Abstract
The parent-of-origin specific expression of imprinted genes relies on DNA methylation of CpG-dinucleotides at differentially methylated regions (DMRs) during gametogenesis. To date, four paternally methylated DMRs have been identified in screens based on conventional approaches. These DMRs are linked to the imprinted genes H19, Gtl2 (IG-DMR), Rasgrf1 and, most recently, Zdbf2 which encodes zinc finger, DBF-type containing 2. In this study, we applied a novel methylated-DNA immunoprecipitation-on-chip (meDIP-on-chip) method to genomic DNA from mouse parthenogenetic- and androgenetic-derived stem cells and sperm and identified 458 putative DMRs. This included the majority of known DMRs. We further characterized the paternally methylated Zdbf2/ZDBF2 DMR. In mice, this extensive germ line DMR spanned 16 kb and possessed an unusual tripartite structure. Methylation was dependent on DNA methyltransferase 3a (Dnmt3a), similar to H19 DMR and IG-DMR. In both humans and mice, the adjacent gene, Gpr1/GPR1, which encodes a G-protein-coupled receptor 1 protein with transmembrane domain, was also imprinted and paternally expressed. The Gpr1-Zdbf2 domain was most similar to the Rasgrf1 domain as both DNA methylation and the actively expressed allele were in cis on the paternal chromosome. This work demonstrates the effectiveness of meDIP-on-chip as a technique for identifying DMRs.
INTRODUCTION
Genomic imprinting describes the expression of a subset of mammalian genes from one parental chromosome (1). Many imprinted genes play developmentally important roles particularly during embryogenesis and also in the adult animal (2,3). The majority of imprinted genes reside within complex domains. Although the domain itself remains imprinted throughout the life of the organism, individual genes within the domain can be expressed in tissue- and developmentally specific patterns and some also show temporal or spatial differences in their imprinted status.
Imprinted domains are established in the germ line and the epigenetic profile of germ cells changes dynamically during development (4). Most strikingly, the DNA methylation of CpG-dinucleotides at differentially methylated regions (DMRs) is erased as the primordial germ cells migrate from the base of the allantois to the genital ridge and differentially re-established during oogenesis and spermatogenesis (5). In the female neonatal mouse, methylation is acquired asynchronously in a gene-specific manner in oocytes arrested at prophase I and during the transition from primordial to antral follicles in the postnatal growth phase (post-pachytene) (6–8). In contrast, methylation is initiated at paternal DMRs prenatally during embryonic germ cell development and completed by the pachytene phase of postnatal spermatogenesis (9–12). The gametic imprints are maintained stably after fertilization despite overall epigenetic reprogramming, and persist during development and into adulthood.
Methyl-substrates and DNA methyltransferases (Dnmts) are required for both the acquisition and the maintenance of DNA methylation. In mice, Dnmt3a and the accessory protein, Dnmt3l, establish imprinted DNA methylation in the germ line (13–15). Dnmt3a has a central role in the de novo methylation process at the paternally methylated H19, Gtl2 (intergenic DMR; IG-DMR) and Rasgrf1 loci, while the role of Dnmt3b appears to be specific to the Rasgrf1 locus (15,16). Dnmt3l has a plant homeodomain (PHD)-like motif but lacks DNA methylation activity (14,17). Instead, Dnmt3l cooperates with Dnmt3a to de novo methylate DNA (18,19). It may serve to activate the functional Dnmts and/or play a role in recognizing the target sequence (20,21). Germ line conditional knockout mice that lack either Dnmt3a or Dnmt3l do not acquire the maternal or paternal methylation imprints (15,16).
To date, DNA methylation is acquired on the paternal allele at 4 DMRs and on the maternal allele at 18 DMRs (22–26). There are additional DMRs where allele-specific methylation is acquired after fertilization. Disruption of the methylating machinery in the germ line primarily results in global loss of imprinting (14,27,28), while loss of the maintenance DNA methylase can affect the expression of a subset of imprinted genes within a domain (29–31).
The number of known imprinted genes is ∼100 but the total number is unknown. A number of approaches have been used to identify new candidates (32). A drawback of expression-based approaches is in the identification of genes expressed at different stages of development or ones that are imprinted only in a subset of tissues. In contrast, approaches based on detecting regions of allele-specific epigenetic marks between the maternal and paternal genomes are applicable to all tissue types at all time points. Tiling array technology and chromatin immunoprecipitation (ChIP-on-chip) has been successfully applied to decipher chromatin structure (33–35). In this study, we applied this technology in combination with the methylated DNA binding column technique (36,37) using the antibody against 5-methyl-cytosine (methylated-DNA immunoprecipitation; meDIP) to determine how effectively we could identify known and novel DMRs.
MATERIALS AND METHODS
Mouse strains and the preparations of DNA and RNA
Derivation of PG-, AG-derived stem and TS cells was described previously in detail (38). C57BL/6 (B6) females were mated with JF1 (39) males to generate B6/JF1 mice and reciprocally crossed to generate JF1/B6 mice. The mature sperm and MII oocytes were obtained from B6 and ICR mice, respectively. Blastocysts were obtained from B6/JF1 mice. Genomic DNAs from mature sperm, MII oocytes, blastocysts and TS cells was prepared as previously described (6,40). Genomic DNA and total RNA were obtained from various organs from B6/JF1 and JF1/B6 mice at embryonic day (E) 13.5, E18.5 and adult stages. For human polymorphic analysis, DNA and RNA were prepared from umbilical cord blood after delivery and from their mothers’ peripheral blood using standard protocols. Total RNA was prepared using ISOGEN (Nippon Gene, Tokyo, Japan), treated with DNase I (Promega, WI, USA) to remove genomic DNA. The absence of genomic DNA contamination was confirmed by the lack of genomic DNA amplification of Gapdh/GAPDH by polymerase chain reaction (PCR).
The isolation of Dnmt3a-deficient and wild-type prospermatogonia
To obtain Dnmt3a-deficient and wild-type prospermatogonia, male germ cells were isolated from E14.5, E16.5, E18.5 and Postnatal day (P) 7 testes from B6 mice and from P7 testes of the conditional Dnmt3a knockout mice by fluorescence activated cell sorting (FACS) as previously described in detail (16).
MeDIP-on-chip analysis
DNA extracted from PG- and AG-derived cells and mature sperm was fragmented to ∼200–1000 bp by sonication (Sonics & Materials, Connecticut, USA). Fragment size was checked on 1% agarose gels. Immunoprecipitation was carried out using a specific antibody for 5-methyl-cytosine (AbD Serotec, Oxfordshire, UK). Input and bound DNA was amplified by GenomePlex Complete Whole Genome Amplification kit (Sigma–Aldrich, Missouri, USA). The relative enrichment of DMRs was determined by sequence-specific real-time PCR analyses using a 7500 Real Time PCR System (Applied Biosystems Japan, Tokyo, Japan) and SYBR Premix Ex Taq II (Perfect Real Time) (Takara Bio, Kyoto, Japan). Primers and PCR conditions are described in Supplementary Table S1.
For the tiling arrays, input DNA was labeled with a cyan-3 dye and bound DNA was labeled with cyan-5. DNAs were hybridized to the mouse whole genome tiling array (Agilent Technologies Japan, Tokyo, Japan). The methylated sequences were compared between PG- and AG-derived cells and sperm DNA using ChIP Analytics 1.3 software (Agilent Technologies Japan, Tokyo, Japan).
Bisufhite-PCR methylation assay
The methylation assay was performed at the DMRs of H19, IG-DMR (Gtl2), Rasgrf1, Zdbf2, Nespas, Gnas1A, Peg10, Peg1, Peg3, Snrpn, Lit1, Zac1, U2af1-rs1, Igf2r (DMR2) and Impact. The Zdbf2 methylated regions were analyzed by both combined bisulfite-PCR restriction analysis (COBRA) and bisulfite-PCR sequencing (11). Each DNA sample (MII oocytes, sperm and several organs tissues) was treated with sodium bisulfite using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) and amplified by PCR as follows: a PCR reaction mix containing 0.5 µM of each of the primer sets, 200 µM dNTPs, 1 × PCR buffer, 1.25 U of Ex Taq Hot Start DNA Polymerase (Takara Bio, Kyoto, Japan) in a total volume of 20 µl. Primers used and PCR conditions are listed in Supplementary Table S1.
COBRA was carried out on bisulfite-treated PCR samples with the following enzymes: TaqI for the DMR of H19, IG-DMR (Gtl2), Nespas, Zac1, Igf2r (DMR2) and Zdbf2; HpyCH4IV for the DMR of Gnas1A, Peg10, Peg1, Peg3, Snrpn, Lit1, U2af1-rs1, Impact and Zdbf2. Samples were electrophoresed on 2% agarose gels. The PCR products were purified and cloned into the pGEM-T Easy vector (Promega, WI, USA) and individual clones were sequenced using T7 or SP6 primer and an automated ABI Prism 3130xl Genetic Analyzer (Applied Biosystems Japan, Tokyo, Japan). An average of 20 clones for each individual was sequenced. At least two separate sodium modification treatments were carried out for each DNA sample, and at least three independent amplification experiments were performed for each individual.
Reverse transcription PCR analysis
Monoallelic expression of Gpr1/GPR1 was investigated by RT-PCR. DNA-free total RNA (1 µg) from mouse and human tissues was reverse-transcribed into cDNA using AMV reverse transcriptase (Roche Diagnostics, Basel, Switzerland) with either a sense or antisense primer in order to determine the direction of transcription. RT products were then amplified using the specified PCR primers.
In situ hybridization analysis
cDNA probes for mouse Zdbf2 and Gpr1 were generated by PCR (Supplementary Table S1) and used to prepare sense and antisense riboprobes by in vitro transcription using the DIG RNA labeling kit (Boehringer Mannheim, Mannheim, Germany). Sagittal sections of 8 μm from paraffin embedding mouse embryos and placentas at E13.5 were used for in situ hybridization as described previously (41). Sections were counterstained with eosin.
Sequence analysis
Nucleotide similarities between mouse DMR1 and human DMRh1 were calculated using the GENETYX software version 11.0 (GENETYX, Tokyo, Japan). Dot-matrix analysis was performed on mouse DMR1 and human DMRh1 to detect homologous regions using Harrplot Ver. 2.0 as part of the computer software GENETYX package.
