Abstract
Electrospray mass spectrometry was used to investigate the mechanism of tetramolecular G-quadruplex formation by the DNA oligonucleotide dTG5T, in ammonium acetate. The intermediates and products were separated according to their mass (number of strands and inner cations) and quantified. The study of the temporal evolution of each species allows us to propose the following formation mechanism. (i) Monomers, dimers and trimers are present at equilibrium already in the absence of ammonium acetate. (ii) The addition of cations promotes the formation of tetramers and pentamers that incorporate ammonium ions and therefore presumably have stacked guanine quartets in their structure. (iii) The pentamers eventually disappear and tetramers become predominant. However, these tetramers do not have their four strands perfectly aligned to give five G-quartets: the structures contain one ammonium ion too few, and ion mobility spectrometry shows that their conformation is more extended. (iv) At 4°C, the rearrangement of the kinetically trapped tetramers with presumably slipped strand(s) into the perfect G-quadruplex structure is extremely slow (not complete after 4 months). We also show that the addition of methanol to the monomer solution significantly accelerates the cation-induced G-quadruplex assembly.
INTRODUCTION
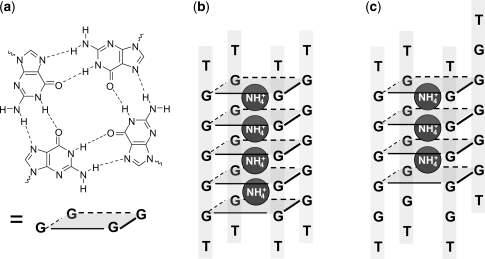
G-quadruplexes encompass all nucleic acid structures involving the G-quartet motif (Figure 1a), in which four guanines are assembled in a square planar structure by Hoogsteen hydrogen bonds (1,2). G-quadruplexes can be formed by biologically relevant DNA and RNA sequences (3–6), and by synthetic G-rich nucleic acid strands. Tetramolecular G-quadruplexes constituted from four identical strands arranged in parallel orientation have been extensively studied (7,8), because they constitute a model for G-quadruplex structure, formation and interaction with small molecules. Furthermore, intermolecular G-quadruplexes are also a particularly interesting scaffold for the design of artificial nanostructures (1,9–11), owing to their stability, rigidity and electronic properties (12,13). Several synthetic quadruplex-forming oligonucleotides, either dimeric (14–16) or tetrameric (17,18), have been reported for their therapeutic properties, such as anti-cancer (19) or anti-HIV (15–17,20) treatment.
Figure 1.
(a) Structure of a guanine quartet (or G-quartet), (b) the perfectly aligned tetramolecular G-quadruplex formed by the oligonucleotide dTG5T contains five G-quartets and four ammonium ions, (c) a tetramolecular G-quadruplex with one strand slipped by one base contains four G-quartets and three ammonium ions.
Of particular interest are the mechanisms leading to G-quadruplex assembly. G-quadruplex structures are stabilized by their G-quartets (Figure 1a), and by cation interactions with these G-quartets. Sodium has a tendency to sit in the G-quartet plane, while ammonium and potassium have a tendency to sit between adjacent G-quartets (21–23). Intermolecular G-quadruplexes involve several strands and several cations. For example, the most stable G-quadruplex formed by the oligonucleotide sequence dTG5T in ammonium ions is a tetramolecular structure (four strands) with all guanines aligned so as to form five G-quartets, with four ammonium ions selectively bound (Figure 1b). The formation pathway of these G-quadruplex structures must necessarily involve multiple steps. Understanding and controlling these steps are of great importance for the design and control of G-quadruplex-based structure fabrication. Tetramolecular G-quadruplexes offer a good model system for characterizing the assembly mechanism because their assembly is much slower than bimolecular and intramolecular quadruplex formation (7), and can be followed on the time scale accessible by several techniques like size-exclusion chromatography, gel electrophoresis, UV spectrophotometry, circular dichroism or NMR.
The simplest mechanism that can account for the tetramolecular G-quadruplex formation is the monomer–dimer–tetramer pathway. To account for the pseudo-fourth order association rate of dTG4T and analogs (24) and for the fact that no dimer can be detected by size exclusion chromatography or spectrophotometry, one must hypothesize a rapid equilibium between monomer and dimer, and a comparatively slow conversion of two dimers into tetramer (8,17). Another possible mechanism is the stepwise strand addition, i.e. the monomer–dimer–trimer–tetramer pathway. No trimer could be evidenced directly to date, but an NMR study of the kinetics of quadruplex formation showed that the reaction order is closer to three in KCl, suggesting trimer formation as the rate-limiting step in the tetramer formation (25). Stefl et al. tested by molecular modeling which intermediates could be sufficiently stable and likely to be involved in the formation mechanism. They found that dimers and trimers could be stable if they have a crossed structures, but not parallel H-bonded duplex structures (26). Another family of possible intermediate evidenced by the modeling study by Stefl et al. (26) are tetramers with partly slipped strands and incomplete cation incorporation (Figure 1c). The NMR study by Bardin and Leroy (25) also revealed some tetramers that are different from the canonical G-quadruplex and that can be kinetically trapped for several days.
The objective of our study was to use electrospray mass spectrometry (ESI-MS) to follow the assembly of tetramolecular G-quadruplexes, in order to confirm or invalidate some of the intermediates that have been previously proposed, but not demonstrated unambiguously. When the instrumental conditions are carefully controlled in order to avoid any dissociation in the mass spectrometer, the non-covalent nucleic acid complexes present in solution are detected intact by ESI-MS (27–29). In the particular case of G-quadruplexes, ESI-MS allows counting the number of strands (30–32) and the number of cations (33) inside each structure. Moreover, with the high sensitivity of ESI-MS combined with the mass spectral separation of each stoichiometry, we might detect some intermediates that have never been unambiguously detected or even proposed before.
