Abstract
Mitochondrial encephalomyopathies are disturbingly complex and devastating diseases, reflecting the underlying importance of the affected organelle. Therapeutic approaches for these diseases remain limited due to a poor understanding of disease pathogenesis due in large part to a lack of tractable model systems in which to study these diseases. This is especially so for disease conditions resulting from mutations directly affecting the mitochondrial genome. Recent studies using Drosophila to develop genetic models with endogenous mitochondrial mutations suggest the fruit fly will contribute significantly to our understanding of mitochondrial disease pathogenesis and the development of novel therapeutic avenues.
Mitochondria are complex organelles that perform a diverse array of important cellular functions, including: synthesis of ATP via oxidative phosphorylation (OXPHOS), calcium homeostasis and regulation of cell viability. Mitochondrial disease refers to a diverse constellation of conditions resulting from dysfunction of this essential organelle. These diseases are relatively common (~1 in 10,000) and are devastating human conditions for which therapeutic options are limited and the prognoses remain extraordinarily poor. Mitochondrial encephalomyopathies are a common form of mitochondrial disease that results from mutations affecting genes encoding essential mitochondrial functions. Class I disorders are those directly affecting mitochondrial DNA (mtDNA) or nuclear genes encoding OXPHOS proteins (Leonard and Schapira, 2000a). In contrast, class II disorders affect nuclear genes encoding mitochondrial proteins not directly functioning in OXPHOS or those encoding non-mitochondrial proteins associated with OXPHOS defects (Leonard and Schapira, 2000b).
Recently, it has become evident that numerous neurological and neurodegenerative diseases, including Parkinson’s disease (PD) and Huntington’s disease (HD), are class II mitochondrial disorders (Burchell et al., 2010a; Burchell et al., 2010b). Within the last year, research examining PINK and PARKIN pathogenesis has strongly supported the once controversial notion that PD is essentially a form of mitochondrial disease. Recent reviews have been written describing Drosophila research that has provided insight into PD pathogenesis (Park et al., 2009). Since this excellent review it was discovered that PINK and PARKIN regulate protein turnover of Mfn (mitofusin; (Poole et al., 2010; Ziviani et al., 2010), explaining how these proteins alter mitochondrial morphology, which likely underlies mitochondrial dysfunction and disease pathogenesis. Additionally, numerous other mutants and transgenes have been generated, which affect mitochondrial localized proteins that model class II mitochondrial diseases in Drosophila, including: tko (Mitoribosomal protein;(Fernandez-Ayala et al., 2010; Royden et al., 1987; Toivonen et al., 2003), sdhA (complex II; (Mast et al., 2008), pol γ (DNA polymerase subunit; (Martinez-Azorin et al., 2008), tafazzin (acyl transferase; (Xu et al., 2006), LetM1 (mitochondrial Ca++/H+ antiporter; (Jiang et al., 2009), and porin (VDAC; (Graham et al., 2010). Many of these models have already been reviewed and, due to space limitations, will not be covered further here (Fernandez-Moreno et al., 2007; Jacobs et al., 2004; Oliveira et al., 2010; Sanchez-Martinez et al., 2006).
Class I mitochondrial encephalomyopathies are extraordinarily difficult to study and, to date, these have largely been examined in cellular and cell cybrid systems. Recently it has been appreciated that these diseases may require a system approach within an intact animal to fully understand pathogenesis and to investigate potential therapeutics (Wallace et al.). The progressive nature of these diseases and complexity of the bioenergetic pathways under conditions of altered carbohydrate and lipid/ketone metabolism support the assertion that system approaches within tractable animal systems will be productive. This review will principally cover the primary research in Drosophila that aims to develop this tractable genetic model system for the study of class I mitochondrial encephalomyopathies. The research reviewed here strongly suggests Drosophila will emerge as a model system that will prove useful for the study of these complex mitochondrial diseases and contribute to the development and testing of novel therapeutic approaches.