RESULTS
MeDIP-on-chip screen for the DMRs
To identify novel DMRs, we applied the meDIP-on-chip method to DNA extracted from parthenogenetic (PG)-derived stem cells (two copies of the maternal genome), androgenetic (AG)-derived stem cells (two copies of the paternal genome) and genomic DNA prepared from mature sperm. We first confirmed that the stem cell genomic DNA had the characteristic epigenetic profile of PG- and AG-genomes by analyzing the methylation status at the DMRs of the imprinted genes H19, IG-DMR (Gtl2), Rasgrf1, Nespas, Gnas1A, Peg10, Peg1/Mest, Peg3, Snrpn, Lit1/Kcnq1ot1, Zac1/Plagl1, U2af1-rs1/Zrsr1, Igf2r (DMR2) and Impact. Representative results for one paternal DMR, IG-DMR (Gtl2), and one maternal DMR, Lit1, are shown (Supplementary Figure S1A). Both stem cells maintained the correct DNA methylation marks all the DMRs except the H19 DMR, which was hypermethylated in both genomes.
Next, we used antibodies specific for 5-methyl-cytosine to isolate methylated DNA from mouse PG- and AG-derived cells and also from sperm. We used quantitative real-time-polymerase chain reaction to assay for the presence of the known DMRs within the immunoprecipitated material using input DNA as a control. The paternal DMRs of H19, IG-DMR (Gtl2) and Rasgrf1 were amplified by real-time-PCR from both AG-derived cell and sperm meDIP samples (Supplementary Figure S1B). The H19 DMR was amplified from both the AG- and the PG-derived cell samples. The maternal DMRs of Nespas, Peg10, Peg1, Peg3, Lit1, U2af1-rs1 and Igf2r (DMR2) were amplified from the materials of the meDIP PG-derived cells. We additionally examined sequences where both maternal and paternal alleles were methylated, including Nanog, Rest, Aicda, Tdrd12, Gdf3 and Slc2a3 (Aicda and Tdrd12 were unmethylated in sperm) and where both alleles were unmethylated, Utf1 (42). In total, the monoparental stem cells maintained the correct parental methylation pattern at over 94% (16/17) of the loci examined. These data indicated that meDIP was effective at isolating known DMRs.
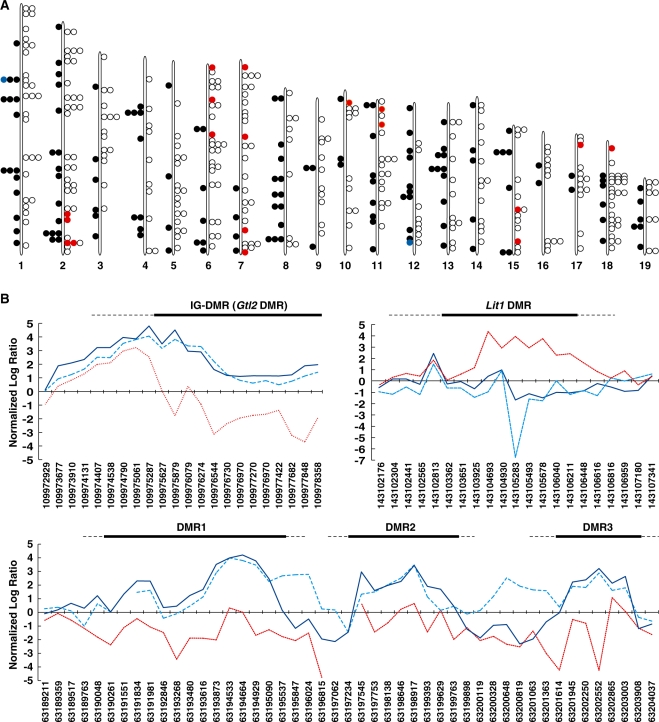
We next performed meDIP-on-chip by applying the meDIP samples to mouse whole-genome tiling arrays. The fixed quantity value that had been obtained from this array analysis corrected the reference value. We looked for the regions under the following conditions: (i) at least three adjoined methylated probes (using neighborhood model supplied by Agilent Technologies Japan, p (Xbar) < 0.07) and (ii) a similar methylation pattern between AG-derived cells and sperm but dissimilar to PG-derived cells (normalized log ratio of the PG-derived cells probe < 0.5). We identified 458 candidate DMRs in the mouse genome. 141 were paternally methylated DMRs and 317 were maternally methylated DMRs (Figure 1, Tables 1 and 2). Of these, 20 were known DMRs. We correctly identified the IG-DMR (Gtl2) and Lit1 DMRs using the tiling arrays for mouse chromosome 7 and 12 (Figure 1B, upper panel). Using the tiling array for chromosome 1, we found the evidence of three closely linked paternally methylated DMRs (Figure 1B, lower panel) that lay within a 60 kb region between the imprinted Zdbf2 (zinc finger, DBF-type containing 2) gene and the uncharacterized gene, Gpr1 (G-protein-coupled receptor 1) (GenBank accession number NM146250) (Figure 2A). We had previously identified Zdbf2 as an imprinted gene linked to a DMR in a parallel study isolating imprinted genes based on their expression status (26). Not all the known DMRs were identified. The Ras/Grf1 DMR could not be identified because the sequence for this region had been excluded from the mouse tiling array due to its highly repetitive sequence. The H19 DMR was also not identified in the screen and this was most likely because the H19 DMR was methylated in both ADS and PDS material, as determined by COBRA, and therefore amplified from both meDIP samples. Nonetheless, these data on known DMRs indicated that meDIP would be an effective technique for identifying novel DMRs.
Figure 1.
Whole mouse genome meDIP-on-chip and genome tiling array screen for DMRs using PG- and AG-derived stem cells and sperm. (A) Chromosome map shows the position of all paternally methylated and maternally methylated DMRs. Red circles to the left-hand side of each chromosome indicate known maternal DMRs and blue circles to the right-hand side of each chromosome indicate known paternal DMRs. Open circles indicate novel maternally methylated DMRs and closed circles indicate novel paternally methylated DMRs. (B) The methylation pattern of the meDIP-on-chip assay. (Upper panel) IG-DMR (Gtl2) paternal DMR (left) and Lit1 maternal DMR (right). (Lower panel) Three paternally methylated DMRs between Gpr1 and Zdbf2: DMR1, DMR2 and DMR3. The longitudinal axes indicate normalized log ratio (log2 Cy5-labeling meDIP DNA fragments/Cy3-labeling whole genome DNA fragments DNA), which represents the methylation degree. The numbers of horizontal axes indicate 5′-flanking base position of the tiling array probe in mouse genome browser mm8 assembly which were obtained from the build 36 ‘essentially complete’ assembly by National Center for Biotechnology Information and the Mouse Genome Sequencing Consortium. Blue solid, aqua broken and red dotted lines represent meDIP-on-chip data of sperm, AG- and PG-derived cells samples, respectively. Black lines indicate the position of IG-DMR (Gtl2) and Lit1 DMR.
Table 1.
Paternal-allele methylated DMR candidates
| Candidate No. | Position (mm8) | Size (kb) | P (Xbar) | Candidate No. | Position (mm8) | Size (kb) | P (Xbar) |
|---|---|---|---|---|---|---|---|
| 1 | chr1:033685996-033687689 | 1.7 | 0.027230699 | 72 | chr8:095022687-095024090 | 1.4 | 0.05286681 |
| 2 | chr1:063193268-063195587 | 2.3 | 0.0218758 | 73 | chr8:111610219-111613561 | 3.3 | 0.057668444 |
| 3 | chr1:063197753-063199822 | 2.1 | 0.042529337 | 74 | chr8:123394925-123395984 | 1.1 | 0.03046366 |
| 4 | chr1:063201945-063203062 (A) | 1.1 | 0.038459964 | 75 | chr8:123960459-123965990 | 5.5 | 0.035588805 |
| 5 | chr1:064666213-064668088 | 1.9 | 0.042587895 | 76 | chr8:124005188-124009612 | 4.4 | 0.038051125 |
| 6 | chr1:075329727-075332982 | 3.3 | 0.022909729 | 77 | chr9:061121907-061124402 | 2.5 | 0.036127605 |
| 7 | chr1:078022275-078024393 | 2.1 | 0.027050465 | 78 | chr9:061182468-061184945 | 2.5 | 0.05035278 |
| 8 | chr1:090499895-090501007 | 1.1 | 0.028617026 | 79 | chr9:119419980-119420779 | 0.8 | 0.025072549 |
| 9 | chr1:133799192-133800261 | 1.1 | 0.02696383 | 80 | chr10:011096967-011097931 | 1.0 | 0.03095065 |
| 10 | chr1:134023790-134025706 | 1.9 | 0.030393751 | 81 | chr10:056062305-056063184 | 0.9 | 0.02961503 |
| 11 | chr1:134149907-134150736 | 0.8 | 0.03685552 | 82 | chr10:060206859-060208255 | 1.4 | 0.04117605 |
| 12 | chr1:138490891-138491572 | 0.7 | 0.04051165 | 83 | chr11:003237473-003238310 | 0.8 | 0.027867135 |
| 13 | chr1:154855403-154856207 | 0.8 | 0.032803357 | 84 | chr11:007322890-007324637 | 1.7 | 0.03812329 |
| 14 | chr1:169125711-169127401 | 1.7 | 0.021216722 | 85 | chr11:032858949-032860475 | 1.5 | 0.02232835 |
| 15 | chr1:182770196-182772623 | 2.4 | 0.03033735 | 86 | chr11:063713890-063716353 | 2.5 | 0.068963565 |
| 16 | chr1:186798464-186800148 | 1.7 | 0.025820382 | 87 | chr11:069415271-069416977 | 1.7 | 0.030176075 |
| 17 | chr2:010252113-010252960 | 0.8 | 0.034892585 | 88 | chr11:084344242-084346009 | 1.8 | 0.06344608 |
| 18 | chr2:032446637-032448262 | 1.6 | 0.055148385 | 89 | chr11:095079200-095080564 | 1.4 | 0.029354626 |
| 19 | chr2:044359572-044360465 | 0.9 | 0.04610098 | 90 | chr11:098780556-098782087 | 1.5 | 0.048574876 |
| 20 | chr2:052779932-052781936 | 2.0 | 0.0356707 | 91 | chr11:120056577-120057507 | 0.9 | 0.03450692 |