MATERIALS AND METHODS
All oligonucleotides were purchased from Eurogentec (Seraing, Belgium), solubilized in water doubly distilled in house, and the 400 µM stock solutions were stored at −20°C. The solvents used include methanol (absolute, HPLC grade, Biosolve, Valkenswaard, The Netherlands), bi-distilled water and aqueous ammonium acetate (5 M stock solution from Fluka, diluted with bi-distilled water). Before G-quadruplex formation, the dTG5T aqueous stock solution was heated at 85°C for 5–10 min. Without heating, ESI-MS of the aqueous stock solution revealed the presence of pre-formed tetramer including ammonium ions, which are due to the purification process after synthesis (which involves ammonia) and the lyophilization of the sample. Heating during at least 5 minutes is required to remove all pre-formed G-quadruplex, and less than 10 minutes is required to avoid degradation of the strands, as controlled by ESI-MS.
ESI-MS experiments were performed on a Q-TOF Ultima Global mass spectrometer (Waters, Manchester, UK). The electrospray ion source was operated in negative ion mode with a capillary voltage of −2.2 kV. The source pressure was 3.4 mbar, the cone voltage was 100 V and the RF lens 1 voltage was 140 V. The voltage entry in the hexapole collision cell is 10 V. The instrument is externally calibrated with sodium iodide and internally calibrated (lock mass) using 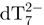 (m/z of monoisotopic peak = 1032.1760).
(m/z of monoisotopic peak = 1032.1760).
To study the G-quadruplex formation pathway, single-stranded dTG5T is first diluted in water or water/methanol, the single strand dT7 is added to serve as internal standard (see below). Various volume percentages of methanol were tested, from 0% to 50%. Aqueous ammonium acetate (final concentration 150 mM) is then added to initiate G-quadruplex formation, and the ESI-MS recording is started simultaneously. The sample is agitated, loaded into the syringe and the spray is initiated as quickly as possible by pushing the syringe piston manually. The flow rate is then set at 2 µl/min for the rest of the experiment. The dead time between ammonium acetate addition and flow stabilization is about 1 min. All kinetics experiments are carried out with the sample at room temperature.
The internal standard dT7 is used to measure the variation of the intensity of each component of the mixture relative to a fixed reference, as a function of time. By using a recently developed method using the internal standard and the mass balance equation for the total strand concentration, the relative ESI-MS response of each component of the mixture can be determined (34). Briefly, for each stoichiometry adopted by the TG5T strand, the ratio between the area of a peak attributable unambiguously to that stoichiometry and the area of a peak of the internal standard (here 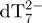 ), is calculated at each time-point. By assuming that the electrospray response of each TG5T stoichiometry and dT7 are proportional throughout the experiment, and by using the mass balance equation applied to TG5T (the total strand concentration being equal to 80 µM), the relative response factors of each stoichiometry can be determined. These response factors are then used to recalculate the concentration of each stoichiometry at each time point.
), is calculated at each time-point. By assuming that the electrospray response of each TG5T stoichiometry and dT7 are proportional throughout the experiment, and by using the mass balance equation applied to TG5T (the total strand concentration being equal to 80 µM), the relative response factors of each stoichiometry can be determined. These response factors are then used to recalculate the concentration of each stoichiometry at each time point.
Ion mobility spectrometry experiments were performed on a Synapt HDMS spectrometer (Waters, Manchester, UK), in negative ion mode electrospray ionization. Samples were prepared in the same way as for the Q-TOF experiments; the capillary voltage was set to −2.2 kV; cone voltage = 40 V; extraction cone = 2 V; source pressure = 3.05 mbar; source and desolvation temperatures = 40°C and 70°C, respectively; trap and transfer voltages = 6 V and 4 V, respectively. The Synapt HDMS is similar to the Q-TOF instrument, except that the hexapole collision cell is replaced by a travelling wave (T-wave) cell (35) that allows separating the ions according to their ion mobility. The cell is filled with N2 at 0.535 mbar, and an electric field is applied to the cell in the form of waves (wave height = 9 V) that pass through the cell at 300 m/s. The bias voltage for entering in the T-wave cell was 10 V. Within an ion packet of a given mass-to-charge ratio, more compact ions have a higher ion mobility because of their smaller collision cross-section, and are more influenced by the electric field caused by the waves. Therefore, more compact ions exit the T-wave cell earlier than ions of more extended conformations.
RESULTS
Identification of the intermediates: dimer, trimer, tetramer(s) and pentamer
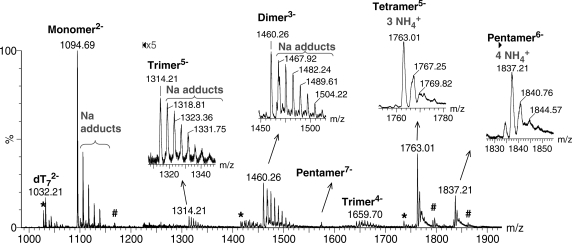
ESI-MS allows detecting intact non-covalent assemblies. In the case of quadruplex nucleic acids, it allows counting the number of strands and of incorporated ammonium ions in each component of a complex mixture (33). Figure 2 shows all species detected on the course of G-quadruplex formation by the single strand dTG5T, from 0 to 80 min after ammonium acetate addition. The experiment is run in 10% methanol. Methanol addition is common in ESI-MS because it enhances the signal-to-noise ratio. We will show below that the methanol content also influences the reaction kinetics. However, from 100% water to 50/50 water/methanol, the nature of the detected species is the same.
Figure 2.