Human Class I Mitochondrial Encephalomyopathies
Mitochondrial encephalomyopathies are a diverse group of disorders with complex clinical features. These multisystem diseases manifest with profound neurological, cardiac, and muscular dysfunction that is often complicated by renal, endocrine, and hepatic involvement (See reviews: (DiMauro and Schon, 2003; Hart PE, 2002; Schapira, 1999). Numerous mitochondrial mutations affecting various protein coding, tRNA or rRNA genes result in various forms of mitochondrial diseases collectively referred to as encephalomyopathies or cardiomyopathies (See Table). Because there are numerous mutagenic targets, these mitochondrial disorders have an extremely high prevalence (10–15 per 100,000) that is similar to muscular dystrophies and ALS (DiMauro and Schon, 2003). Among the most severe mitochondrial encephalomyopathies are those resulting from mutations affecting tRNAs or the protein coding mtATP6 gene. tRNA mutations dramatically diminish the ability of mitochondria to synthesize proteins and result in severe mitochondrial dysfunction. The mtATP6 protein is an essential component of the mitochondrial F1Fo-ATP synthase (Complex V) that functions as a hydrogen ion channel serving to couple ion transport with rotary ATP catalysis (Boyer, 1997; Mitchell and Moyle, 1967; Stock et al., 2000; Walker, 1995) (Boyer, 1997; Mitchell and Moyle, 1967; Stock et al., 2000; Walker, 1995). There are at least nine distinct mutations within the mtATP6 gene resulting in NARP, MILS or FBSN diseases including three mutations that have been extensively characterized: mtATP6W109R (De Meirleir et al., 1995) mtATP6L156R (de Vries et al., 1993; Holt et al., 1990), and mtATP6L217R (Holt et al., 1990), as well as a common 5kb deletion that affects several loci and removes the entire mtATP6 gene (Mita et al., 1990; Schon et al., 1989). Although there are numerous neurological features associated with mtATP6 mutations, striated muscle degeneration and cardiomyopathy are also a common result of mtATP6 dysfunction (Finsterer, 2004; Fosslien, 2003; Kelly and Strauss, 1994; Marin-Garcia et al., 2001; Ozawa, 1994; Ventura-Clapier et al., 2004).
Table.
Archetypal mitochondrial encephalomyopathies & associated diseases.
| Disease/Initialism | Affected gene(s) | References |
|---|---|---|
| MELAS | tRNA-L, ND1, ND5, ND4 | Campos et al., 1997a; Goto et al., 1990; Kirby et al., 2004; Lertrit et al., 1992; Santorelli et al., 1997a |
| MERRF | tRNA-K | Shoffner et al., 1990 |
| NARP | ATP6 | Castagna et al., 2007; Holt et al., 1990; Lopez-Gallardo et al., 2009; Sciacco et al., 2003 |
| MILS | ATP6, tRNA-K, | Campos et al., 1997b; Carrozzo et al., 2001; de Vries et al., 1993; Moslemi et al., 2005; Pastores et al., 1994; Santorelli et al., 1994; Santorelli et al., 1993; Sciacco et al., 2003; Shtilbans et al., 2000 |
| FBSN | ATP6 | De Meirleir et al., 1995; Thyagarajan et al., 1995 |
| LHON | ND1, ND4, ND6, ATP6*, CYB | Howell et al., 1991; Johns and Neufeld, 1991; Johns et al., 1992; Lamminen et al., 1995; Wallace et al., 1988 |
| MIDD | tRNA-L, tRNA-S, 12SrRNA | Prezant et al., 1993; Reid et al., 1994; Sue et al., 1999; van den Ouweland et al., 1992 |
| Infantile encephalomyo. | ND3 | McFarland et al., 2004 |
| Cardiomyopathy | tRNA-I, tRNA-L, tRNA-G, ATP6 | Manouvrier et al., 1995; Merante et al., 1994; Pastores et al., 1994; Silvestri et al., 1994; Taniike et al., 1992; Zeviani et al., 1991 |
| Myopathy/diabetes | tRNA-E, tRNA-L | Hao et al., 1995; Katagiri et al., 1994 |
| Exercise intolerance | CYB | Andreu et al., 1999 |
| KSS | Large deletion(s), tRNA-L, tRNA-K | Mita et al., 1990; Moraes et al., 1989; Nishigaki et al., 2003; Schapira, 2006; Schon et al., 1989 |
| PEO | Large deletion(s) | Mita et al., 1990; Moraes et al., 1989; Schon et al., 1989 |
| Myopathy | Large deletion(s) | Holt et al., 1989 |
| Pearson’s syndrome | Large deletion(s) | Rotig et al., 1990 |
NARP is a single disease defined by different authors as either Neuropathy, Ataxia, Retinitis Pigmentosa or Neurogenic muscle weakness, Ataxia, Retinitis Pigmentosa. MILS is Maternally Inherited Leigh Syndrome. HC is Hypertrophic Cardiomyopathy. PEO is Progressive External Opthalomoplegia. KSS is Kearns-Sayre Syndrome. LHON is Leber Hereditary Optic Neuropathy. FBSN is Familial Bilateral Striatal Necrosis. MELAS is Mitochondrial myopathy, Encephalopathy, Lactic Acidosis and Stroke-like episodes. MERRF is Myoclonic Epilepsy and Ragged-Red Fibers. MIDD is Maternally Inherited Diabetes and Deafness.