| 21 | chr2:071545340-071547095 | 1.8 | 0.027983196 | 92 | chr12:009601787-009602759 | 1.0 | 0.04254318 |
| 22 | chr2:101555546-101557244 | 1.7 | 0.03754241 | 93 | chr12:029403118-029403923 | 0.8 | 0.029047519 |
| 23 | chr2:105485539-105487500 | 2.0 | 0.033533122 | 94 | chr12:040508958-040510065 | 1.1 | 0.04656238 |
| 24 | chr2:115764618-115765843 | 1.2 | 0.023765821 | 95 | chr12:045433938-045434465 | 0.5 | 0.018067254 |
| 25 | chr2:118589797-118591157 | 1.4 | 0.044992935 | 96 | chr12:070676298-070678203 | 1.9 | 0.035833355 |
| 26 | chr2:143855825-143857111 | 1.3 | 0.043339927 | 97 | chr12:073969064-073971035 | 2.0 | 0.01724685 |
| 27 | chr2:157666102-157666938 | 0.8 | 0.025987396 | 98 | chr12:076990926-076992593 | 1.7 | 0.038976375 |
| 28 | chr2:164902535-164903153 | 0.6 | 0.024327435 | 99 | chr12:076999983-077001402 | 1.4 | 0.010916126 |
| 29 | chr2:165569294-165572201 | 2.9 | 0.014741534 | 100 | chr12:104876896-104879970 | 3.1 | 0.061366655 |
| 30 | chr2:165589044-165591976 | 2.9 | 0.019366147 | 101 | chr12:108768086-108769476 | 1.4 | 0.04249084 |
| 31 | chr2:168529651-168530604 | 1.0 | 0.021744832 | 102 | chr12:109975627-109980439 (B) | 4.8 | 0.020096103 |
| 32 | chr2:172816442-172817394 | 1.0 | 0.027761048 | 103 | chr13:019379713-019380663 | 1.0 | 0.04339324 |
| 33 | chr3:032093257-032094758 | 1.5 | 0.0346579 | 104 | chr13:040724758-040725660 | 0.9 | 0.04800732 |
| 34 | chr3:088058689-088060627 | 2.0 | 0.053832173 | 105 | chr13:044715633-044717473 | 1.8 | 0.028829992 |
| 35 | chr3:102212197-102213932 | 1.7 | 0.053714477 | 106 | chr13:044593235-044595857 | 2.6 | 0.039124887 |
| 36 | chr3:127453428-127458227 | 4.8 | 0.04294138 | 107 | chr13:053240493-053244189 | 3.7 | 0.04368776 |
| 37 | chr3:131091297-131092327 | 1.0 | 0.03170784 | 108 | chr13:053424451-053429206 | 4.8 | 0.06249129 |
| 38 | chr3:147900692-147901491 | 0.8 | 0.037227307 | 109 | chr13:053443843-053444926 | 1.1 | 0.033136632 |
| 39 | chr4:044273005-044274686 | 1.7 | 0.0342338 | 110 | chr13:060625134-060626135 | 1.0 | 0.041249607 |
| 40 | chr4:045675124-045675950 | 0.8 | 0.02793303 | 111 | chr13:077617432-077618221 | 0.8 | 0.041074943 |
| 41 | chr4:045785001-045787233 | 2.2 | 0.016951019 | 112 | chr13:098168444-098169681 | 1.2 | 0.038340382 |
| 42 | chr4:045790862-045793054 | 2.2 | 0.05020369 | 113 | chr13:115171670-115172595 | 09 | 0.020752199 |
| 43 | chr4:064738195-064739382 | 1.2 | 0.027920863 | 114 | chr14:010460554-010462803 | 2.2 | 0.049559623 |
| 44 | chr4:128217514-128218988 | 1.5 | 0.04806063 | 115 | chr14:047675151-047676493 | 1.3 | 0.021633925 |
| 45 | chr4:129251321-129253174 | 1.8 | 0.04603652 | 116 | chr14:054031699-054033685 | 2.0 | 0.034764 |
| 46 | chr4:134438654-134439691 | 1.0 | 0.036609355 | 117 | chr14:098222686-098223268 | 0.6 | 0.035441127 |
| 47 | chr4:139097297-139098542 | 1.2 | 0.031863578 | 118 | chr14:121390692-121394556 | 3.9 | 0.025694156 |
| 48 | chr5:023937045-023939114 | 2.1 | 0.06268672 | 119 | chr15:007427165-007427914 | 0.7 | 0.025462002 |
| 49 | chr5:093884506-093886995 | 2.5 | 0.048833344 | 120 | chr15:025697417-025700122 | 2.7 | 0.016209736 |
| 50 | chr5:131881178-131882657 | 1.5 | 0.04750135 | 121 | chr15:025706271-025707476 | 1.2 | 0.01838927 |
| 51 | chr5:147464697-147466931 | 2.2 | 0.022693422 | 122 | chr15:025718408-025718933 | 0.5 | 0.027751224 |
| 52 | chr6:052109421-052111561 | 2.1 | 0.030017477 | 123 | chr15:053006538-053007526 | 1.0 | 0.027212601 |
| 53 | chr6:054026130-054027411 | 1.3 | 0.035516605 | 124 | chr15:102070682-102072718 | 2.0 | 0.06376759 |
| 54 | chr6:097066893-097068534 | 1.6 | 0.034522403 | 125 | chr15:102830007-102831360 | 1.4 | 0.031679183 |
| 55 | chr6:122672646-122673998 | 1.4 | 0.03949519 | 126 | chr16:030125324-030126481 | 1.2 | 0.04029876 |
| 56 | chr6:140298497-140300873 | 2.4 | 0.025882334 | 127 | chr16:043719336-043720459 | 1.1 | 0.024739949 |
| 57 | chr6:144207949-144209008 | 1.1 | 0.032060467 | 128 | chr17:035110303-035112872 | 2.6 | 0.04365454 |
| 58 | chr6:145073228-145074452 | 1.2 | 0.043128457 | 129 | chr17:046081814-046082662 | 0.8 | 0.032995045 |
| 59 | chr7:099433113-099434110 | 1.0 | 0.021295292 | 130 | chr18:034674166-034677415 | 3.2 | 0.023786515 |
| 60 | chr7:118306647-118310770 | 4.1 | 0.05059889 | 131 | chr18:036410061-036411625 | 1.6 | 0.04972436 |
| 61 | chr7:132646656-132648366 | 1.7 | 0.03798946 | 132 | chr18:038504756-038506451 | 1.7 | 0.028145561 |
| 62 | chr7:140825063-140826533 | 1.5 | 0.033876684 | 133 | chr18:053592000-053594251 | 2.3 | 0.048128698 |
| 63 | chr8:008487401-008490250 | 2.8 | 0.035363488 | 134 | chr18:064469013-064469974 | 1.0 | 0.048328057 |
| 64 | chr8:012503705-012505149 | 1.4 | 0.041198887 | 135 | chr18:081142034-081143948 | 1.9 | 0.038805358 |
| 65 | chr8:046469489-046470833 | 1.3 | 0.04832795 | 136 | chr19:010297511-010299422 | 1.9 | 0.04160669 |
| 66 | chr8:060216840-60219015 | 2.2 | 0.054875467 | 137 | chr19:025669277-025670970 | 1.7 | 0.045235418 |
| 67 | chr8:074455057-074456587 | 1.5 | 0.03862939 | 138 | chr19:037879058-037880854 | 1.8 | 0.057981245 |
| 68 | chr8:074616339-074617293 | 1.0 | 0.04343615 | 139 | chr19:041064820-041068690 | 3.9 | 0.034352854 |
| 69 | chr8:087664692-087665701 | 1.0 | 0.051280405 | 140 | chr19:042462156-042463219 | 1.1 | 0.02959308 |
| 70 | chr8:087667382-087671399 | 4.0 | 0.046456236 | 141 | chr19:057171672-057172217 | 0.5 | 0.03625847 |
| 71 | chr8:094959358-094962722 | 3.4 | 0.044347655 |
P (Xbar) indicates p (Xbar) value of the most significant probe in its region. (A) Zdbf2 DMR and (B) IG-DMR (Gtl2).
Table 2.
Maternal-allele methylated DMR candidates
| Candidate No. | Position (mm8) | Size (kb) | P (Xbar) | Candidate No. | Position (mm8) | Size (kb) | P (Xbar) |
|---|---|---|---|---|---|---|---|
| 1 | chr1:004677405-004680223 | 2.8 | 0.0237986 | 81 | chr4:056833933-056836223 | 2.3 | 0.022042342 |
| 2 | chr1:005905383-005908212 | 2.8 | 0.018068576 | 82 | chr4:060007834-060010504 | 2.7 | 0.027354108 |
| 3 | chr1:013109934-013114495 | 4.6 | 0.019053403 | 83 | chr4:082007129-082010469 | 3.3 | 0.022314303 |
| 4 | chr1:014902821-014905844 | 3.0 | 0.022682365 | 84 | chr4:103116841-103119256 | 2.4 | 0.031934537 |
| 5 | chr1:024538601-024542424 | 3.8 | 0.020918498 | 85 | chr4:107903819-107903877 | 1.6 | 0.026745262 |
| 6 | chr1:032116554-032116613 | 1.8 | 0.026415968 | 86 | chr4:125827000-125827059 | 1.6 | 0.033597253 |
| 7 | chr1:036626972-036628483 | 1.5 | 0.022979708 | 87 | chr4:153741937-153743571 | 1.6 | 0.029771574 |
| 8 | chr1:036715035-036716800 | 1.8 | 0.021054856 | 88 | chr4:154495756-154498309 | 2.6 | 0.022843158 |
| 9 | chr1:040269834-040271457 | 1.6 | 0.021841165 | 89 | chr5:037624391-037627129 | 2.7 | 0.030124942 |
| 10 | chr1:058656604-058658693 | 2.1 | 0.021467961 | 90 | chr5:064090611-064092787 | 2.2 | 0.032423064 |
| 11 | chr1:063133112-063135558 | 2.4 | 0.018698972 | 91 | chr5:071978637-071980584 | 1.9 | 0.03554431 |
| 12 | chr1:069309424-069312191 | 2.8 | 0.019993572 | 92 | chr5:077929980-077934343 | 4.4 | 0.027385928 |
| 13 | chr1:069759227-069760723 | 1.5 | 0.024084114 | 93 | chr5:078188911-078192010 | 3.1 | 0.045278415 |
| 14 | chr1:074749647-074750852 | 1.2 | 0.02189668 | 94 | chr5:082095599-082097977 | 2.4 | 0.027626283 |
| 15 | chr1:074883540-074885912 | 2.4 | 0.023577677 | 95 | chr5:101886786-101889296 | 2.5 | 0.028704671 |
| 16 | chr1:082165204-082169216 | 4.0 | 0.021599824 | 96 | chr5:106800163-106801479 | 1.3 | 0.027590053 |
| 17 | chr1:091763096-091766857 | 3.8 | 0.0240105 | 97 | chr5:107927323-107929481 | 2.2 | 0.033987135 |
| 18 | chr1:093630290-093634089 | 3.8 | 0.02066035 | 98 | chr5:110582368-110584177 | 1.8 | 0.029948255 |
| 19 | chr1:122427617-122432992 | 5.4 | 0.01444563 | 99 | chr5:116343153-116344638 | 1.5 | 0.02797624 |