Identification of all intermediates: 80 µM dTG5T monomer, 10 µM dT7 internal standard, water/methanol 90/10, sum of all scans from 0 to 80 min after 150 mM NH4OAc addition. The spectrum was background subtracted, and peaks from 1225 to 1950 Da were magnified five times on the full spectrum. The zooms clarify the nature of the cation adducts on the dimer, trimer, tetramer and pentamer. Asterisk and hash symbols indicate impurities in the monomer solution (*monomer –133 Da, #monomer +146 Da), or multimers incorporating one of these modified monomers.
The monomer is detected at charges state 2− (m/z = 1094.69) and 3− (not shown), with some sodium ions non-specifically bound, but no ammonium ions bound. Sodium was not purposefully added in our experiment, and is a common contamination in DNA samples and glassware. Dimers and trimers of dTG5T are also detected, as shown in the zooms of the peak distributions issuing at m/z = 1314.21 (trimer5−) and 1460.26 (dimer3−). Like the monomer, the dimer and trimer are detected with a non-specific distribution of sodium adducts, and no ammonium ions bound. A tetramer is unambiguously detected at the charge state 5−. The expected tetramolecular G-quadruplex contains five G-quartets and therefore four ammonium ions (Figure 2b). Surprisingly, the peak at m/z = 1763.01 corresponds to four strands and three ammonium ions (no sodium). The presence of specifically retained ammonium ions indicates that the structure contains stacked G-quartets, and can therefore be called a G-quadruplex, but this tetramolecular G-quadruplex is different from the expected one. Another unanticipated result is the detection of a pentameric species at charge states 6− and 7−. The pentamer contains predominantly four ammonium ions, and therefore presumably five stacked G-quartets. The internal standard is detected only as monomer: it does neither self-aggregate nor bind to the different forms of dTG5T.
We also investigated the G-quadruplex formation by dTGnT sequences with n = 3–6, to compare with the dTG5T results discussed above. For all sequences, dimers were detected both in pure water and in the early stages after ammonium acetate addition. Trimers could be detected for dTG6T. Note that the ESI response decreases as n decreases, so the trimer signal in dTG4T and dTG3T might be under our detection threshold. Regarding tetramers and pentamers, in the case of dTG4T, ESI-MS spectra taken at different time points after ammonium acetate addition show only the presence of tetramers with three ammonium ions, i.e. the expected G-quadruplex with four G-quartets. For dTG6T, the initially formed tetramer is detected with four ammonium ions. The pentamer of dTG6T is also detected, with five ammonium ions bound. In summary, for dTGnT sequences with n > 4, the perfectly aligned tetrameric G-quadruplex should contain n G-quartets and n − 1 ammonium ions, but the species initially formed are predominantly tetramers with n − 2 ammonium ions (presumably n − 1 G-quartets) and pentamers with n − 1 ammonium ions (presumably n G-quartets).
Kinetics
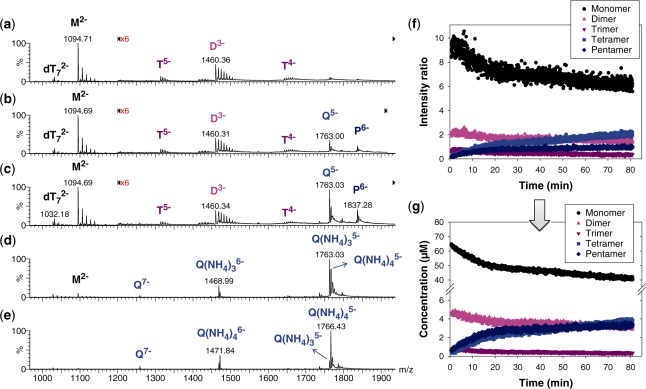
Before discussing the nature of the irregular tetramer and of the pentamer, we shall examine the temporal evolution of the composition of the solution. The kinetics of G-quadruplex formation was followed online during 80 min, in the presence of an internal reference (oligonucleotide dT7). Figure 3 shows some selected mass spectra at different time points after ammonium acetate addition, in 10% methanol. In the earliest spectrum (2 min, Figure 3a), the monomer (M), dimer (D) and trimer (T) are detectable. A control experiment with the monomer diluted to the same concentration without ammonium acetate shows that the monomer, dimer and trimer are also present in water, with similar non-specific sodium adduct distribution. We can therefore conclude that monomers, dimers and trimers are in equilibrium prior to ammonium acetate addition.
Figure 3.
Kinetics of G-quadruplex formation by 80 µM dTG5T in 150 mM NH4OAc and 10% methanol. (a–e) Electrospray mass spectra recorded 2 min (a), 10 min (b), 60 min (c), 25 days (d) and 164 days (e) after ammonium acetate addition. M stands for monomer, D for dimer, T for trimer, Q for tetramer, P for pentamer. Note the magnification from 1200 to 2000 m/z in spectra (a–c). (f) Intensity ratio measured between a characteristic peak of each stoichiometry (M2− for monomer, D3− for the dimer, T5− for the trimer, Q5− for the tetramer and P6− for the pentamer) and the reference peak 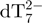 , as a function of time elapsed after ammonium addition. (g) Time evolution of the concentration of each stoichiometry, obtained after correcting for the relative response factor of each characteristic peak (note the break on the y-axis).
, as a function of time elapsed after ammonium addition. (g) Time evolution of the concentration of each stoichiometry, obtained after correcting for the relative response factor of each characteristic peak (note the break on the y-axis).
