were as follows: PRGP1, AL356858; PRGP2, AC011495; TMG3, AF274854; and TMG4, Hs11 2049.
Determination of Fetal and Adult Tissue Distribution.
PCR was performed on normalized first-strand cDNA preparations from fetal and adult human tissues (CLONTECH). Fetal cDNA was prepared from pooled male and female fetuses at gestational weeks 16 to 36. All PCRs were performed by using a standard thermal cycling program: 94°C, 1 min (1 cycle); 94°C, 30 sec, 68°C, 1.5 min (N cycles); and 68°C, 5 min (1 cycle). The number of annealing/extension cycles (N) for each set of reactions was as follows: PRGP1, 40; PRGP2, 40; TMG3, 40; TMG4, 36; Gas6, 40; prothrombin, 35; MGP, 32; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 25. N was adjusted for each set of reactions such that the most intense amplimer band of the set was of subsaturating intensity. PCR primers were designed to anneal to cDNAs at positions corresponding to different exons to prevent false positive signals arising from contamination of template cDNAs with genomic DNA. Because the genomic sequence of Gas6 has not yet been published, exon boundaries were predicted based on those of protein S, a structurally similar protein. The following PCR primers were used: PRGP1, 5′-TTCCTCACGGGAGAAAAAGCCAATTCC and 5′-CCAGTCACTTCCTCGGTTACTCTC; PRGP2, 5′-CTGGCAGCCTCTGGGGTACACGAC and 5′-CCAGGACACGGGAACACGTTGCCG; TMG3, 5′-GAGGCCAAGGATGCCCATTCGGTC and 5′-CACATACATGGCATCTGAGCTCTG; TMG4, 5′-GGATTACTTGGCTACTATCTTTGTATC and 5′-CTGTTCATAAGAAGGTAATCCTGCATC; Gas6, 5′-GCCCCCGACCTCCGTGCCGTGCCTCTC and 5′-AGCCTCCGGTTGACCTCCAGTGTCATG; prothrombin, 5′-GGGCGGGTGACAGGCTGGGGCAACCTG and 5′-CAGGCGGAACACATGTGTGTAGAAGCC; MGP, 5′-CCCCTCAGCAGAGATGGAGAGCTAAAG and 5′-ATGTTGACTCTCCTTTGACCCTCACTG. GAPDH primers were supplied by the manufacturer.
Results and Discussion
TMG3.
In a previous investigation (30), we reported that the 5′ end of a cDNA sequence for the rabbit aortic cyclic nucleotide gated channel (GenBank no. X59668) (33) appeared to encode a Gla domain with the characteristic pattern of glutamate and cysteine residues, but this sequence was present in a different reading frame than that of the channel protein. By using the predicted amino acid sequence of this apparently out-of-frame Gla domain, we identified an EST (GenBank no. AI266282) from a human ovarian carcinoma cDNA library that appeared to be the human ortholog of the rabbit Gla protein. PCR primers derived from this EST sequence were used to amplify the TMG3 cDNA from an adult human spinal cord cDNA library by RACE. The resulting cDNA (GenBank no. AF326350) was 1237 bp in length and consisted of a 33-bp 5′ untranslated region (UTR), a 696-bp DNA sequence coding for 231 amino acids, and a 508-bp 3′ UTR. With the exception of the Gla domain-encoding region, this cDNA bore no resemblance to that of the rabbit cyclic nucleotide gated channel protein. A polyadenylation signal was not present at the 3′ end of this cDNA, suggesting that the cDNA was not full-length. Comparison of the cDNA sequence to human genomic DNA (GenBank no. AF274854) revealed an adenine-rich sequence in the genomic DNA adjacent to the region that corresponded to the 3′ end of the cDNA that was isolated. Therefore, it appeared that our cDNA sequence was prematurely truncated within the 3′ UTR because of mispriming of the oligo(dT) primer used in the construction of the cDNA library. This genomic sequence was derived from the X chromosome, band p28.