| 20 | chr1:122440765-122442941 | 2.2 | 0.018751323 | 100 | chr5:119921542-119924672 | 3.1 | 0.03576451 |
| 21 | chr1:123354835-123356653 | 1.8 | 0.023314446 | 101 | chr5:120063522-120065663 | 2.1 | 0.028085513 |
| 22 | chr1:136329040-136330902 | 1.9 | 0.018591803 | 102 | chr5:127531574-127533303 | 1.7 | 0.028648317 |
| 23 | chr1:155966454-155968446 | 2.0 | 0.015548464 | 103 | chr5:135534407-135535387 | 1.0 | 0.020641953 |
| 24 | chr1:156487452-156488820 | 1.4 | 0.01905871 | 104 | chr5:136172390-136173572 | 1.2 | 0.022459375 |
| 25 | chr1:161880204-161881805 | 1.6 | 0.038172938 | 105 | chr5:139072890-139074587 | 1.7 | 0.016678514 |
| 26 | chr1:166956508-166959120 | 2.6 | 0.016881404 | 106 | chr5:146893529-146896066 | 2.5 | 0.02011744 |
| 27 | chr1:174212164-174214785 | 2.6 | 0.022220261 | 107 | chr6:004695854-004698573 | 2.7 | 0.019778943 |
| 28 | chr1:189305059-189311596 | 6.5 | 0.017962102 | 108 | chr6:007503950-007507696 | 3.8 | 0.023743302 |
| 29 | chr2:009813042-009815047 | 2.0 | 0.022884484 | 109 | chr6:017229969-017232226 | 2.3 | 0.025952324 |
| 30 | chr2:011207696-011209104 | 1.4 | 0.01718148 | 110 | chr6:017979125-017983000 | 3.9 | 0.017993594 |
| 31 | chr2:024916269-024918291 | 2.0 | 0.022236267 | 111 | chr6:021165838-021167511 | 1.7 | 0.030279916 |
| 32 | chr2:025016418-025018399 | 2.0 | 0.019263456 | 112 | chr6:021935187-021937334 | 2.1 | 0.02010118 |
| 33 | chr2:025397984-025402571 | 4.6 | 0.014632969 | 113 | chr6:030682464-030689996 | 7.5 | 0.01629422 |
| 34 | chr2:029573050-029574474 | 1.4 | 0.026143976 | 114 | chr6:034850411-034851986 | 1.6 | 0.017477673 |
| 35 | chr2:033788267-033789456 | 1.2 | 0.019592382 | 115 | chr6:036317254-036319844 | 2.6 | 0.016221117 |
| 36 | chr2:035104192-035104243 | 1.1 | 0.028701099 | 116 | chr6:043237998-043240078 (E) | 2.1 | 0.03237432 |
| 37 | chr2:051793296-051796677 | 3.4 | 0.018992666 | 117 | chr6:058835661-058837219 (F) | 1.6 | 0.027194222 |
| 38 | chr2:061604035-061608674 | 4.6 | 0.020744983 | 118 | chr6:065310430-065311666 (G) | 1.2 | 0.021420863 |
| 39 | chr2:062211510-062213411 | 1.9 | 0.023512473 | 119 | chr6:067304657-067306131 | 1.5 | 0.03576346 |
| 40 | chr2:062446005-062447593 | 1.6 | 0.020700894 | 120 | chr6:067460849-067466104 | 5.3 | 0.026164446 |
| 41 | chr2:069685551-069687058 | 1.5 | 0.030491233 | 121 | chr6:067463404-067466104 | 2.7 | 0.015576691 |
| 42 | chr2:070367063-070369555 | 2.5 | 0.018729487 | 122 | chr6:071303159-071304733 | 1.6 | 0.020830877 |
| 43 | chr2:070919022-070920942 | 1.9 | 0.01798238 | 123 | chr6:073398337-073400658 | 2.3 | 0.041336037 |
| 44 | chr2:076138655-076140328 | 1.7 | 0.01826188 | 124 | chr6:079950693-079953626 | 2.9 | 0.025820898 |
| 45 | chr2:105024797-105026056 | 1.3 | 0.018905096 | 125 | chr6:082367687-082369045 | 1.4 | 0.023050921 |
| 46 | chr2:105460287-105463427 | 3.1 | 0.016755583 | 126 | chr6:083983935-083985438 | 1.5 | 0.023546984 |
| 47 | chr2:110162692-110165055 | 2.4 | 0.02837651 | 127 | chr6:085090127-085092702 | 2.6 | 0.022739487 |
| 48 | chr2:113164875-113167167 | 2.3 | 0.023015061 | 128 | chr6:096129315-096132554 | 3.2 | 0.025416961 |
| 49 | chr2:122328928-122330175 | 1.2 | 0.02046392 | 129 | chr6:097024399-097026153 | 1.8 | 0.029560687 |
| 50 | chr2:125363369-125365775 | 2.4 | 0.032801084 | 130 | chr6:125286598-125287941 | 1.3 | 0.01435962 |
| 51 | chr2:127813988-127816711 | 2.7 | 0.012370981 | 131 | chr6:126128327-126130786 | 2.5 | 0.025953725 |
| 52 | chr2:130267142-130269115 | 2.0 | 0.022524204 | 132 | chr6:136482254-136483949 | 1.7 | 0.018379996 |
| 53 | chr2:132635416-132638005 | 2.6 | 0.013028574 | 133 | chr6:136770656-136774801 | 4.1 | 0.031868864 |
| 54 | chr2:146527611-146531637 | 4.0 | 0.018155145 | 134 | chr6:142658106-142660237 | 2.1 | 0.016816275 |
| 55 | chr2:148097997-148100499 | 2.5 | 0.009921394 | 135 | chr7:006332204-006336197 (H) | 4.0 | 0.013280352 |
| 56 | chr2:152377240-152379677 (A) | 2.4 | 0.010118501 | 136 | chr7:016103614-016106029 | 2.4 | 0.014163033 |
| 57 | chr2:157250325-157252581 (B) | 2.3 | 0.008028563 | 137 | chr7:016150455-016153988 | 3.5 | 0.014222109 |
| 58 | chr2:170218228-170220655 | 2.4 | 0.0194618 | 138 | chr7:018211612-018213729 | 2.1 | 0.017967263 |
| 59 | chr2:173928712-173931558 | 2.8 | 0.009727121 | 139 | chr7:024238192-024242555 | 4.4 | 0.016059337 |
| 60 | chr2:173937217-173941051 (C) | 3.8 | 0.009931667 | 140 | chr7:024293881-024297190 | 3.3 | 0.019919932 |
| 61 | chr2:173968974-173971383 (D) | 2.4 | 0.007812512 | 141 | chr7:029830009-029831950 | 1.9 | 0.012292666 |
| 62 | chr2:178336086-178338064 | 2.0 | 0.019697197 | 142 | chr7:035245768-035246777 | 1.0 | 0.021291312 |
| 63 | chr2:181599626-181601824 | 2.2 | 0.013717825 | 143 | chr7:043474646-043476273 | 1.6 | 0.012648921 |
| 64 | chr3:007594593-007598083 | 3.5 | 0.016771285 | 144 | chr7:046243531-046246986 | 3.4 | 0.01994469 |
| 65 | chr3:034831468-034834969 | 3.5 | 0.031780172 | 145 | chr7:049626297-049628167 | 1.9 | 0.02512963 |
| 66 | chr3:036862151-036862198 | 1.8 | 0.024218604 | 146 | chr7:059882077-059882135 (I) | 2.5 | 0.014637309 |
| 67 | chr3:046541933-046543604 | 1.7 | 0.02516645 | 147 | chr7:067677277-067678725 | 1.4 | 0.01958729 |
| 68 | chr3:054719962-054723493 | 3.5 | 0.023525374 | 148 | chr7:068137247-068139281 | 2.0 | 0.034075223 |
| 69 | chr3:055269547-055271782 | 2.2 | 0.020089474 | 149 | chr7:075181242-075183250 | 2.0 | 0.014702246 |
| 70 | chr3:055867538-055872400 | 4.9 | 0.02141071 | 150 | chr7:089507231-089508988 | 1.8 | 0.03743501 |
| 71 | chr3:065753115-065756368 | 3.3 | 0.033186894 | 151 | chr7:100051451-100053814 | 2.4 | 0.014619963 |
| 72 | chr3:067551215-067551270 | 1.5 | 0.02275754 | 152 | chr7:110417089-110421405 | 4.3 | 0.019979531 |
| 73 | chr3:079927589-079929496 | 1.9 | 0.02613796 | 153 | chr7:111948575-111950637 | 2.1 | 0.015058612 |
| 74 | chr3:079971586-079973377 | 1.8 | 0.026952958 | 154 | chr7:114353321-114354903 | 1.6 | 0.023320151 |
| 75 | chr3:084389767-084391322 | 1.6 | 0.02515726 | 155 | chr7:122461720-122463398 | 1.7 | 0.026265722 |
| 76 | chr3:087881593-087883247 | 1.7 | 0.027132021 | 156 | chr7:128478998-128481285 (J) | 2.3 | 0.020451799 |
| 77 | chr3:108023194-108024627 | 1.4 | 0.024553038 | 157 | chr7:126327181-126328933 | 1.8 | 0.018874127 |
| 78 | chr3:126354778-126357616 | 2.8 | 0.02315601 | 158 | chr7:139434397-139437287 | 2.9 | 0.016238498 |
| 79 | chr4:021856794-021857823 | 1.0 | 0.030982286 | 159 | chr7:139829080-139831032 | 2.0 | 0.027838653 |
| 80 | chr4:044730990-044732806 | 1.8 | 0.022573303 | 160 | chr7:140772576-140774926 | 2.4 | 0.019183591 |
| 161 | chr7:141142723-141144797 | 2.1 | 0.02639694 | 240 | chr14:083252269-083257004 | 4.7 | 0.011833122 |
| 162 | chr7:143103651-143106675 (K) | 3.0 | 0.016338563 | 241 | chr14:102728834-102730676 | 1.8 | 0.015235411 |
| 163 | chr8:004204874-004207016 | 2.1 | 0.022798432 | 242 | chr14:104990563-104993500 | 2.9 | 0.019385036 |
| 164 | chr8:026413587-026415169 | 1.6 | 0.03148531 | 243 | chr14:107793504-107797494 | 4.0 | 0.016853297 |
| 165 | chr8:027459206-027460587 | 1.4 | 0.032607652 | 244 | chr14:116969080-116973383 | 4.3 | 0.013358023 |
| 166 | chr8:038199721-038202173 | 2.5 | 0.026130743 | 245 | chr15:006526293-006528390 | 2.1 | 0.020619694 |
| 167 | chr8:044803368-044806622 | 3.3 | 0.043445475 | 246 | chr15:006821080-006822833 | 1.8 | 0.012574255 |
| 168 | chr8:071204755-071207122 | 2.4 | 0.021933837 | 247 | chr15:010121536-010124016 | 2.5 | 0.016022267 |
| 169 | chr8:122877933-122878642 | 0.7 | 0.030691648 | 248 | chr15:044616875-044618897 | 2.0 | 0.014585087 |
| 170 | chr8:124158092-124159752 | 1.7 | 0.027747734 | 249 | chr15:061866956-061868107 | 1.2 | 0.015759477 |
| 171 | chr8:124353331-124355356 | 2.0 | 0.027783262 | 250 | chr15:072372513-072373586 | 1.1 | 0.019629903 |