The spectra recorded after 10 and 60 min (Figure 3b and c, respectively) show the increase of the peak corresponding to the previously described G-quadruplex (Q) and pentamer (P). In all our experiments (from 80 µM to 120 µM dTG5T, from 0% to 50% methanol), the pentamer is still present at the end of the recording (typically 80 min). Aliquots of the solutions studied online for 80 min were left at 4°C. ESI-MS spectra were also recorded 25 days (Figure 3d) and more than 4 months (Figure 3e) after ammonium acetate addition. After 25 days (Figure 3d), the pentamer signal disappeared, and the monomer signal dramatically reduced compared to the quadruplex. However, the dominant signal of the quadruplex is still with three ammonium ions. The relative signal of the expected quadruplex with four ammonium ions has increased only slightly. Spectra recorded at the time of this writing, with the same sample aged more than 4 months at 4°C, show that the predominant peak is now of the expected G-quadruplex with four ammonium ions.
From the 80-min online experiment, we can monitor the relative intensities of peaks representing each stoichiometry (M2− for monomer, D3− for the dimer, T5− for the trimer, Q5− for the tetramer and P6− for the pentamer). The intensity ratios between each of these peaks and the internal standard 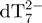 are first plotted as a function of time (Figure 3f). Then, after determination of the relative response factors of each species (34), the intensity ratios are converted into absolute concentrations (Figure 3g). The starting monomer concentration is around 65 µM, the rest being dimer (5 ± 1 µM) and trimer (<1 µM). The equilibrium dissociation constant of the dimer can therefore be estimated at ≈0.8 mM, and the Kd for trimer dissociation into dimer and monomer at >0.3 mM. Similar intensity ratios are obtained in the absence of ammonium acetate (low ionic strength). As the reaction time increases, the monomer, dimer and trimer concentrations decrease, while the tetramer and pentamer concentrations increase. One can therefore assume a pre-equilibrium between monomer, dimer and trimer, in fast interconversion compared to tetramer and pentamer formation.
are first plotted as a function of time (Figure 3f). Then, after determination of the relative response factors of each species (34), the intensity ratios are converted into absolute concentrations (Figure 3g). The starting monomer concentration is around 65 µM, the rest being dimer (5 ± 1 µM) and trimer (<1 µM). The equilibrium dissociation constant of the dimer can therefore be estimated at ≈0.8 mM, and the Kd for trimer dissociation into dimer and monomer at >0.3 mM. Similar intensity ratios are obtained in the absence of ammonium acetate (low ionic strength). As the reaction time increases, the monomer, dimer and trimer concentrations decrease, while the tetramer and pentamer concentrations increase. One can therefore assume a pre-equilibrium between monomer, dimer and trimer, in fast interconversion compared to tetramer and pentamer formation.
Influence of methanol on the kinetics
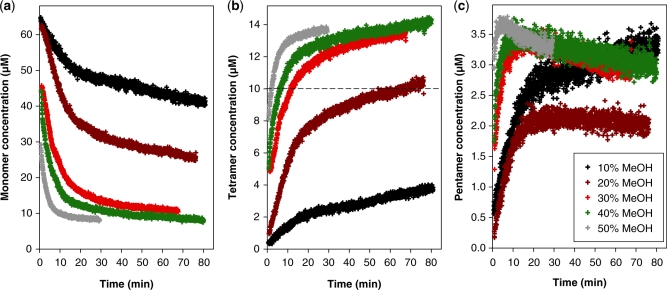
As mentioned earlier, ESI-MS experiments are often carried out with methanol addition to the injected solution in order to increase the signal-to-noise ratio. Having obtained several unexpected results in the presence of methanol (incomplete incorporation of ammonium ions in the tetramer, pentamer formation), we verified that these species were not due to the use of methanol. Experiments in pure water confirmed the formation of the same species as in 10% methanol. However, the rate of G-quadruplex formation is much lower in pure water (about 1–2% of quadruplex formed after 80 min), in agreement with previously published formation rate constants in ammonium acetate (33). This prompted us to study the influence of the volume percentage of methanol on the kinetics. The monomer, tetramer and pentamer concentrations were recalculated as a function of time for the kinetics experiments carried out at 10–50% methanol, and the results are displayed in Figure 4.
Figure 4.
Influence of the volume percentage of methanol on the reaction kinetics with 80 µM total dTG5T strand concentration and 150 mM total NH4OAc concentration. Time evolution of the concentration of (a) monomer, (b) tetramer and (c) pentamer.
The alcohol content has a dramatic influence on the rate of G-quadruplex formation (G-quadruplex here means all species with stacked G-quartets and incorporated ammonium ions, i.e. the tetramers and pentamers): the higher the methanol content, the faster the G-quadruplex assembly. The time required for the tetramer concentration to reach 10 µM (50% of the maximum possible, starting with 80 µM single strand) is 2 min at 50% MeOH, 6 min at 40% MeOH, 12 min at 30% MeOH, 68 min at 20% MeOH and outside the time scale of the experiment in 10% MeOH or pure water (see dashed line in Figure 4b). Figure 4 also clearly shows that the initial rate of pentamer formation (Figure 4c) is faster than the initial rate of tetramer formation (Figure 4b). Also, at 30%, 40% and 50% methanol, we can observe within the time scale of our experiment an initial increase of the pentamer concentration, followed by a slower decrease.
Another interesting aspect of the time evolution of the concentrations is that, on the time scale of our experiment, the concentrations of each species asymptotically reach values that are different from those expected with the complete formation of the quadruplex including four ammonium ions (Figure 3e). In conclusion, several different steps in the G-quadruplex formation pathway can be distinguished, that proceed on different time scales: (i) the monomer–dimer–trimer equilibrium does not need ammonium acetate to be established; (ii) the formation of the pentamer and of the first tetrameric forms requires ammonium acetate, proceeds on the hour time scale, and does not consume all the monomer; (iii) the conversion to the perfect quadruplex including four ammonium ions and five G-quartets proceeds on a much slower time scale and eventually consumes all the monomer.