not a highly conserved residue.
Difficulties in Studying Mitochondrial Encephalomyopathies
The archetypal mitochondrial encephalomyopathies (Table) are complex, multi-system diseases that are proving difficult to study for a myriad of reasons, most notable: 1) their progressive nature, 2) tissue-specific pathogenesis, 3) our rudimentary understanding of the compensatory mechanisms resulting from altered metabolism, 4) heteroplasmy of mitochondrial mutations, 5) limited techniques to manipulate the mitochondrial genome, and 6) a lack of tractable models of mitochondrial disease. Innovative techniques have been used to model mitochondrial diseases in cellular and cell cybrid models and much of our current understanding of these diseases is derived from such approaches. It is important to note, however, that no clear understanding of the associated pathogenesis has resulted, suggesting the inherent difficulty in using cellular models to study these multisystem, progressive diseases. This sentiment was exemplified in a review on mitochondrial encephalomyopathies, “The thus far insurmountable problem of obtaining animal models for mtDNA mutations has prevented detailed studies of pathogenesis” (DiMauro and Andreu, 2000).
The clinical complexity of mitochondrial encephalomyopathies is exemplified by those resulting from mtATP6 dysfunction. In humans, mtATP6 dysfunction results in three different diseases with overlapping clinical manifestations, however, differences between these diseases and even between patients diagnosed with the same disease are evident. Thus, the genotype/phenotype relationship remains ambiguous and the heterogeneous clinical presentation of these diseases is well known (Lopez-Gallardo et al., 2009; Santorelli et al., 1997). It is not evident why numerous mutations all affecting the same biochemical process, presumably resulting in bioenergetic impairment in the affected tissues, result in a veritable plethora of clinical manifestations that are not fully explained by the heteroplasmy of the mutations (DiMauro and Andreu, 2000; Schon et al., 1997).
In humans, 9 mutations in the mtATP6 gene, 8 of which are missense, cause NARP, FBSN and MILS diseases. MILS can be caused by the same mutations as NARP but MILS is a more severe disease often with infantile onset and generally results when there is a higher mutant heteroplasmy (Tatuch et al., 1992). Importantly, many patients with complex V dysfunction are normal at birth and exhibit onset of the disease after many months to decades dependent upon the severity of the mutation and percent heteroplasmy. Even the most severe molecular lesion, an insertion at codon 32 that is almost certainly a null (truncates the protein at codon 64), was found at a high mutant heteroplasmy (85%), which presented in a patient at age 4 as a “developmental delay” that became more serious in advanced childhood (Lopez-Gallardo et al., 2009). Both late onset and childhood onset cases of complex V deficiency have been described with high mutant heteroplasmy ~ 90%. The same mutation at similar heteroplasmy may have a different age of onset or different severity dependent upon the presence of other stressors such as a febrile condition or severe viral infection (Castagna et al., 2007; Santorelli et al., 1997; Tsao et al., 2001). In an atypical case, a NARP patient presented at the age of 4 with >95% mutant heteroplasmy of the T8993C mutation. Despite childhood respiratory distress, muscle weakness and neurological abnormalities the condition subsequently resolved by the age of 18 at which time the patients condition was noted to be stable (Debray et al., 2007). The clinical heterogeneity, often late childhood early adulthood onset and occasional atypical mild cases suggest that metazoans are able to compensate for very severe impairment of complex V and that the study of an animal model system of this disease will help us understand these endogenous coping mechanisms and why they eventually fail in the face of stressors or with age.