| 172 | chr9:021087872-021089270 | 1.4 | 0.03632289 | 251 | chr15:072636322-072638800 (O) | 2.5 | 0.017643908 |
| 173 | chr9:037116789-037118411 | 1.6 | 0.020918619 | 252 | chr15:075807678-075810850 | 3.2 | 0.023505678 |
| 174 | chr9:037176612-037180135 | 3.5 | 0.027288798 | 253 | chr15:079431697-079433748 | 2.1 | 0.030031314 |
| 175 | chr9:039941858-039944124 | 2.3 | 0.036523227 | 254 | chr15:085473004-085473063 | 1.6 | 0.020536428 |
| 176 | chr9:050502657-050504704 | 2.0 | 0.03374793 | 255 | chr15:096882333-096884933 (P) | 2.6 | 0.017182048 |
| 177 | chr9:057300197-057303815 | 3.6 | 0.018607844 | 256 | chr15:101240038-101241546 | 1.5 | 0.03318241 |
| 178 | chr9:076107910-076110030 | 2.1 | 0.026395189 | 257 | chr15:101971141-101973609 | 2.5 | 0.020543724 |
| 179 | chr9:078180813-078182487 | 1.7 | 0.025889887 | 258 | chr16:017489943-017489987 | 0.9 | 0.01920948 |
| 180 | chr9:082941778-082943974 | 2.2 | 0.025293902 | 259 | chr16:021246116-021248115 | 2.0 | 0.026626676 |
| 181 | chr9:102361832-102363332 | 1.5 | 0.020740993 | 260 | chr16:029128734-029130726 | 2.0 | 0.027809 |
| 182 | chr9:108197365-108199028 | 1.7 | 0.025672207 | 261 | chr16:030429910-030429969 | 1.2 | 0.0295966 |
| 183 | chr10:012779656-012782167 (L) | 2.5 | 0.019184785 | 262 | chr16:031853152-031855686 | 2.5 | 0.023718907 |
| 184 | chr10:020413715-020416568 | 2.9 | 0.01953244 | 263 | chr16:036479910-036481593 | 1.7 | 0.020951904 |
| 185 | chr10:021379027-021380617 | 1.6 | 0.0238754 | 264 | chr16:091108135-091109901 | 1.8 | 0.037090495 |
| 186 | chr10:022333858-022335802 | 1.9 | 0.017312726 | 265 | chr16:091112310-091115789 | 3.5 | 0.030529456 |
| 187 | chr10:022505019-022510288 | 5.3 | 0.017806638 | 266 | chr16:091947817-091948515 | 0.7 | 0.039174825 |
| 188 | chr10:024283881-024285477 | 1.6 | 0.016753925 | 267 | chr16:092582773-092585646 | 2.9 | 0.028216336 |
| 189 | chr10:073216320-073218166 | 1.8 | 0.047033958 | 268 | chr16:097764922-097766838 | 1.9 | 0.024059681 |
| 190 | chr10:082295933-082297899 | 2.0 | 0.03255051 | 269 | chr17:005374631-005376776 | 2.1 | 0.03132879 |
| 191 | chr10:097122222-097124647 | 2.4 | 0.025589569 | 270 | chr17:009511762-009513773 | 2.0 | 0.024981726 |
| 192 | chr10:099187071-099189870 | 2.8 | 0.02582734 | 271 | chr17:012584437-012586532 (Q) | 2.1 | 0.02412774 |
| 193 | chr10:106887829-106890671 | 2.8 | 0.024832647 | 272 | chr17:031917288-031918966 | 1.7 | 0.036958188 |
| 194 | chr10:126580305-126584057 | 3.8 | 0.0229678 | 273 | chr17:034564671-034568817 | 4.1 | 0.036491122 |
| 195 | chr11:003794275-003795370 | 1.1 | 0.03017051 | 274 | chr17:036596043-036597688 | 1.6 | 0.029813139 |
| 196 | chr11:011925467-011927317 (M) | 1.9 | 0.026753291 | 275 | chr17:036804142-036806133 | 2.0 | 0.032254983 |
| 197 | chr11:016407175-016408969 (N) | 1.8 | 0.028730938 | 276 | chr17:044937669-044938910 | 1.2 | 0.02387914 |
| 198 | chr11:022872000-022874562 | 2.6 | 0.0290943 | 277 | chr17:046191541-046193261 | 1.7 | 0.03355552 |
| 199 | chr11:031268290-031272176 | 3.9 | 0.02519847 | 278 | chr17:047558119-047560713 | 2.6 | 0.03126954 |
| 200 | chr11:053300081-053301551 | 1.5 | 0.015159988 | 279 | chr17:049759295-049760847 | 1.6 | 0.022615742 |
| 201 | chr11:053305814-053310528 | 4.7 | 0.011801407 | 280 | chr17:079521685-079524587 | 2.9 | 0.036173925 |
| 202 | chr11:053870758-053872961 | 2.2 | 0.024789684 | 281 | chr17:093502830-093507935 | 5.1 | 0.020683607 |
| 203 | chr11:057642798-057648684 | 5.9 | 0.01782747 | 282 | chr18:013114801-013118408 (R) | 3.6 | 0.03690424 |
| 204 | chr11:062391207-062393390 | 2.2 | 0.022354733 | 283 | chr18:020143702-020145888 | 2.2 | 0.020389104 |
| 205 | chr11:062604615-062606649 | 2.0 | 0.037107296 | 284 | chr18:034372092-034373598 | 1.5 | 0.0217988 |
| 206 | chr11:069619339-069621611 | 2.3 | 0.015448221 | 285 | chr18:034703079-034704303 | 1.2 | 0.045119096 |
| 207 | chr11:072177596-072178833 | 1.2 | 0.011860856 | 286 | chr18:035787290-035788030 | 0.7 | 0.037418738 |
| 208 | chr11:084772811-084774719 | 1.9 | 0.017397704 | 287 | chr18:037071868-037074704 | 2.8 | 0.019358814 |
| 209 | chr11:085702850-085705929 | 3.1 | 0.019243628 | 288 | chr18:037085878-037089320 | 3.4 | 0.018705504 |
| 210 | chr11:088814584-088816685 | 2.1 | 0.03279886 | 289 | chr18:037117819-037121107 | 3.3 | 0.028756998 |
| 211 | chr11:101961201-101964545 | 3.3 | 0.017420538 | 290 | chr18:037426316-037429037 | 2.7 | 0.02729652 |
| 212 | chr12:037616718-037620453 | 3.7 | 0.00981845 | 291 | chr18:037931616-037934633 | 3.0 | 0.018353485 |
| 213 | chr12:040643123-040643182 | 2.7 | 0.014325851 | 292 | chr18:039899306-039901593 | 2.3 | 0.017372293 |
| 214 | chr12:084898561-084900520 | 2.0 | 0.016897557 | 293 | chr18:042076444-042078040 | 1.6 | 0.022621712 |
| 215 | chr12:096086762-096090337 | 3.6 | 0.009172818 | 294 | chr18:046336482-046338773 | 2.3 | 0.01798738 |
| 216 | chr12:099134675-099136350 | 1.7 | 0.014613676 | 295 | chr18:046480672-046483104 | 2.4 | 0.02476409 |
| 217 | chr12:102219226-102221216 | 2.0 | 0.01636928 | 296 | chr18:046589690-046591475 | 1.8 | 0.019116558 |
| 218 | chr12:105623102-105624458 | 1.4 | 0.014140618 | 297 | chr18:046851351-046854314 | 3.0 | 0.03013274 |
| 219 | chr12:111719113-111720871 | 1.8 | 0.008191496 | 298 | chr18:054120897-054122418 | 1.5 | 0.026858354 |
| 220 | chr13:019400293-019403773 | 3.5 | 0.040705923 | 299 | chr18:062303490-062306672 | 3.2 | 0.02011947 |
| 221 | chr13:019629918-019631961 | 2.0 | 0.022918101 | 300 | chr18:066582220-066586790 | 4.6 | 0.017490398 |
| 222 | chr13:031621242-031624357 | 3.1 | 0.01656243 | 301 | chr18:066984440-066986686 | 2.2 | 0.02194144 |
| 223 | chr13:047204189-047206166 | 2.0 | 0.012271536 | 302 | chr18:069320834-069324481 | 3.6 | 0.022667281 |
| 224 | chr13:065271696-065274388 | 2.7 | 0.017073411 | 303 | chr18:070654510-070657602 | 3.1 | 0.038828205 |
| 225 | chr13:075554829-075557522 | 2.7 | 0.009975607 | 304 | chr18:072474614-072477050 | 2.4 | 0.028672088 |
| 226 | chr13:092214744-092216480 | 1.7 | 0.012622342 | 305 | chr18:077060298-077063598 | 3.3 | 0.0280942 |
| 227 | chr13:096349727-096351467 | 1.7 | 0.012113783 | 306 | chr18:080524705-080527164 | 2.5 | 0.02553843 |
| 228 | chr13:096543879-096546003 | 2.1 | 0.025604498 | 307 | chr18:082539721-082542232 | 2.5 | 0.02821235 |
| 229 | chr13:099736482-099737805 | 1.3 | 0.022977684 | 308 | chr18:086846350-086848475 | 2.1 | 0.032687873 |
| 230 | chr13:113332691-113335190 | 2.5 | 0.023172792 | 309 | chr19:006991711-006994911 | 3.2 | 0.026918497 |
| 231 | chr13:116052081-116054730 | 2.6 | 0.017533854 | 310 | chr19:007678835-007680562 | 1.7 | 0.032486826 |
| 232 | chr14:011077128-011078632 | 1.5 | 0.01759021 | 311 | chr19:009201186-009203180 | 2.0 | 0.031132337 |
| 233 | chr14:015035019-015037201 | 2.2 | 0.033699445 | 312 | chr19:016501850-016504566 | 2.7 | 0.030156422 |
| 234 | chr14:028837272-028839252 | 2.0 | 0.027515797 | 313 | chr19:018816356-018817936 | 1.6 | 0.027075935 |
| 235 | chr14:039801943-039804245 | 2.3 | 0.031648647 | 314 | chr19:037760893-037765203 | 4.3 | 0.025636315 |
| 236 | chr14:054047482-054049683 | 2.2 | 0.03008235 | 315 | chr19:038589068-038591035 | 2.0 | 0.034372117 |
| 237 | chr14:068123705-068125678 | 2.0 | 0.023587078 | 316 | chr19:059520244-059523338 | 3.1 | 0.028666519 |
| 238 | chr14:068289885-068291193 | 1.3 | 0.023119137 | 317 | chr19:060521572-060524423 | 2.9 | 0.029403668 |
| 239 | chr14:073375085-073376336 | 1.3 | 0.03023008 |
P (Xbar) indicates p (Xbar) value of the most significant probe in its region. (A) Mcts2 DMR, (B) Nnat DMR, (C) Nespas DMR, (D) Gnas1A DMR, (E) Peg10 DMR, (F) Peg1 DMR, (G) Nap1l5 DMR, (H) Peg3 DMR, (I) Snrpn DMR, (J) Inpp5f_v2 DMR, (K) Lit1 DMR, (L) Zac1 DMR, (M) Meg1 DMR, (N) U2af1-rs1 DMR, (O) Peg13 DMR, (P) Slc38a4 DMR, (Q) Igf2r DMR2 and (R) Impact DMR.