Tetrameric intermediates have a more extended conformation than the canonical G-quadruplex
Based on the number of ammonium ions incorporated, we can conclude that the tetramers are present in several forms. The presence of non-canonical, slipped G-quadruplexes has been suggested by the modeling studies by Stefl et al. (26), and by the NMR studies of Bardin and Leroy (25). The 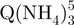 − tetramers observed by ESI-MS could correspond to these slipped G-quadruplexes, because less than five G-quartets are formed (Figure 1c). To verify this interpretation, we performed ion mobility spectrometry (IMS) experiments (Figure 5). Ion mobility allows separating ions of a given mass-to-charge ratio based on their ion mobility, which for a given charge is directly related to the ion’s collision cross section. The regular G-quadruplexes being more compact, they will travel faster in the ion mobility cell than the slipped G-quadruplex structures. The experiments not only confirm that the
− tetramers observed by ESI-MS could correspond to these slipped G-quadruplexes, because less than five G-quartets are formed (Figure 1c). To verify this interpretation, we performed ion mobility spectrometry (IMS) experiments (Figure 5). Ion mobility allows separating ions of a given mass-to-charge ratio based on their ion mobility, which for a given charge is directly related to the ion’s collision cross section. The regular G-quadruplexes being more compact, they will travel faster in the ion mobility cell than the slipped G-quadruplex structures. The experiments not only confirm that the 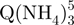 − tetramers (red triangles) have indeed a more extended conformation than the
− tetramers (red triangles) have indeed a more extended conformation than the 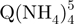 − tetramers (green squares), but also reveal that the degree of compactness of the
− tetramers (green squares), but also reveal that the degree of compactness of the 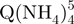 − tetramers increases with formation time. Each stoichiometry therefore corresponds to an ensemble of conformations. The arrival time distribution of the whole-tetramer adduct distribution (black circles) is shifting to shorter times (more compact conformations) as the formation time increases from 2 min (Figure 5a) to 120 min (Figure 5b) and 165 days (Figure 5c).
− tetramers increases with formation time. Each stoichiometry therefore corresponds to an ensemble of conformations. The arrival time distribution of the whole-tetramer adduct distribution (black circles) is shifting to shorter times (more compact conformations) as the formation time increases from 2 min (Figure 5a) to 120 min (Figure 5b) and 165 days (Figure 5c).
Figure 5.
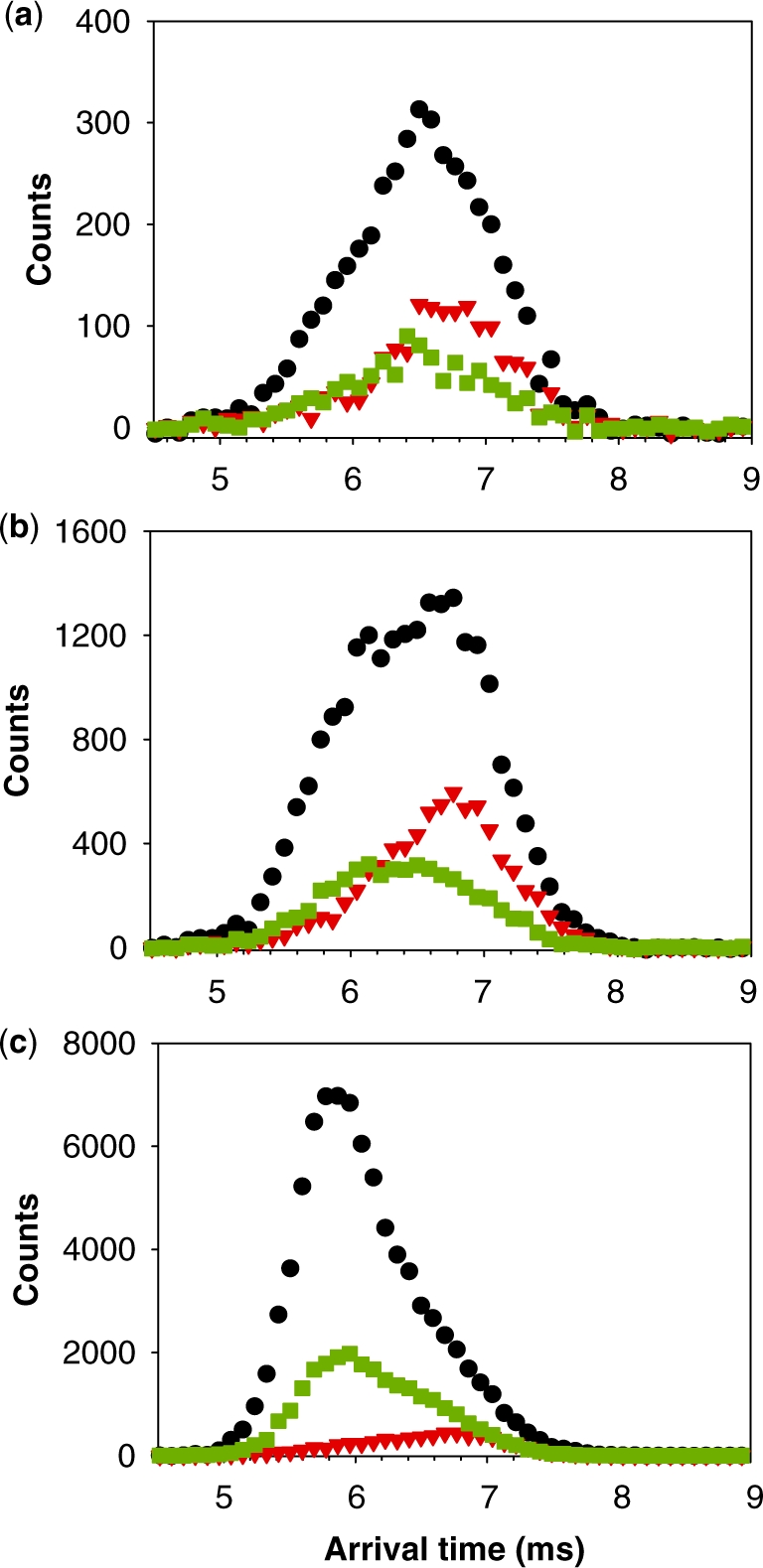
Ion mobility spectrometry of the tetramer formed from 80 µM TG5T in 20% MeOH and 150 mM NH4OAc after (a) 10 min, (b) 2 h and (c) 4 months. Red triangles, arrival time distribution of 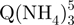 − (m/z = 1762–1765). Green squares, arrival time distribution of
− (m/z = 1762–1765). Green squares, arrival time distribution of 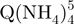 − (m/z = 1765–1768). Black circles, arrival time distribution of the sum of all G-quadruplex adduct forms (m/z = 1762–1775). Shorter arrival times indicate more compact conformation.