TMG4.
A query of the completed human genome sequence database with the amino acid sequence of the PRGP1 Gla domain identified another Gla domain-encoding sequence within a genomic clone (GenBank no. AL035400) that mapped to chromosome 11, band p13. A subsequent search of the EST database dbEST with this genomic sequence identified an EST from a human choriocarcinoma library (GenBank no. BE732028). Full-length TMG4 cDNAs were then isolated by RACE from a human T47D breast carcinoma library and from an MCF7 breast cancer cell line library. The longest cDNA (GenBank no. AF326351) was 1108 bp in length and consisted of a 109-bp 5′ UTR, a 681-bp DNA sequence coding for 227 amino acids, and a 318-bp 3′ UTR. Two tandem polyadenylation sequences (AATAAA) were present in the sequence beginning 15 bp from the 3′ end.
Sequence Analysis of TMG3.
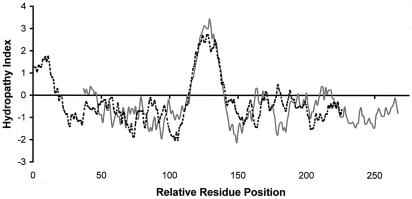
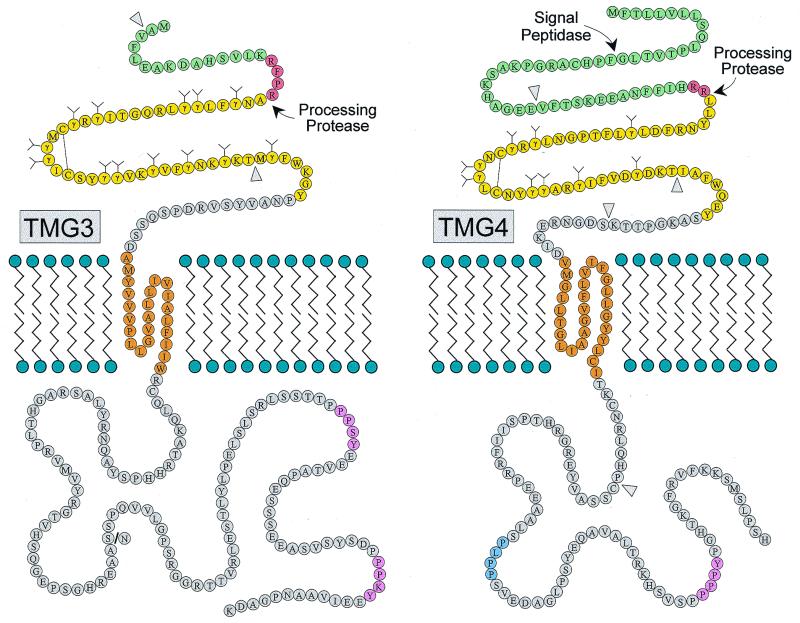
The protein encoded by the TMG3 cDNA has a predicted molecular mass of 25.8 kDa and lacked a signal peptide, a characteristic shared with PRGP1. Sequence alignment with other Gla proteins suggested that TMG3 possessed a Gla domain with up to 13 Gla residues preceded by a conserved propeptide implicated in the recognition by γ-glutamyl carboxylase, with a −1, −4 dibasic motif at the predicted propeptide cleavage site (Fig. 1). The predicted mature protein consisted of 212 amino acid residues with a molecular mass of 23.7 kDa after cleavage of the propeptide. As with other members of this protein family, hydropathy analysis indicated that TMG3 was a single-pass transmembrane protein (Fig. 2). This hydrophobic region (residues 60 to 82) was flanked by acidic residues (Asp-53 and Asp-59) on the N-terminal side and by basic residues (Arg-83, Lys-88, and Arg-91) on the C-terminal side, suggesting that TMG3 was oriented in the membrane such that the C-terminal 129 amino acids were exposed to the cytoplasm (ref. 35; Fig. 3 Left). Such an orientation would allow exposure of the N-terminal domain of TMG3 to the γ-glutamyl carboxylase during the posttranslational carboxylation reaction occurring within the lumen of the endoplasmic reticulum (4).