Studies of mitochondrial disease pathogenesis that do not capture the progressive nature of these diseases could be uninformative or worse provide misleading “insight” into pathogenesis. The finding that humans with severe mitochondrial impairment remain asymptomatic for extended periods of time (months to decades) and disease onset often follows an additional stressor, demonstrates we have the capacity to tolerate marked mitochondrial dysfunction via various compensatory mechanisms. A comprehensive understanding of these endogenous compensatory mechanisms and elucidating how acute stressors derail these mechanisms and typically lead irreversibly to the disease state will certainly lead to a more complete understanding of disease pathogenesis and novel therapeutic approaches.
Most class I mitochondrial encephalomyopathies result from mtDNA mutations, of which there are more than 150 described pathogenic mutations (mostly point mutations or DNA deletions/rearrangements). The inability to use traditional gene manipulation strategies available for nuclear DNA has severely limited the development of genetic animal models. This is further complicated by the mutant heteroplasmy of the mtDNA mutations, which typically must reach >70–85% within a tissue to be pathogenic. The identification of pathogenic mtDNA mutations with a high mutant heteroplasmy (Celotto et al., 2006) and the ability to select for and manipulate the heteroplasmy of mtDNA mutations (Xu et al., 2008) in an amenable genetic model system such as Drosophila represent important advances toward overcoming the key difficulties in studying these complex disease conditions.
Endogenous Mitochondrial ATP6 Mutant in Drosophila
An endogenous mitochondrial mtATP61 mutation was discovered in Drosophila as a maternally inherited enhancer of sesB1, which encodes ANT (adenine nucleotide transporter; Celotto et al., 2006). The missense mutation results in a glycine (G) to glutamate (E) alteration at position 116 in the mtATP6 protein, which encodes the hydrogen ion channel component of the Fo portion of complex V (F1Fo ATP synthase). mtATP6 performs the important function of coupling hydrogen ion transport with ATP synthesis derived from OXPHOS. Numerous human missense mutations affecting mtATP6 result in mitochondrial encephalomyopathies including NARP, MILS, or FBSN. The mtATP61 mutation presumably derived from the known mutator effect resulting from ANT dysfunction, which is also responsible for autosomal dominant progressive external opthalmoplegia (adPEO; (Kaukonen et al., 2000). The finding that the mtATP61 mutation had a high degree of heteroplasmy (98% mutant) suggests that ANT dysfunction, in addition to mutating mtDNA, likely also selected for the mutation, accounting for the high mutant heteroplasmy and remarkable stability of the mutation in a sesB1 genetic background. Although this is a difficult detail to formally demonstrate, we have observed that the mtATP61 mutation does not erode in a sesB1 background (stable at least 6 years), whereas, when maintained in a sesB+ background the phenotype erodes leading to revertant animals in less than a year (unpublished data).
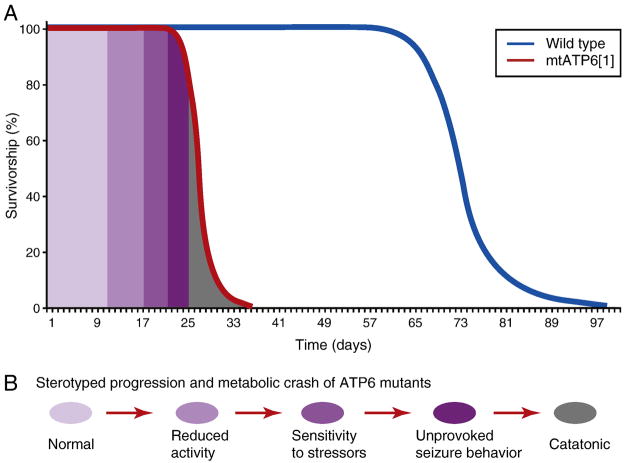
mtATP61 animals exhibit striking phenotypes including reduced longevity, progressive locomotor impairment and muscular degeneration, stress-induced behavioral impairment, seizures and neural dysfunction (Celotto et al., 2006). Additionally, the neural dysfunction associated with mtATP61 profoundly accelerates the onset and severity of sesB1neurodegeneration (Celotto et al., 2006). Importantly, mtATP61 mutants model mitochondrial encephalomyopathy and exhibit a stereotyped phenotypic progression that is highly reminiscent to the human disease condition (Figure 1). Mutant mtATP61 animals were found to have markedly reduced ATP synthase activity, but interestingly, these animals exhibited essentially normal respiration. Although these results were initially controversial, it has since been demonstrated that cells lacking a complex V (mtDNA–less rho0 cells) with a restored ETS (electron transport system) by expression of AXO and NDI1 have near normal respiration independent of proton pumping (Perales-Clemente et al., 2008). Collectively these findings demonstrate that animal respiration is a highly buffered trait, even in the presence of pathogenic mutations that significantly impair complex V.