− (m/z = 1765–1768). Black circles, arrival time distribution of the sum of all G-quadruplex adduct forms (m/z = 1762–1775). Shorter arrival times indicate more compact conformation.
DISCUSSION
ESI-MS experiments have therefore unambiguously confirmed the presence of several stoichiometries (dimers, trimers, tetramers with incomplete ammonium ion incorporation), and revealed others (pentamers). The presence of ammonium ions is related to the number of stacked G-quartets in the structures. Ion mobility spectrometry experiments have shown that incomplete ammonium incorporation in the tetramers was associated with more extended conformations. Moreover, the temporal evolution of the composition of the solution can be characterized. These results help to refine the mechanism of tetramolecular G-quadruplex formation (Figure 6).
Figure 6.
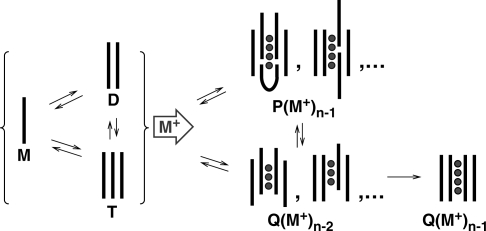
Mechanism of tetramolecular G-quadruplex formation by dTGnT. The strands are represented by black lines and the cations are represented by grey circles. M stands for monomer, D for dimer, T for trimer, Q for tetramer and P for pentamer.
The most simple assembly mechanism accounting for the pseudo-fourth order association kinetics is the pre-equilibrium between monomers and dimers, followed by the formation of tetramers. Another proposed mechanism is the stepwise addition of monomer to form dimer, trimer and tetramer. Due to their low abundance, dimers and trimers cannot be detected by size exclusion chromatography or spectrophotometric techniques. By NMR, the presence of dimers was suggested by small proton shifts of the monomer, and the dimer dissociation constant was estimated between 2 mM and 6 mM for dTG4T (25). Here, we have unambiguously proved that dimers and trimers are present in equilibrium with the monomer, and could estimate a Kd ≈0.8 mM for dTG5T for the dimer formation and >0.3 mM for the trimer formation from a dimer and a monomer, in 150 mM NH4OAc. These dimers and trimers are not detected with specific ammonium ions retained in the electrosprayed species. This does not necessarily indicate that ammonium ions are not coordinated to dimers and trimers in solution, but it indicates that ammonium ions are not retained in the gas-phase. Past ESI-MS studies of G-quadruplexes has shown that only ammonium ions coordinated between two G-quartets are bound tightly enough to be retained in the gas phase (30,33). We therefore interpret the absence of ammonium ion adduct as an indication of the absence of at least two stacked G-quartets formed in these structures. Monomers, dimers and trimers have instead a high affinity for the residual sodium present in solution. Finally, monomer, dimers and trimers are also detected with a similar intensity ratio in pure water (no ammonium acetate).
The further addition of strands is initiated by the addition of ammonium acetate. We observe the simultaneous formation of two mass spectrometrically distinguishable species: a tetramer with incomplete incorporation of ammonium ions and which has more extended conformations than the canonical tetrameric G-quadruplex; and a pentamer, which is also incorporating ammonium ions. The ammonium count and ion mobility experiments suggest that the tetramers detected are tetramers with some degree of strand slippage. Tetrameric intermediates with an incomplete set of G-quartets due to strand slippage have been predicted by molecular modeling (26), and detected by NMR (25).
Regarding the pentamer formation, two different pathways are possible: (i) trimer+dimer, and (ii) tetramer+monomer. The ‘dimer+trimer’ pathway is possible because we detect that trimers are indeed present. However, if this pathway is the major one, it would mean that ammonium-induced dimer–trimer interaction is more likely to result in the formation of nucleation G-quartets than dimer–dimer interactions. It would therefore mean that the lower frequency of dimer–trimer encounter must be overcome by the higher probability that, during this encounter, enough guanines are in a favorable conformation to form the nucleation G-quartets. In terms of favorable encounter conformation for the reaction to occur, this is plausible because there are more guanines in five strands than in four. Actually, the same probabilistic (or, in other words, entropic) arguments also explain that the rate of cation-induced G-quartet formation increases as the length of the G-tract increases in dTGnT sequences (7,24). The ‘tetramer+monomer’ pathway is also possible. However, we found that the formation of pentamer is faster than the formation of tetramers with three ammonium ions, and that the pentamer eventually converts into the latter (Figure 3d). The tetramer from which the pentamer forms must therefore be a different kind of tetramer structure, presumably with even more strand slippage than the tetramers detected here. The capture of a monomer from these tetramers would then be favorable because it allows several new G-quartets to form, to reach a total of five G-quartets (four ammonium ions).