Figure 1.
Amino acid sequence alignment of the signal/propeptides and Gla domains of Gla proteins. Highly conserved residues are shaded. Residues within the propeptide implicated in recognition by the γ-glutamyl carboxylase are denoted with an asterisk. Positions at which γ-carboxylation of glutamic acid residues is either known to occur or may occur are indicated by γ. The propeptidase cleavage site is indicated by ↓. The position of the disulfide loop within the Gla domain is also indicated.
Figure 2.
Hydropathy analysis of TMG3 (solid gray line) and TMG4 (dashed black line) by using a window size of 7 residues.
Figure 3.
Predicted topology and structures of TMG3 (Left) and TMG4 (Right). Notable features include the signal/propeptide (green), predicted propeptidase recognition site (red), Gla domain (yellow), transmembrane region (orange), potential SH3 domain-binding motif PXXP (blue), and potential WW domain-binding motifs PPXY (pink). The predicted propeptidase cleavage sites and the predicted signal peptidase cleavage site of TMG4 are indicated with arrows. The positions corresponding to exon boundaries are indicated by wedges (▿). A potential polymorphic site within the cytoplasmic domain of TMG3 is indicated with a slash (Left).
The putative cytoplasmic domain of TMG3 bears two PPXY motifs, a common feature of all four transmembrane Gla proteins from this family. This sequence constitutes the minimal binding motif of WW domains of approximately 40 amino acid residues. WW domains are present in a variety of proteins involved in cytoskeletal interactions, protein turnover, signal transduction, and cell-cycle progression (31). Like PRGP1, TMG3 has two PPXY motifs. At present, little is known about how the sequences flanking PPXY motifs confer specificity for particular WW domains. However, comparison of the extended PPXY motifs of TMG3 and PRGP1 revealed a consensus sequence D/T-P-P-P-X-Y-E-E/D-Ψ (where X is variable and Ψ is hydrophobic). These adjacent sequences may also be important in determining the specificity of WW domain interactions.
Comparison of the TMG3 cDNA to genomic DNA revealed a single base difference within the region encoding the cytoplasmic domain of the protein. Base number 491 in the cDNA sequence was guanine, whereas the corresponding position in the genomic DNA was adenine. This corresponds to a substitution of asparagine (AAC) for serine (AGC) at residue number 134 in the mature protein (Fig. 3 Left). Several independent cloning experiments failed to produce a cDNA clone with an A at this position. Thus, it remains unclear whether this difference was the result of a mutation or a sequencing mistake in the genomic sequence, or whether it represented a naturally occurring dimorphism.
Sequence Analysis of TMG4.
The protein encoded by the TMG4 cDNA contains nine potential Gla residues and has a predicted molecular mass of 25.4 kDa. Like PRGP2, TMG4 has an N-terminal hydrophobic sequence predicted to be a signal peptide by the method of Nielsen et al. (ref. 36; Fig. 2; Fig. 3 Right). The signal peptide is followed by a propeptide that differs from the typical propeptides of the other vitamin K-dependent proteins in that it has an isoleucine at the −4 position and lacks the canonical −1, −4 dibasic motif at the presumed site of propeptide cleavage (Fig. 1). The predicted mature protein consists of 177 amino acid residues with a molecular mass of 19.9 kDa after cleavage of the signal peptide and propeptide.
As with the other three transmembrane Gla proteins, the putative cytoplasmic domain of TMG4 possessed a PPXY motif. In this regard, TMG4 most closely resembles PRGP2. Both proteins possess a single PPXY motif within a consensus sequence P-P-P-P-Y, a sequence that was originally identified in proteins interacting with the WW domain-containing protein yes kinase-associated Protein (37). The extended motif within one of these proteins, WBP-2, exactly matches the sequence P-P-P-P-Y-P-G in TMG4, suggesting that TMG4 may interact with yes kinase-associated protein. TMG4 also contains a single copy of the sequence PXXP, the minimal binding motif for proteins bearing Src homology (SH) 3 domains (38).