Figure 1. Reduced longevity and behavioral progression of disease state.
A) Depiction of the lifespan defect observed in mtATP6 mutants relative to wild type controls at 25 C. B) mtATP6 mutants exhibit a stereotyped progression akin to that experienced by many mitochondrial disease patients.
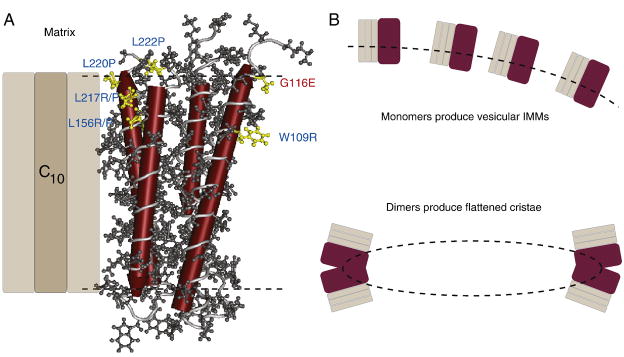
Tomographic TEM analysis of mtATP61 mutant tissues revealed an altered inner mitochondrial membrane (IMM) structure (Celotto et al., 2006). Interestingly, the IMM in mtATP61 mutants exhibits a dramatic loss of normal cristae and the membranes are highly interconnected and vesicular in appearance. Recent studies suggest that complex V, specifically the mtATP6 protein of complex V, is a key determinant of mitochondrial structure and is required to form flattened cristae (Celotto et al., 2006; Goyon et al., 2008; Paumard et al., 2002; Rak et al., 2007; Strauss et al., 2008; Wittig and Schagger, 2008; Wittig et al., 2008). Furthermore, the missense mutation resulting from mtATP61 is structurally near human mtATP6W109R both of which are predicted to affect protein dimerization (Figure 2). These findings suggest that the Drosophila mtATP61 and human mtATP6W109R mutations may cause dysfunction via the production of mtATP6 monomers that are unable to dimerize, possibly explaining both the altered physiology and ultrastructure. Further studies are being performed to fully test this hypothesis.
Figure 2. ATP synthase dimers alter mitochondrial structure.
A) The structure of ATP synthase ATP6 subunit of the Fo derived from published structural data (Rastogi and Girvin, 1999). Four of the predicted helices of ATP6 are shown in red. Residues highlighted in yellow are affected by pathogenic mutations in human (blue) and the fly (red). The fly mtATP61 mutation in located opposite ATP synthase c subunits and near the W109R residue both of which are predicted to be key sites near at the dimer interface. B) Monomers of ATP synthase would not bend the IMM (dashed line), whereas, dimers are believed to bend the membrane at the apex producing flattened cristae.
Collectively the data thus far suggest that Drosophila mtATP61 mutants capture many relevant features of mitochondrial encephalomyopathy and enable the study of neuromuscular system pathogenesis resulting from altered OXPHOS within intact animals. Furthermore, the identification and study of endogenous compensatory mechanisms, and elucidation of the mechanism underlying stress-sensitivity and the basis for the progressive nature of the disease are feasible and should lead to novel therapeutic avenues.