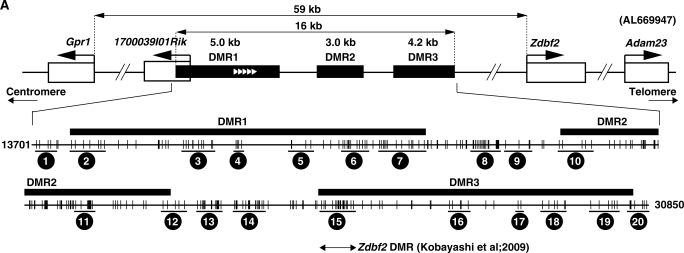
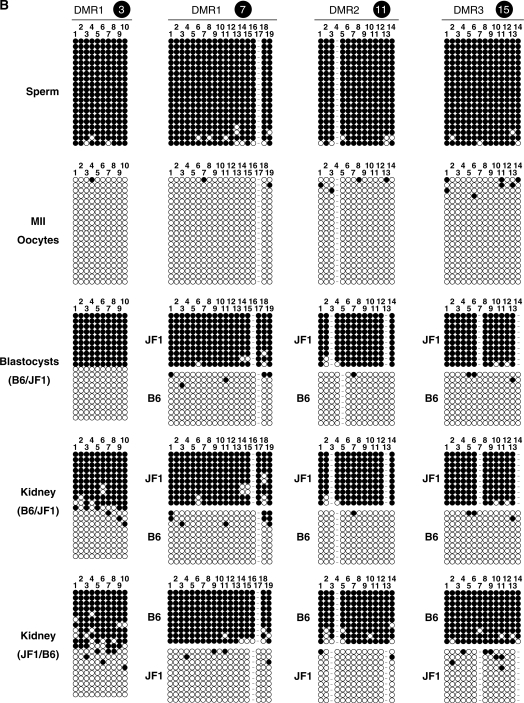
Figure 2.
Three paternally methylated DMRs on the Gpr1-Zdbf2 imprinted domain. (A) Physical map of the mouse Gpr1-Zdbf2 locus (upper panel). Black boxes represent the position of three paternally methylated DMRs, called DMR1 (5.0 kb), DMR2 (3.0 kb) and DMR3 (4.2 kb). Arrows above genes (white boxes) show the direction of transcription. White arrowheads indicate five times repeats of the 37 bp repetitive sequence. Close-up of the three paternally methylated DMRs (lower panel). The vertical bar represents a CpG site. The regions we analyzed bisulfite-PCR methylation sequencings were indicated 1–20. (B) Bisulfite-PCR sequencing results for four regions (regions 3, 7, 11 and 15) on genomic DNA prepared from sperm, MII oocytes, blastocysts from B6/JF1 mice and the kidney from B6/JF1 and reciprocal JF1/B6 mice. Each row represents a unique methylation profile within the pool of 20 clones sequenced. Closed and open circles represent methylated and unmethylated CpGs, respectively.
Paternally methylated DMRs in the Gpr1-Zdbf2 imprinted domain
Within our tiling array, there were three separate regions of differential methylation on chromosome 1 in close proximity. In our very recent study on Zdbf2, we identified one paternally methylated region on chromosome 1 in this vicinity (26). We chose to characterize these three DMRs in greater detail in order to determine their relationship to the Zdbf2 DMR. Using the combined bisulfite-PCR restriction analysis (COBRA) and bisulfite-PCR sequencing, we confirmed that these three DMRs were methylated in genomic DNA from mature sperm and unmethylated in metaphase II (MII) oocytes DNA and differentially methylated in blastocysts DNA from B6/JF1 mice (Figure 2B). Genomic DNA from somatic tissues from B6/JF1 and JF1/B6 embryos at E13.5 and adult mice was assayed by the same methylation protocol. All of the tissues of both adult and embryo, including the liver, lung, heart, kidney, spleen and brain, were differentially methylated and the methylation was reprogrammed in the next generation and stably maintained after tissue differentiation (Supplementary Figure S2A).
We called these paternal DMRs DMR1, DMR2 and DMR3. They were 5.0, 3.0 and 4.2 kb, respectively. None of the DMRs would be defined as CpG islands using the following standard criteria: minimum length 100 bp; GC content > 50%; Obs/Exp CpG > 0.6. Instead, they exhibited a low G + C content (43.5%, 46.6% and 42.4%, respectively) and a low frequency of CpG dinucleotides (CpG observed/expected = 0.22, 0.34 and 0.19, respectively). Analysis of the primary sequence of the DMR1 region revealed five repeats of the 37 bp repetitive sequence. Many imprinted DMRs are characterized by repeat sequence elements. DMR3 contained the 341 bp sequence of the Zdbf2 DMR that we reported previously (26). Further analysis demonstrated that the three DMRs were closely linked within a 16 kb region separated by regions the lacked allele-specific methylation (Supplementary Figures S2B-1, 8, 14 and 20).
The de novo methylation of the DMRs linked to Gpr1-Zdbf2 is dependent on methyltransferase Dnmt3a
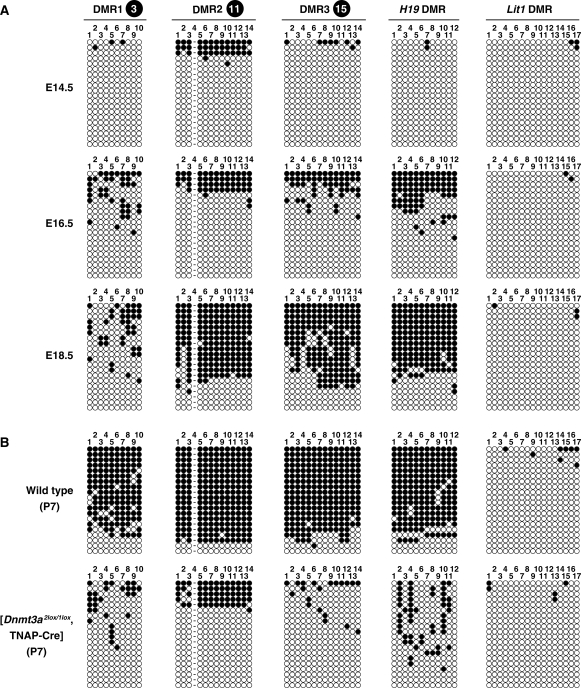
To investigate the developmental changes in methylation at the paternally methylated DMRs in the Gpr1-Zdbf2 domain, we carried out bisulfite-PCR methylation analysis in genomic DNA isolated from male germ cells at E14.5, E16.5 and E18.5. The paternally methylated H19 and the maternally methylated Lit1 DMRs were included as controls. The regions we analyzed and the CpG sites in this study are shown in Figure 2A.
In E14.5 prospermatogonia, DMR2 was ∼15% methylated while DMR1 and DMR3 were unmethylated (Figure 3A). In contrast, the paternally methylated H19 DMR was unmethylated in E14.5 prospermatogonia. This was different to the Kato’s et al. (16) paper that reported that the H19 DMR was hypomethylated (5–15%) in E14.5 prospermatogonia. The maternally methylated Lit1 DMR was almost unmethylated. In E16.5 prospermatogonia, methylation at DMR2 had increased, methylation was observed at DMR1 but methylation at DMR3 was mosaic. In E18.5, methylation of DMR2 and DMR3 further increased but DMR1 methylation was still mosaic. These data suggested that the DMR2 region was the first to acquire DNA methylation followed by DMR3 and then DMR1.
Figure 3.
Methylation acquisition during spermatogenesis and absence of methylation imprints in Dnmt3a-difecient spermatogonia. (A) Methylation status of three Gpr1-Zdbf2 DMRs in E14.5, E16.5 and E18.5 prospermatogonia. As a control, the paternally methylated H19 DMR and the maternally methylated Lit1 DMR were included. The regions we analyzed and the CpG sites in this study are shown in Figure 2A. (B) Methylation profile of DMRs in normal and Dnmt3a-deficient prospermatogonia. Male germ cells at P7 were isolated from testes of normal and the conditional Dnmt3a knockout mice by FACS. The bisulfite-PCR-based assays for the three paternally methylated Gpr1-Zdbf2 and the H19 DMRs and maternally methylated Lit1 DMR.
The de novo methylation of H19 DMR and IG-DMR (Gtl2) is mediated by the de novo methyltransferase Dnmt3a (16). We asked whether the Zdbf2 DMRs were also dependent on Dnmt3a by examining normal and Dnmt3a-deficient prospermatogonia. Male germ cells at P7 were isolated from the testes of the conditional Dnmt3a knockout mice by FACS as previously described (16). We performed the bisulfite-PCR based assays for the paternally methylated DMRs on this material. The degree of methylation in Dnmt3a-deficient prospermatogonia was decreased compared to wild type prospermatogonia (Figure 3B). Similar to H19 DMR and IG-DMR (Gtl2), establishment of the Zdbf2 DMR was dependent on Dnmt3a.