Organization of the TMG Genes and Their Chromosomal Localization.
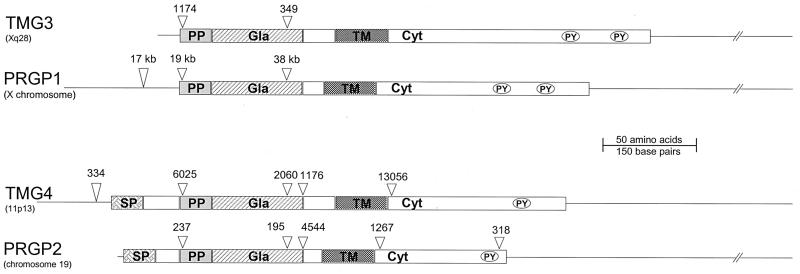
Human genomic sequences corresponding to all four transmembrane Gla proteins were used to assign chromosome localization and determine gene organization. The chromosome assignments were as follows: PRGP1, X chromosome; PRGP2, chromosome 19; TMG3, Xq28; and TMG4, 11p13. Comparison of the intron/exon boundaries (Fig. 4) revealed similarities between the gene organization for PRGP1 and TMG3 and between PRGP2 and TMG4. The C-terminal portion of the Gla domain, transmembrane domain, cytoplasmic domain, and 3′ UTR of PRGP1 and TMG3 were all encoded by a single exon (Fig. 3 Left). In PRGP2 and TMG4, the Gla domain was encoded by two exons, the transmembrane domain by one exon, and either most (PRGP2) or all (TMG4) of the cytoplasmic domain by one exon. Although the sizes of the protein themselves were approximately equal, the sizes of their corresponding genes varied widely, from approximately 2.8 kb (TMG3) to greater than 78 kb (PRGP1). The genes encoding PRGP2 and TMG4 were of intermediate size (7.7 kb and 24 kb, respectively).
Figure 4.
Organization of the transmembrane Gla protein genes. The positions of intron/exon boundaries are indicated by wedges (▿). Numbers indicate the length (in bp) of intervening introns. The lengths of the introns in the PRGP1 gene (GenBank no. AL356858) are approximate because unresolved gaps of indeterminate length are present in the genomic sequence. Structural features are as follows: SP, signal peptide; PP, propeptide; Gla, Gla domain; TM, transmembrane region; Cyt, cytoplasmic domain; PY, PPXY motif. Chromosome assignments are in parentheses.
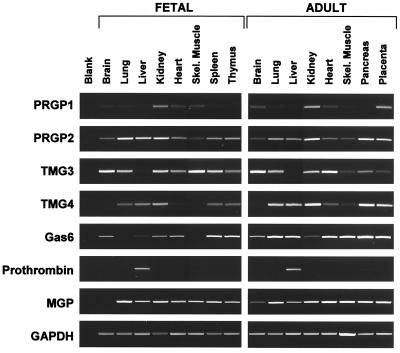
Fetal and Adult Tissue Distribution.
PCRs were performed on panels of first-strand cDNA preparations from a variety of fetal and adult human tissues that had been normalized relative to the abundance of several housekeeping genes (Fig. 5). For the PRGPs and TMGs, the fetal and adult expression pattern appeared to be similar for a given protein, with a few notable exceptions. First, the expression of TMG3 in skeletal muscle was markedly decreased in the adult relative to the fetus, but this pattern is reversed for fetal and adult heart. Second, whereas the overall pattern of TMG4 expression was similar in the fetus and in the adult, there was a significant increase in adult tissues relative to their fetal counterparts. This appeared to be the case for MGP as well. This pattern contrasted with that of Gas6, which showed little or no correlation between fetal and adult expression profiles.