Engineering Endogenous Mitochondrial Mutations
Ideally we would be able to engineer specific mitochondrial mutations for study within a well-characterized animal model. This would require a mechanism of mutagenizing the mitochondrial genome and, importantly, a powerful means of selection within a metazoan. Although this has not been accomplished to date, recent studies have demonstrated the ability to use mitochondrial-targeted restriction enzymes to select for alterations in the mitochondrial genome (Xu et al., 2008). The premise of this technique is that the mitochondrial genome is small (~16.5 kb in human and ~19kb in Drosophila) and numerous restriction endonucleases have a single recognition sequence within the wild type genome. A mitochondrial-targeted restriction enzyme (mtRE) directs the enzyme to the mitochondria, preventing toxicity to the nuclear genome. Expression of the mtRE will result in efficient cleavage of wild-type mtDNAs. This is extraordinarily toxic and, in fact, most animals will die upon expression of the mtRE. However, resistant mitochondrial genomes that alter the recognition sequence then repopulate the organism in response to the mtRE selection resulting in the occasional “escaper” animal. Such a process was used with an Xho I mtRE to isolate 3 independent escaper animals with distinct mutations in the cytochrome c oxidase I gene (mtCoI). The mtCoIA302T animals were healthy, the mtCoIR301Q animals were male sterile and the mtCoIR301S animals exhibited numerous phenotypes consistent with mitochondrial disease, such as reduced lifespan, muscle atrophy and neurodegenearation (Xu et al., 2008). Similarly mutations affecting the mtND2 locus were isolated using Bgl II mtRE as selection (Xu et al., 2008). It is not clear whether the mutations that were selected for by mtRE expression are naturally occurring within the existing mitochondrial genomes at a very low level of heteroplasmy, are caused by mtRE activity or result from the high mutation rate that likely results owing to the severe mitochondrial dysfunction resulting from mtDNA cleavage. Nonetheless, these results demonstrate mtDNA selection within a metazoan and represent a tremendous technological advance within the field. In theory, such a process could be used with numerous distinct mtREs to isolate a series of mtDNA mutations for modeling mitochondrial encephalomyopathies. The major limitations of this method are the inability to generate intermediate mutant heteroplasmy (the animals selected for have essentially 100% mutant homoplasmy) and an inability to generate a precise mutation – any mutation that alters the restriction recognition signal can result in an escaper animal and some of these will be silent or otherwise non-informative.
Future Directions
The research reviewed here lays the foundation for studies of pathogenesis associated with endogenous mitochondrial mutations in an amenable genetic animal model. Studies of the mtATP6 mutation suggest that examining disease progression in aging animals and elucidating the compensatory mechanisms associated with mitochondrial impairment are promising avenues of research that could identify novel therapeutic pathways. Furthermore, this well-characterized model of mitochondrial encephalomyopathy is ideal for the development and testing of allotopic expression (expressing recoded mitochondrial genes from the nucleus) as a gene therapy for devastating diseases such as NARP and MILS. Lastly, efforts are underway to combine the powerful selection of mtREs with a transformation mechanism to introduce an engineered mitochondrial genome (O’Farrell, personal communication): such an accomplishment will overcome the difficulties with this approach and revolutionize the study of mitochondrial diseases in animal systems.
Acknowledgments
I thank Dr. Alicia Celotto for comments on this review and Drs. Russell Saneto, Wayne Van Voorhies and Carmen Mannella for helpful discussions about mitochondrial disease pathogenesis. I also thank the reviewers for their critiques, which allowed me to improve the manuscript. NIH R01AG025046, NIH R01AG027453, UMDF 70455, and American Heart Association 0630344N grants supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyer PD. The ATP synthase--a splendid molecular machine. Annu Rev Biochem. 1997;66:717–49. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- Burchell VS, et al. Targeting mitochondrial dysfunction in neurodegenerative disease: Part I. Expert Opin Ther Targets. 2010a;14:369–85. doi: 10.1517/14728221003652489. [DOI] [PubMed] [Google Scholar]
- Burchell VS, et al. Targeting mitochondrial dysfunction in neurodegenerative disease: Part II. Expert Opin Ther Targets. 2010b;14:497–511. doi: 10.1517/14728221003730434. [DOI] [PubMed] [Google Scholar]