After the formation of the tetramers and pentamers that we detect on the minute time scale, several slow rearrangements occur. We can therefore imagine the following formation mechanism: in the pentamer, the degree of strand slippage is relatively high and the stem of G-quartets incorporates five strands instead of four, allowing for a greater enthalpic stabilization than the corresponding tetramer with the same degree of strand slippage. Then, the degree of strand slippage diminishes by internal rearrangements to minimize the Gibbs free energy, until one strand is ejected from the pentamer to lead to the tetramer. At some point, one extra monomer is lost but in the resulting tetramer the number of G-quartets reduced from n to n − 1, and the number of ammonium ions from n − 1 to n − 2. The final step in canonical G-quadruplex formation is the rearrangement leading to the suppression of the last strand slippage and the re-incorporation of the last cation. As demonstrated by NMR and mass spectrometry, this ultimate step is extremely slow.
In summary, the ESI-MS results, together with previous studies, lead us to propose the following formation mechanism. There is a pre-equilibrium between monomer, dimer and trimer, with Kd in the range 0.1–1 mM, which exists also in pure water. The monomer, dimer and trimer have non-specific interaction with trace amounts of sodium. The addition of ammonium acetate initiates the formation of higher order structures: tetramers and pentamers. These pentamers are not well-ordered, probably contain several slipped strands, but those formed on the minute time scale incorporate ammonium ions so as to maximize the number of G-quartets and gain enthalpic stabilization. The rearrangements tend to align four of the strands with each other, and at some point one of the strands of the pentamer is expelled to form a slipped tetramer. This step is enthalpically disfavored because one G-quartet is lost, but entropically favored by the release of one cation and one strand. Rearrangements continue in the tetramer until the four strands are eventually strictly parallel and the last ammonium ion is trapped in the stem. The rearrangements in the tetramer are extremely slow at low temperature. We have seen that for dTG5T at 4°C the tetramer is not yet fully converted to the canonical structure after more than 4 months.
This is in entire agreement with the NMR results of Bardin and Leroy (25), who evidenced the formation of kinetically trapped tetramers for dTG5T at room temperature and below, and for dTG6T below 42°C. Kinetically trapped structures were also evidenced for the bimolecular G-quadruplexes formed by the sequences dG4T4G4 and dG4T4G3 (36). The formation of pentamers has however never been reported before. Note that these pentamers are transient species, and that their abundance remains low throughout the formation pathway (max. 3.5 µM starting from 80 µM monomer concentration). Their UV, CD or NMR spectroscopic signal might be undistinguishable from the expected tetramer signal. It is also possible that, once isolated in size exclusion chromatography or gel electrophoresis, these pentamers dissociate into tetramer and monomer and can remain undetectable by these techniques. Only electrospray mass spectrometry in native conditions currently allows their detection. The possible formation of pentamers has only once been suggested before. Based on spectroscopy results on the sequence dTG4, the formation of pentamers or hexamers has been suggested by Hardin et al. (37), who later proposed a mechanism of tetramolecular quadruplex formation involving the disproportionation of hexamers (38). However, when we performed ESI-MS experiments on the sequence dTG4 in ammonium acetate, we found that this sequence forms a mixture of tetramers and octamers at equilibrium (data not shown), so their interpretation of the formation of pentamers during tetramolercular quadruplex formation, although now proven entirely right by our results, could actually have been based on an artifact.
The detailed knowledge of the mechanism of intermolecular G-quadruplex formation is of prime importance for the design and fabrication of quadruplex-based nanostructures or quadruplex-based therapeutic agents. Depending on the application, kinetically trapped intermediates should be avoided, or could be desirable. For example, intermediates with slipped strands and pentamers could serve as a nucleation platform for the addition of subsequent strands and the formation of larger structures, because they already provide some template for G-quartet stacking. Also, understanding and controlling the formation of multimeric G-quadruplexes of therapeutic potential is crucial for their safe use. ESI-MS should become a method of choice to reveal the self-assembly mechanisms, higher order structures (32), and the kinetically trapped intermediates of many other sequences. The influence of methanol on the rates of self-assembly is also of prime interest for the fabrication of quadruplex-based structures. Our results show that methanol can be extremely useful to accelerate the assembly process, while still leading to the expected G-quadruplex product.
FUNDING
University of Liège (grant D 08/10 to V.G.); Fonds de la Recherche Scientifique-FNRS (postdoctoral research fellowship to F.R., research associate position to V.G., CC 1.5.286.09.F to V.G. and FRFC 2.4.589.08.F to E.D.P.). Funding for open access charge: University of Liège (grant FRSD-08/10).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Dr Jiři Šponer for useful discussion on the nature of the pentamers, and undergraduate student Jennie Roberts for skillfully contributing some experiments.