Figure 5.
Tissue distribution of mRNAs for various Gla proteins in fetal and adult tissues. PCR was performed on first-strand cDNAs under nonsaturating conditions as described in Materials and Methods.
It is tempting to speculate that inadequate carboxylation of the Gla proteins, shown here to be expressed in the fetus, is partly responsible for the teratogenesis associated with warfarin exposure in utero. For example, the insufficient carboxylation of Gas6, which has been previously shown to be expressed in the fetus (39), may underlie at least part of the fetopathology resulting from maternal warfarin prophylaxis. However, a definitive answer will have to await the results of other experiments, such as the targeted disruption of the Gas6 gene in mice. Other likely candidates include PRGP1 and -2, and TMG3 and -4, but such speculation will require a more substantive understanding of the functions of these proteins.
Acknowledgments
We thank Dominic Chung for his helpful advice in the preparation of this manuscript, and Betty Haldeman and Don Foster for their generous gift of a collection of human RACE-ready cDNA libraries. This work was supported, in part, by Research Grant HL-16919 from the National Institutes of Health. J.D.K. was supported by a National Science Foundation Graduate Fellowship.
Abbreviations
- CNS
central nervous system
- EST
expressed sequence tag
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- Gla
γ-carboxyglutamic acid
- MGP
matrix Gla protein
- PRGP
proline-rich Gla protein
- RACE
rapid amplification of cDNA ends
- TMG
transmembrane Gla protein
- UTR
untranslated region
- YAP
yes kinase-associated protein
Footnotes
References
- 1.Stenflo J. J Biol Chem. 1974;249:5527–5535. [PubMed] [Google Scholar]
- 2.Nelsestuen G L, Zytkovicz T H, Howard J B. J Biol Chem. 1974;249:6347–6350. [PubMed] [Google Scholar]
- 3.Wu S M, Cheung W F, Frazier D, Stafford D W. Science. 1991;254:1634–1636. doi: 10.1126/science.1749935. [DOI] [PubMed] [Google Scholar]
- 4.Bristol J A, Ratcliffe J V, Roth D A, Jacobs M A, Furie B C, Furie B. Blood. 1996;88:2585–2593. [PubMed] [Google Scholar]
- 5.Kurachi K, Davie E W. Proc Natl Acad Sci USA. 1982;79:6461–6464. doi: 10.1073/pnas.79.21.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgensen M J, Cantor A B, Furie B C, Brown C L, Shoemaker C B, Furie B. Cell. 1987;48:185–191. doi: 10.1016/0092-8674(87)90422-3. [DOI] [PubMed] [Google Scholar]
- 7.Foster D C, Rudinski M S, Schach B G, Berkner K L, Kumar A A, Hagen F S, Sprecher C A, Insley M Y, Davie E W. Biochemistry. 1987;26:7003–7011. doi: 10.1021/bi00396a022. [DOI] [PubMed] [Google Scholar]
- 8.Furie B, Furie B C. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 9.Mann K G, Nesheim M E, Church W R, Haley P, Krishnaswamy S. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 10.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 11.Varnum B C, Young C, Elliot G, Garcia A, Bartley T D, Fridell Y W, Hunt R W, Trail G, Clogston C, Toso R J, Yanagihara D, Bennett L, Sylber M, Merewether L A, Tseng A, Escobar E, Liu E T, Yamane H K. Nature (London) 1995;373:623–626. doi: 10.1038/373623a0. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi K, Nagata K, Toshima J, Nakano T, Arita H, Tsuda H, Suzuki K, Mizuno K. J Biol Chem. 1995;270:22681–22684. doi: 10.1074/jbc.270.39.22681. [DOI] [PubMed] [Google Scholar]