- Castagna AE, et al. Late onset Leigh syndrome and ataxia due to a T to C mutation at bp 9,185 of mitochondrial DNA. Am J Med Genet A. 2007;143A:808–16. doi: 10.1002/ajmg.a.31637. [DOI] [PubMed] [Google Scholar]
- Celotto AM, et al. Mitochondrial encephalomyopathy in Drosophila. J Neurosci. 2006;26:810–20. doi: 10.1523/JNEUROSCI.4162-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meirleir L, et al. Bilateral striatal necrosis with a novel point mutation in the mitochondrial ATPase 6 gene. Pediatr Neurol. 1995;13:242–6. doi: 10.1016/0887-8994(95)00184-h. [DOI] [PubMed] [Google Scholar]
- de Vries DD, et al. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann Neurol. 1993;34:410–2. doi: 10.1002/ana.410340319. [DOI] [PubMed] [Google Scholar]
- Debray FG, et al. Long-term outcome of Leigh syndrome caused by the NARP-T8993C mtDNA mutation. Am J Med Genet A. 2007;143A:2046–51. doi: 10.1002/ajmg.a.31880. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Andreu AL. Mutations in mtDNA: are we scraping the bottom of the barrel? Brain Pathol. 2000;10:431–41. doi: 10.1111/j.1750-3639.2000.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–68. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ayala DJ, et al. Gene expression in a Drosophila model of mitochondrial disease. PLoS One. 2010;5:e8549. doi: 10.1371/journal.pone.0008549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moreno MA, et al. Drosophila melanogaster as a model system to study mitochondrial biology. Methods Mol Biol. 2007;372:33–49. doi: 10.1007/978-1-59745-365-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J. Mitochondriopathies. Eur J Neurol. 2004;11:163–86. doi: 10.1046/j.1351-5101.2003.00728.x. [DOI] [PubMed] [Google Scholar]
- Fosslien E. Review: Mitochondrial medicine--cardiomyopathy caused by defective oxidative phosphorylation. Ann Clin Lab Sci. 2003;33:371–95. [PubMed] [Google Scholar]
- Goyon V, et al. Yeast cells depleted in Atp14p fail to assemble Atp6p within the ATP synthase and exhibit altered mitochondrial cristae morphology. J Biol Chem. 2008;283:9749–58. doi: 10.1074/jbc.M800204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BH, et al. Neurologic dysfunction and male infertility in Drosophila porin mutants: a new model for mitochondrial dysfunction and disease. J Biol Chem. 2010;285:11143–53. doi: 10.1074/jbc.M109.080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PEDD, Schapira AHV. In: Mitochondrial disorders in neurology. Schapira A, DiMauro S, editors. Vol. 2. Butterworth Heinemann; Boston: 2002. [Google Scholar]
- Holt IJ, et al. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46:428–33. [PMC free article] [PubMed] [Google Scholar]
- Jacobs HT, et al. Mitochondrial disease in flies. Biochim Biophys Acta. 2004;1659:190–6. doi: 10.1016/j.bbabio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Jiang D, et al. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326:144–7. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukonen J, et al. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–5. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Strauss AW. Inherited cardiomyopathies. N Engl J Med. 1994;330:913–9. doi: 10.1056/NEJM199403313301308. [DOI] [PubMed] [Google Scholar]
- Leonard JV, Schapira AH. Mitochondrial respiratory chain disorders I: mitochondrial DNA defects. Lancet. 2000a;355:299–304. doi: 10.1016/s0140-6736(99)05225-3. [DOI] [PubMed] [Google Scholar]
- Leonard JV, Schapira AH. Mitochondrial respiratory chain disorders II: neurodegenerative disorders and nuclear gene defects. Lancet. 2000b;355:389–94. doi: 10.1016/s0140-6736(99)05226-5. [DOI] [PubMed] [Google Scholar]
- Lopez-Gallardo E, et al. NARP syndrome in a patient harbouring an insertion in the MT-ATP6 gene that results in a truncated protein. J Med Genet. 2009;46:64–7. doi: 10.1136/jmg.2008.060616. [DOI] [PubMed] [Google Scholar]
- Marin-Garcia J, et al. Mitochondrial pathology in cardiac failure. Cardiovasc Res. 2001;49:17–26. doi: 10.1016/s0008-6363(00)00241-8. [DOI] [PubMed] [Google Scholar]
- Martinez-Azorin F, et al. Over-expression of the catalytic core of mitochondrial DNA (mtDNA) polymerase in the nervous system of Drosophila melanogaster reduces median life span by inducing mtDNA depletion. J Neurochem. 2008;105:165–76. doi: 10.1111/j.1471-4159.2007.05122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast JD, et al. Reactive oxygen species act remotely to cause synapse loss in a Drosophila model of developmental mitochondrial encephalopathy. Development. 2008;135:2669–79. doi: 10.1242/dev.020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, et al. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 1990;18:561–7. doi: 10.1093/nar/18.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Moyle J. Chemiosmotic hypothesis of oxidative phosphorylation. Nature. 1967;213:137–9. doi: 10.1038/213137a0. [DOI] [PubMed] [Google Scholar]