Imprinted genes near Zdbf2
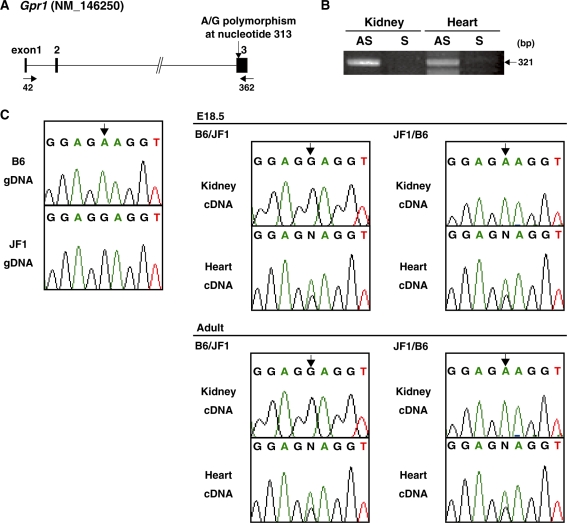
Imprinted genes are commonly clustered within the genome. We therefore sought to determine the imprinting status of the nearby Gpr1 gene. We identified a single nucleotide polymorphism (SNP) in exon 3 of Gpr1 between the B6 and JF1 strains of mice (Figure 4A). We performed allele-specific reverse transcription-PCR (RT-PCR) sequencing analysis using E18.5 tissues obtained from reciprocal crosses between these strains and also adult material. The transcriptional direction of the RT-PCR products was determined by using either sense or antisense primers as primers for cDNA synthesis (Figure 4B, Supplementary Figure S3A). Only the paternal Gpr1 allele was detected in kidney cDNA but in brain, lung, liver, heart, spleen, testis and the placenta Gpr1 was biallelically expressed (Figure 4C and Supplementary Figure S3). We also examined the expressed sequence tag (EST), 1700039l01Rik (GenBank accession number XM 001478509), located ∼40 kb upstream of Gpr1 and overlapping DMR1. This EST consisted of three exons. Using a similar SNP-based assay, we found that the transcript was biallelically expressed in the testis (data not shown). Adam23 (a disintegrin and metallopeptidase domain 23) (GenBank accession number NM 011780), a gene located ∼140 kb downstream of Zdbf2, showed biallelic expression in the all tissues which we examined (data not shown).
Figure 4.
Tissue-specific imprinted expression of mouse Gpr1. (A) Structure of the mouse Gpr1 gene. Exons are shown as filled boxes and primers are indicated by arrows. The DNA polymorphism between B6 and JF1 is indicated by the vertical arrow. (B) Direction expression analysis of mouse Gpr1 gene. The first cDNA strands syntheses were performed using either the sense (S) or the antisense (AS) primer of the mouse Gpr1 gene. Arrow indicates the cDNA product of Gpr1 gene amplified by RT-PCR on right. (C) Allele-specific expression analysis of mouse Gpr1 gene. cDNA and genomic PCR products were amplified and sequenced directly from E18.5 embryos and adult material obtained from a B6/JF1 and reciprocal crossed JF1/B6 mice.
Expression of mouse Zdbf2 and Gpr1
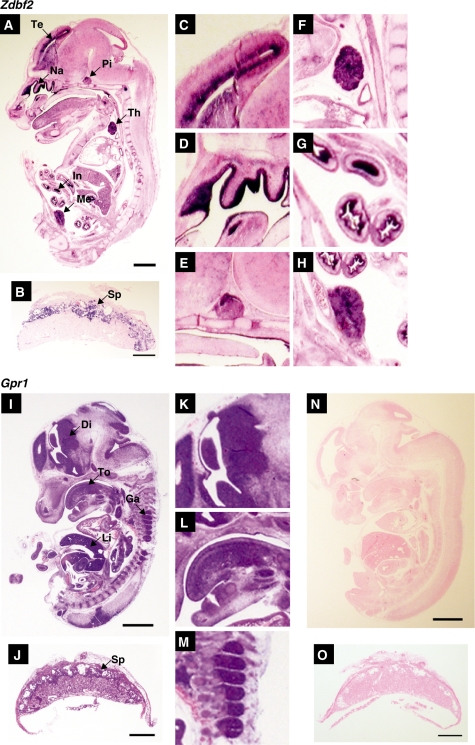
In order to determine whether Zdbf2 and Gpr1 were co-expressed in the same tissues, we examined their expression pattern in E13.5 mouse embryos by in situ hybridization. Zdbf2 was strongly expressed in the mesencephalon, pituitary gland, nasal epithelium, thymus, intestinal epithelium, the mesonephrum in the mouse and in the spongiotrophoblast layer of the placenta (Figure 5A–H). Despite the tissue-specific monoallelic expression of Gpr1 gene, the gene was widely expressed with the strongest expression being in the diencephalon, dorsal root ganglion, tongue, liver in the mouse embryo and in the spongiotrophoblast layer of the placenta (Figure 5I–M).
Figure 5.
Expression of mouse Zdbf2 and Gpr1. The expression of Zdbf2 and Gpr1 were examined in sagittal sections of E13.5 embryo (A and I) and placenta (B and J) by in situ hybridization. Telencephalon: Te (C), nasal epithelium: Na (D), pituitary gland: Pi (E), thymus: Th (F), intestinal epithelium: In (G), mesonephrum: Me (H), the spongiotrophoblast layer of the placenta: Sp (B), diencephalon: Di (K), tongue: To (L), dorsal root ganglion: Ga (M), liver and the spongiotrophoblast layer of the placenta: Sp (J). No signal was seen with the Gpr1 sense probe (N and O). Scale bars indicate 1 mm.
Characterizing the GPR1-ZDBF2 human imprinted domain
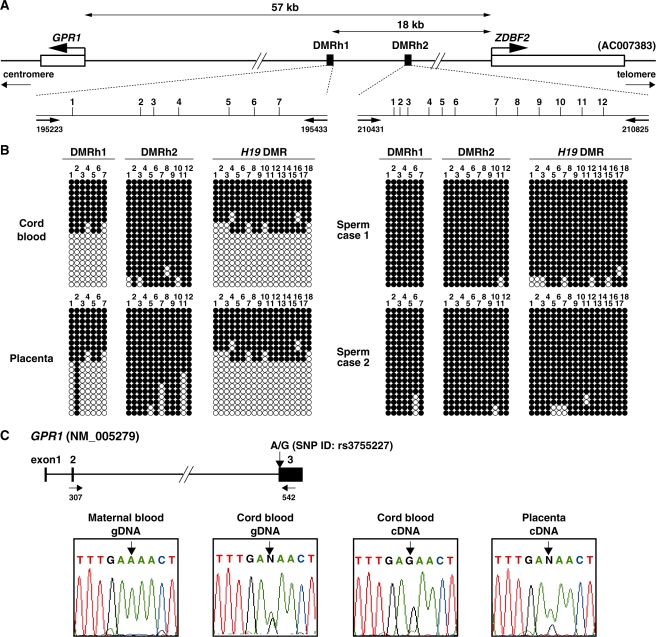
We applied the meDIP-on-chip method to human sperm DNA to isolate paternally methylated human DMRs. We isolated two regions, which we called DMRh1 and DMRh2 (data not shown, Figure 6A). We examined whether these methylated sequences were DMRs by applying the bisulfite-based PCR methylation assay to genomic DNA isolated from human sperm, blood and placenta. We found that DMRh1 was fully methylated in sperm DNA and ∼50% methylated in umbilical cord blood and placental DNA (Figure 6B). In contrast, DMRh2 was fully methylated in all the samples. Part of the DMRh1 sequence (GenBank accession number AC007383; 194613–195967) was similar to part of the mouse DMR1 (GenBank accession number AL669947; 13006–14276) with a 50.6% nucleotide match indicating that we had identified the human homologue of the mouse Zdbf2 DMR (Supplementary Figure S4).
Figure 6.
Paternal allele-specific methylation at the region between GPR1 and ZDBF2 of human chromosome 2 and imprinting of human GPR1. (A) Structure of the human region between GPR1 and ZDBF2. Two methylated regions, DMRh1 and DMRh2, identified in meDIP-on-chip of normal human sperm indicated by filled boxes. The vertical bars represent CpG sites. The horizontal arrows represent primer positions. The extent of the regions analyzed in this study and Genbank accession numbers are shown over the line. (B) Bisulfite-PCR sequencing of genomic DNA prepared from cord blood, placenta and two cases’ sperm. Each row represents a unique methylation profile within the pool of 20 clones sequenced. Closed and open circles represent methylated and unmethylated CpGs, respectively. (C) The paternal-specific expression of GPR1 in human samples. The A/G polymorphic site (SNP ID: rs3755227) in GPR1 exon 3 is indicated by vertical arrow. Heterozygosity was demonstrated in DNA isolated from cord blood with double peaks in chromatographic sequencing data at the polymorphic residues identified (arrow).
Human ZDBF2 is imprinted and expressed only from the paternal allele (26). To determine the allelic expression of GPR1 (GenBank accession number NM 005279) in human material, we identified an SNP within exon 3 of GPR1. We identified the SNPs 3 of 35 cases. We performed RT-PCR analyses on umbilical cord blood and placenta RNA. GPR1 was expressed from only paternal allele in the three all neonatal leukocytes but not in the placenta (Figure 6C). Both the human and the mouse GPR1 genes were imprinted and expressed from the paternal genome.
DISCUSSION
In this paper, we report on a novel DNA methylation-based screen for imprinted genes that resulted in the identification of 458 putative DMRs. Of these, 20 were previously characterized DMRs. Several methods for systematical searching for imprinted methylation regions within the mouse genome have been reported. The representative method is restriction landmark genomic scanning with methylation-sensitive restriction endonuclease (RLGS-M), which identified the U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit (U2afbp-rs) on mouse chromosome 11 (43), and the Grf1/Cdc25Mm on mouse chromosome 9 (44). Another approach based on DNA methylation is called Methylation-sensitive Representational Difference Analysis (Me-RDA/MS-RDA). With this method, two imprinted genes, maternally expressed Nesp and paternally expressed Gnasxl, were identified at the distal end of mouse chromosome 2 (45,46). In another study using two different methylation-sensitive restriction enzymes, Hin6I (HhaI) and HpaII, three imprinted genes were identified. Interestingly, two of these were located within the intronic regions of other genes (24). Recently, the tiling array technology has been successfully applied to decipher chromatin structure (33,35) using chromatin immunoprecipitation (ChIP-chip) (34). A tiling array approach can provide genome-wide profiling of the methylation pattern in a particular sample when used in combination with a methylated DNA binding column specific to methylated CpG sites (36,37), sodium bisulfite modification (47), and/or the antibody against 5-methylcytosine. In this study, we have demonstrated the power of this technique when applied to studies on genomic imprinting.
The paternally methylated DMRs that we identified on mouse chromosome 1 were located near the imprinted gene, Zdbf2. We and another group previously identified Zdbf2 as an imprinted gene in expression-based screens (26,48) thus validating both approaches. The paternally methylated DMR consisted of three distinct methylated regions interspersed with two non-methylated regions. Similar to the paternal DMRs of H19 and IG-DMR (Gtl2), methylation at the three paternally methylated DMRs was present in the male germline but not in the female germline and was dependent on Dnmt3a, suggesting that all three regions are germ line DMRs. We determined that the Gpr1-Zdbf2 paternally methylated region spanned 16 kb, which is the longest DMR so far reported (23). We identified a direct repeat sequence in the first Gpr1-Zdbf2 DMRs. This type of repeat is associated with other imprinted DMRs but its function is still unknown (49,50). When we further characterized the Zdbf2 domain, we found that Gpr1/GPR1, which lies 60 kb from Zdbf2, was also paternally expressed. At the human locus, we identified a single, paternally methylated DMR between GPR1 and ZDBF2, and showed that the human GPR1 gene was also imprinted and paternally expressed in neonatal leukocytes but not in the placenta.