- 13.Nakano T, Kawamoto K, Higashino K, Arita H. FEBS Lett. 1996;387:78–80. doi: 10.1016/0014-5793(96)00395-x. [DOI] [PubMed] [Google Scholar]
- 14.Nakano T, Higashino K, Kikuchi N, Kishino J, Nomura K, Fujita H, Ohara O, Arita H. J Biol Chem. 1995;270:5702–5705. doi: 10.1074/jbc.270.11.5702. [DOI] [PubMed] [Google Scholar]
- 15.Ishimoto Y, Ohashi K, Mizuno K, Nakano T. J Biochem (Tokyo) 2000;127:411–417. doi: 10.1093/oxfordjournals.jbchem.a022622. [DOI] [PubMed] [Google Scholar]
- 16.Price P A, Poser J W, Raman N. Proc Natl Acad Sci USA. 1976;73:3374–3375. doi: 10.1073/pnas.73.10.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price P A, Urist M R, Otawara Y. Biochem Biophys Res Commun. 1983;117:765–771. doi: 10.1016/0006-291x(83)91663-7. [DOI] [PubMed] [Google Scholar]
- 18.Luo G, Ducy P, McKee M D, Pinero G J, Loyer E, Behringer R R, Karsenty G. Nature (London) 1997;386:78–81. doi: 10.1038/386078a0. [DOI] [PubMed] [Google Scholar]
- 19.Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G. Nature (London) 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 20.Berkner K L, Pudota B N. Proc Natl Acad Sci USA. 1998;95:466–471. doi: 10.1073/pnas.95.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundaram K S, Lev M. J Lipid Res. 1988;29:1475–1479. [PubMed] [Google Scholar]
- 22.Sundaram K S, Fan J H, Engelke J A, Foley A L, Suttie J W, Lev M. J Nutr. 1996;126:2746–2751. doi: 10.1093/jn/126.11.2746. [DOI] [PubMed] [Google Scholar]
- 23.Hall J G, Pauli R M, Wilson K M. Am J Med. 1980;68:122–140. doi: 10.1016/0002-9343(80)90181-3. [DOI] [PubMed] [Google Scholar]
- 24.Pati S, Helmbrecht G D. Reprod Toxicol. 1994;8:115–120. doi: 10.1016/0890-6238(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 25.Zakzouk M S. J Laryngol Otol. 1986;100:215–219. doi: 10.1017/s0022215100099011. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan L C. Teratology. 1985;32:333–337. doi: 10.1002/tera.1420320302. [DOI] [PubMed] [Google Scholar]
- 27.Saxena S P, Fan T, Li M, Israels E D, Israels L G. J Clin Invest. 1997;99:602–607. doi: 10.1172/JCI119202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermeer C. Mol Cell Biochem. 1984;61:17–35. doi: 10.1007/BF00239604. [DOI] [PubMed] [Google Scholar]
- 29.Romero E E, Velazquez-Estades L J, Deo R, Schapiro B, Roth D A. Exp Cell Res. 1998;243:334–346. doi: 10.1006/excr.1998.4151. [DOI] [PubMed] [Google Scholar]
- 30.Kulman J D, Harris J E, Haldeman B A, Davie E W. Proc Natl Acad Sci USA. 1997;94:9058–9062. doi: 10.1073/pnas.94.17.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Einbond A, Sudol M. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 32.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Biel M, Altenhofen W, Hullin R, Ludwig J, Freichel M, Flockerzi V, Dascal N, Kaupp U B, Hofmann F. FEBS Lett. 1993;329:134–138. doi: 10.1016/0014-5793(93)80209-d. [DOI] [PubMed] [Google Scholar]
- 34.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartmann E, Rapoport T A, Lodish H F. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Int J Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 37.Chen H I, Sudol M. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen G B, Ren R, Baltimore D. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 39.Antipatis C, Ashworth C J, Grant G, Lea R G, Hay S M, Rees W D. Am J Physiol. 1998;275:1184–1191. doi: 10.1152/ajplung.1998.275.6.L1184. [DOI] [PubMed] [Google Scholar]