- Oliveira MT, et al. Animal models of mitochondrial DNA transactions in disease and ageing. Exp Gerontol. 2010 doi: 10.1016/j.exger.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T. Mitochondrial cardiomyopathy. Herz. 1994;19:105–18. 125. [PubMed] [Google Scholar]
- Park J, et al. Mitochondrial dysfunction and Parkinson’s disease genes: insights from Drosophila. Dis Model Mech. 2009;2:336–40. doi: 10.1242/dmm.003178. [DOI] [PubMed] [Google Scholar]
- Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–30. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales-Clemente E, et al. Restoration of electron transport without proton pumping in mammalian mitochondria. Proc Natl Acad Sci U S A. 2008;105:18735–9. doi: 10.1073/pnas.0810518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, et al. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, et al. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J Biol Chem. 2007;282:10853–64. doi: 10.1074/jbc.M608692200. [DOI] [PubMed] [Google Scholar]
- Rastogi VK, Girvin ME. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 1999;402:263–8. doi: 10.1038/46224. [DOI] [PubMed] [Google Scholar]
- Royden CS, et al. The tko locus, site of a behavioral mutation in D. melanogaster, codes for a protein homologous to prokaryotic ribosomal protein S12. Cell. 1987;51:165–73. doi: 10.1016/0092-8674(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martinez A, et al. Modeling human mitochondrial diseases in flies. Biochim Biophys Acta. 2006;1757:1190–8. doi: 10.1016/j.bbabio.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorelli FM, et al. Heterogeneous clinical presentation of the mtDNA NARP/T8993G mutation. Neurology. 1997;49:270–3. doi: 10.1212/wnl.49.1.270. [DOI] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial disorders. Biochim Biophys Acta. 1999;1410:99–102. doi: 10.1016/s0005-2728(98)00160-1. [DOI] [PubMed] [Google Scholar]
- Schon EA, et al. Mitochondrial DNA mutations and pathogenesis. J Bioenerg Biomembr. 1997;29:131–49. doi: 10.1023/a:1022685929755. [DOI] [PubMed] [Google Scholar]
- Schon EA, et al. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244:346–9. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- Stock D, et al. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–9. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- Strauss M, et al. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–60. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatuch Y, et al. Heteroplasmic mtDNA mutation (T----G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am J Hum Genet. 1992;50:852–8. [PMC free article] [PubMed] [Google Scholar]
- Toivonen JM, et al. Gene dosage and selective expression modify phenotype in a Drosophila model of human mitochondrial disease. Mitochondrion. 2003;3:83–96. doi: 10.1016/S1567-7249(03)00077-1. [DOI] [PubMed] [Google Scholar]
- Tsao CY, et al. High mitochondrial DNA T8993G mutation (<90%) without typical features of Leigh’s and NARP syndromes. J Child Neurol. 2001;16:533–5. doi: 10.1177/088307380101600716. [DOI] [PubMed] [Google Scholar]
- Ventura-Clapier R, et al. Energy metabolism in heart failure. J Physiol. 2004;555:1–13. doi: 10.1113/jphysiol.2003.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE. Determination of the structures of respiratory enzyme complexes from mammalian mitochondria. Biochim Biophys Acta. 1995;1271:221–7. doi: 10.1016/0925-4439(95)00031-x. [DOI] [PubMed] [Google Scholar]
- Wallace DC, et al. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Structural organization of mitochondrial ATP synthase. Biochim Biophys Acta. 2008;1777:592–8. doi: 10.1016/j.bbabio.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Wittig I, et al. Characterization of domain interfaces in monomeric and dimeric ATP synthase. Mol Cell Proteomics. 2008;7:995–1004. doi: 10.1074/mcp.M700465-MCP200. [DOI] [PubMed] [Google Scholar]
- Xu H, et al. Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science. 2008;321:575–7. doi: 10.1126/science.1160226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, et al. A Drosophila model of Barth syndrome. Proc Natl Acad Sci U S A. 2006;103:11584–8. doi: 10.1073/pnas.0603242103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, et al. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–23. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]