Imprinted genes are regulated by parent-of-origin specific DNA methylation within their DMRs in cis. The DMRs on mouse chromosome 1 are paternally methylated. Paternally methylated DMRs are present at only three other imprinted domains, H19, IG-DMR (Gtl2) and Rasgrf1 DMRs. DMRs function as imprinting centers, controlling the neighboring imprinted genes (51,52). In the case of the H19 DMR, and possibly the IG-DMR (Gtl2), paternal methylation inhibits the expression of the paternal allele via an insulator that operates as a methylation sensitive boundary (53). The Gpr1-Zdbf2 DMRs shows more similarity to the Rasgrf1 DMR as both DNA methylation and active gene expression is from the paternal allele. The imprinted expression of protein coding genes can also be achieved by direct DNA methylation of their promoter (Peg1, Peg3, Zac1) or indirectly, by methylation of the promoter of a long, noncoding antisense RNAs (Lit1, Igf2r) (54). In the latter case, and in the boundary model, imprinting is achieved by an interplay between the maternally and paternally expressed genes. Currently, there is no evidence of a maternally expressed transcript initiating near the Gpr1-Zdbf2 DMRs. However, although the EST (1700039l01Rik) was not imprinted in the tissues and at the time points we tested, we cannot exclude imprinting at a different time point or the presence of other imprinted genes lying at a distance from the DMR for this domain.
This work demonstrates the effectiveness of meDIP-on-chip in identifying DMRs. Chromosome 1 was not identified as containing an imprinted domain based on phenotypic studies in mice with maternal or paternal duplications but two approaches have identified an imprinted locus on this chromosome. Conversely, there are regions on mouse chromosome 2 and 12 where imprinted domains are predicted but for which no candidates have been identified (55). Our method has identified a number of novel DMRs providing candidates for these effects. We still do not know which features of DMRs are the most important for attracting germline methylation. For example, CpG-spacing, the presence of repeats, the genomic context of the DMR or a combination of these factors may be involved. Systematic searches will aid in the characterization of common features of paternal and maternal DMRs. These criteria can then be applied to other genomes, including the human genome, to identify novel DMRs. Our method is also suitable for adaptation for other types of epigenetic modification such as a specific histone modification. The identification of new DMRs and imprinted domains will provide novel insights into the mechanism of imprinting and its biological role in mammals.
This work also identified a new imprinted gene that had not been isolated in any expression-based screen. The mouse Gpr1 encodes a 353 amino acid plasma membrane protein with seven transmembrane domains, which is coupled to the G protein, Gpa2 (56,57). It may therefore play a role in signal transduction. The Gpr1–Gpa2 complex is responsive to glucose and sucrose (58). Several endocrine disorders have been shown to be caused by either loss- or gain-of-function in G proteins or G-protein-coupled receptors (59). GNAS is a complex imprinted locus that produces multiple transcripts. The main transcript derived from GNAS, Gsα, encodes the α-subunit of the stimulatory guanine nucleotide-binding protein. Gsα is expressed biallelically in nearly all tissues and plays essential roles in a multitude of physiologic processes. Other transcripts produced by GNAS are expressed exclusively from either the paternal or the maternal GNAS allele (60,61). The expression in renal proximal tubules occurs predominantly from the maternal allele and this tissue specific imprinting of Gsα is an important role in different kind of pseudohypoparathyroidism (62). We found that the imprinting of Gpr1 was confined to the embryonic and adult kidney. In others tissues, the gene was not imprinted. This might suggest a functional importance for dosage of Gpr1 in the development of the kidney.
In summary, using an meDIP method on the whole mouse genome, we identified 458 regions as putative DMRs. We found that the technique successfully identified the majority of known DMRs. The failure to identify two known DMRs was not related to the technique but due to the nature of these sequences, one being highly repetitive and therefore excluded from the array and the second sequence lacking differential methylation in the stem cell material used in the assay. We also further characterized the mouse Zdbf2 DMR isolated by this technique and found that it had an unusual with a tripartite structure spanning a relatively extensive genomic region. Similar to the H19 DMR and IG-DMR (Gtl2), methylation in the male germline was dependent on Dnmt3a. We have also identified paternal expression of the nearby Gpr1/GPR1 gene in mouse and human. MeDIP is a powerful, cross-genome method for identifying allele-specific epigenetic marks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas ‘Comparative Genomics’ and B from the Ministry of Education, Culture, Sports, Science and Technology of Japan (19390423, 20017003 and 21028003); Japan Science and Technology Agency (JST); Takeda Science Foundation; Children’s Cancer Association of Japan; National Institute of Genetics (NIG) cooperative Research Program (2008-B); Mitsubishi Foundation; and Smoking Research Foundation; Nestle Nutrition Council Japan. The sponsors of the study had no role in study design, data collection, data analysis, data interpretation or writing the report. Funding for open access charge: KAKENHI (Grant-in-Aid for Scientific Research) on B from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21028003).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Mr. H. Furuumi for technical assistant and Clarissa Velayo for support and valuable suggestions.
REFERENCES
- 1.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 2.Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 3.Tilghman SM. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 5.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14(Spec No. 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 6.Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- 7.Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum. Mol. Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- 8.Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 2002;277:5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- 9.Davis TL, Trasler JM, Moss SB, Yang GJ, Bartolomei MS. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics. 1999;58:18–28. doi: 10.1006/geno.1999.5813. [DOI] [PubMed] [Google Scholar]
- 10.Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 11.Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84:952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Ueda T, Abe K, Miura A, Yuzuriha M, Zubair M, Noguchi M, Niwa K, Kawase Y, Kono T, Matsuda Y, et al. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells. 2000;5:649–659. doi: 10.1046/j.1365-2443.2000.00351.x. [DOI] [PubMed] [Google Scholar]
- 13.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 14.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 16.Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 17.Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, Shintani A, Minoshima S, Krohn K, Antonarakis SE, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 18.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 20.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikyo N, Williamson CM, John RM, Barton SC, Beechey CV, Ball ST, Cattanach BM, Surani MA, Peters J. Genetic and functional analysis of neuronatin in mice with maternal or paternal duplication of distal Chr 2. Dev. Biol. 1997;190:66–77. doi: 10.1006/dbio.1997.8681. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- 24.Smith RJ, Dean W, Konfortova G, Kelsey G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 2003;13:558–569. doi: 10.1101/gr.781503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi H, Yamada K, Morita S, Hiura H, Fukuda A, Kagami M, Ogata T, Hata K, Sotomaru Y, Kono T. Identification of the mouse paternally expressed imprinted gene Zdbf2 on chromosome 1 and its imprinted human homolog ZDBF2 on chromosome 2. Genomics. 2009;93:461–472. doi: 10.1016/j.ygeno.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 28.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 29.Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol. Cell. Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 31.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 32.Maeda N, Hayashizaki Y. Genome-wide survey of imprinted genes. Cytogenet. Genome Res. 2006;113:144–152. doi: 10.1159/000090826. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 36.Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat. Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 37.Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP, Freitag M. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa H, Shindo N, Kumagai T, Usami Y, Shikanai M, Jonwn K, Fukuda A, Kawahara M, Sotomaru Y, Tanaka S, et al. Developmental ability of trophoblast stem cells in uniparental mouse embryos. Placenta. 2009;30:448–456. doi: 10.1016/j.placenta.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Koide T, Moriwaki K, Uchida K, Mita A, Sagai T, Yonekawa H, Katoh H, Miyashita N, Tsuchiya K, Nielsen TJ, et al. A new inbred strain JF1 established from Japanese fancy mouse carrying the classic piebald allele. Mamm. Genome. 1998;9:15–19. doi: 10.1007/s003359900672. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum. Mol. Genet. 2007;16:2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 41.Arima T, Drewell RA, Arney KL, Inoue J, Makita Y, Hata A, Oshimura M, Wake N, Surani MA. A conserved imprinting control region at the HYMAI/ZAC domain is implicated in transient neonatal diabetes mellitus. Hum. Mol. Genet. 2001;10:1475–1483. doi: 10.1093/hmg/10.14.1475. [DOI] [PubMed] [Google Scholar]
- 42.Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev. Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashizaki Y, Shibata H, Hirotsune S, Sugino H, Okazaki Y, Sasaki N, Hirose K, Imoto H, Okuizumi H, Muramatsu M, et al. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat. Genet. 1994;6:33–40. doi: 10.1038/ng0194-33. [DOI] [PubMed] [Google Scholar]
- 44.Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, et al. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat. Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- 45.Kelsey G, Bodle D, Miller HJ, Beechey CV, Coombes C, Peters J, Williamson CM. Identification of imprinted loci by methylation-sensitive representational difference analysis: application to mouse distal chromosome 2. Genomics. 1999;62:129–138. doi: 10.1006/geno.1999.6022. [DOI] [PubMed] [Google Scholar]
- 46.Peters J, Wroe SF, Wells CA, Miller HJ, Bodle D, Beechey CV, Williamson CM, Kelsey G. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc. Natl Acad. Sci. USA. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 2008;18:1735–1741. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 49.Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 50.Yoon BJ, Herman H, Sikora A, Smith LT, Plass C, Soloway PD. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 2002;30:92–96. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 52.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 54.Reik W, Walter J. Evolution of imprinting mechanisms: the battle of the sexes begins in the zygote. Nat. Genet. 2001;27:255–256. doi: 10.1038/85804. [DOI] [PubMed] [Google Scholar]
- 55.Beechey CV, Ball ST, Townsend KM, Jones J. The mouse chromosome 7 distal imprinting domain maps to G-bands F4/F5. Mamm. Genome. 1997;8:236–240. doi: 10.1007/s003359900400. [DOI] [PubMed] [Google Scholar]
- 56.Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun CW, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1997;240:287–292. doi: 10.1006/bbrc.1997.7649. [DOI] [PubMed] [Google Scholar]
- 58.Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 59.Spiegel AM. Mutations in G proteins and G protein-coupled receptors in endocrine disease. J. Clin. Endocrinol. Metab. 1996;81:2434–2442. doi: 10.1210/jcem.81.7.8675557. [DOI] [PubMed] [Google Scholar]
- 60.Juppner H, Schipani E, Bastepe M, Cole DE, Lawson ML, Mannstadt M, Hendy GN, Plotkin H, Koshiyama H, Koh T, et al. The gene responsible for pseudohypoparathyroidism type Ib is paternally imprinted and maps in four unrelated kindreds to chromosome 20q13.3. Proc. Natl Acad. Sci. USA. 1998;95:11798–11803. doi: 10.1073/pnas.95.20.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein LS. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc. Natl Acad. Sci. USA. 1998;95:8715–8720. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bastepe M, Juppner H. GNAS locus and pseudohypoparathyroidism. Horm. Res. 2005;63:65–74. doi: 10.1159/000083895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.