REFERENCES
- 1.Davis JT, Spada GP. Supramolecular architectures generated by self-assembly of guanosine derivatives. Chem. Soc. Rev. 2007;36:296–313. doi: 10.1039/b600282j. [DOI] [PubMed] [Google Scholar]
- 2.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 3.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Phan A-T, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mergny J-L, Gros J, De Cian A, Bourdoncle A, Rosu F, Sacca B, Guittat L, Amrane S, Mills M, Alberti P, et al. Energetics, kinetics and dynamics of quadruplex folding. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. The Royal Society of Chemistry, Cambridge; 2006. pp. 31–80. [Google Scholar]
- 8.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alberti P, Bourdoncle A, Sacca B, Lacroix L, Mergny JL. DNA nanomachines and nanostructures involving quadruplexes. Org. Biomol. Chem. 2006;4:3383–3391. doi: 10.1039/b605739j. [DOI] [PubMed] [Google Scholar]
- 10.Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 11.Lena S, Masiero S, Pieraccini S, Spada GP. Guanosine hydrogen-bonded scaffolds: a new way to control the bottom-up realisation of well-defined nanoarchitectures. Chem. Eur. J. 2009;15:7792–7806. doi: 10.1002/chem.200802506. [DOI] [PubMed] [Google Scholar]
- 12.Cohen H, Sapir T, Borovok N, Molotsky T, DiFelice R, Kotlyar AB, Porath D. Polarizability of G4-DNA observed by electrostatic force microscopy measurements. Nano Lett. 2007;7:981–986. doi: 10.1021/nl070013b. [DOI] [PubMed] [Google Scholar]
- 13.Miannay FA, Banyasz A, Gustavsson T, Markovitsi D. Excited states and energy transfer in G-quadruplexes. J. Phys. Chem. C. 2009;113:11760–11765. [Google Scholar]
- 14.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou SH, Chin KH, Wang AHJ. DNA aptamers as potential anti-HIV agents. Trends Biochem. Sci. 2005;30:231–234. doi: 10.1016/j.tibs.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyatt JR, Davis PW, Freier SM. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 18.D'Onofrio J, Petraccone L, Martino L, Di FG, Iadonisi A, Balzarini J, Giancola C, Montesarchio D. Synthesis, biophysical characterization, and anti-HIV activity of glyco-conjugated G-quadruplex-forming oligonucleotides. Bioconjug. Chem. 2008;19:607–616. doi: 10.1021/bc7003395. [DOI] [PubMed] [Google Scholar]
- 19.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Onofrio J, Petraccone L, Martino L, Di FG, Iadonisi A, Balzarini J, Giancola C, Montesarchio D. Synthesis, biophysical characterization, and anti-HIV activity of glyco-conjugated G-quadruplex-forming oligonucleotides. Bioconjug. Chem. 2008;19:607–616. doi: 10.1021/bc7003395. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Leszczynski J. Origin of Na+/K+ selectivity of the guanine tetraplexes in water: the theoretical rationale. J. Phys. Chem. A. 2002;106:529–532. [Google Scholar]
- 22.Hud NV, Schultze P, Sklenar V, Feigon J. Binding sites and dynamics of ammonium ions in a telomere repeat DNA quadruplex. J. Mol. Biol. 1999;285:233–243. doi: 10.1006/jmbi.1998.2327. [DOI] [PubMed] [Google Scholar]
- 23.Sket P, Crnugelj M, Plavec J. Identification of mixed di-cation forms of G-quadruplex in solution. Nucleic Acids Res. 2005;33:3691–3697. doi: 10.1093/nar/gki690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mergny J-L, De Cian A, Gheleb A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardin C, Leroy JL. The formation pathway of tetramolecular G-quadruplexes. Nucleic Acids Res. 2008;36:477–488. doi: 10.1093/nar/gkm1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stefl R, Cheatham TE, Spackova N, Fadrna E, Berger I, Koca J, Sponer J. Formation pathways of a guanine-quadruplex DNA revealed by molecular dynamics and thermodynamic analysis of the substates. Biophys. J. 2003;85:1787–1804. doi: 10.1016/S0006-3495(03)74608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofstadler SA, Griffey RH. Analysis of noncovalent complexes of DNA and RNA by mass spectrometry. Chem. Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 28.Beck J, Colgrave ML, Ralph SF, Sheil MM. Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom. Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 29.Rosu F, De Pauw E, Gabelica V. Electrospray mass spectrometry to study drug-nucleic acid interactions. Biochimie. 2008;90:1074–1087. doi: 10.1016/j.biochi.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Rosu F, Gabelica V, Houssier C, Colson P, De Pauw E. Triplex and quadruplex DNA structures studied by electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 2002;16:1729–1736. doi: 10.1002/rcm.778. [DOI] [PubMed] [Google Scholar]
- 31.Li HH, Yuan G, Du DM. Investigation of formation, recognition, stabilization, and conversion of dimeric G-quadruplexes of HIV-1 integrase inhibitors by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2008;19:550–559. doi: 10.1016/j.jasms.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Smargiasso N, Rosu F, Hsia W, Colson P, Baker ES, Bowers MT, De Pauw E, Gabelica V. G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008;130:10208–10216. doi: 10.1021/ja801535e. [DOI] [PubMed] [Google Scholar]
- 33.Gros J, Rosu F, Amrane S, De Cian A, Gabelica V, Lacroix L, Mergny JL. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabelica V, Rosu F, De Pauw E. A simple method to determine electrospray response factors of noncovalent complexes. Anal. Chem. 2009;81:6708–6715. doi: 10.1021/ac900785m. [DOI] [PubMed] [Google Scholar]
- 35.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequencyonly stacked ring ion guide. Rapid Commun. Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 36.Prislan I, Lah J, Vesnaver G. Diverse polymorphism of G-quadruplexes as a kinetic phenomenon. J. Am. Chem. Soc. 2008;130:14161–14169. doi: 10.1021/ja8026604. [DOI] [PubMed] [Google Scholar]
- 37.Hardin CC, Corregan MJ, Lieberman DV, Brown BA., II Allosteric interactions between DNA strands and monovalent cations in DNA quadruplex assembly: thermodynamic evidence for three linked association pathways. Biochemistry. 1997;36:15428–15450. doi: 10.1021/bi970488p. [DOI] [PubMed] [Google Scholar]
- 38.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]