Abstract
KSRP is a multi-domain RNA-binding protein that recruits the exosome-containing mRNA degradation complex to mRNAs coding for cellular proliferation and inflammatory response factors. The selectivity of this mRNA degradation mechanism relies on KSRP recognition of AU-rich elements in the mRNA 3′UTR, that is mediated by KSRP’s KH domains. Our structural analysis shows that the inter-domain linker orients the two central KH domains of KSRP—and their RNA-binding surfaces—creating a two-domain unit. We also show that this inter-domain arrangement is important to the interaction with KSRP’s RNA targets.
INTRODUCTION
Gene regulation by adenosine-uridine-rich element (ARE)-mediated mRNA decay (AMD) is important for cellular proliferation, immune response and cardiovascular toning. AMD malfunction has been linked to cancer (1) and to inflammatory diseases such as Crohn-like inflammatory bowel disease and inflammatory arthritis (2). The effect of cis-acting AREs on the stability of specific mRNAs is mediated by the binding of regulatory proteins (ARE-binding proteins, ARE-BPs). Some of these proteins (e.g. TTP, BRF1 and KSRP) promote mRNA degradation while others (e.g. HuD and HuR) act as antagonists, stabilizing the mRNA (3).
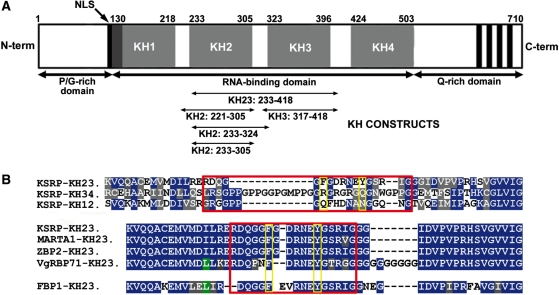
K-homology splicing regulator protein/fuse-binding protein 2 (KSRP/FBP2) is a multi-functional protein of the far upstream element (FUSE)-binding protein (FBP) family that has been involved in several steps of transcriptional and post-transcriptional gene control. In AMD, KSRP binds to a subset of AREs, recruiting the exosome and de-adenylation factors to the mRNA targets (4,5), defining a functional model for ARE-dependent mRNA degradation. The central part of KSRP is organized in four single-stranded nucleic acid-binding domains, the so-called K-homology domains (Figure 1A, KH1–4) which are responsible for its nucleic acid-binding activity. Using cross-linking and mRNA decay assays, we have previously shown that a minimum of two domains of KSRP are required for mRNA binding and degradation (5). Furthermore, in vitro studies (6–8) have confirmed that the isolated domains bind to AU-rich sequences with low affinity (Kd > 100 µM) and that simultaneous binding of at least two domains is necessary to obtain Kd values in the nanomolar-to-micromolar range.
Figure 1.
KSRP protein. (A) Domain organization of KSRP and constructs used in this study. (B) Sequence alignment of the inter-domain KH2–KH3 linker. Top—sequence alignment of KSRP KH1–KH2, KH2–KH3 and KH3–KH4 inter-domain linkers (red box). Bottom—sequence alignment of the KH2–KH3 linker of KSRP (Homo sapiens) with the ones of the MARTA1 (Rattus norvegicus), ZBP2 (Gallus gallus) and VgRBP71 (Xenopus laevis) protein homologues and of the human FUSE-binding protein (FBP1) (red box). Residues conserved in two or more constructs are coloured in green and blue, respectively; conservative substitutions of hydrophobic and polar residues are marked in grey. Yellow boxes highlight the positions of the two F311 and Y317 residues; red boxes define the boundaries of the linker between KH2 and KH3. The KH2–KH3 linker is conserved in the KSRP/FBP family and very different from the KH1–KH2 and KH3–KH4 linkers.
The selectivity of KSRP action in promoting mRNA degradation depends on its ability to recognize its ARE targets. However, the protein interacts with a broad range of AU-rich sequences. We dissected the sequence preference of the single domains showing not only the presence of both positive and negative sequence selectivity, but also that only one of the four domains of KSRP has a preference for AU-rich sequences (7). This hints that sequence specificity is only one of several criteria involved in target discrimination. KSRP-RNA recognition is a multi-domain event and inter-domain interactions, if they take place, could play an important role. The single-stranded regions recognized by the KH domains of KSRP are embedded within different 3′ untranslated regions (UTRs). Inter-domain contacts could modulate RNA recognition by relating orientation/accessibility of the RNA-binding surfaces of the domains to the single-stranded regions of the RNA targets. Thus, we need to understand if the four domains of KSRP act as structurally independent binding units and we need to evaluate the role of inter-domain linkers in protein–RNA interaction.
We have recently shown that the KH1–KH2 and KH3–KH4 domains of KSRP are connected by long and flexible linkers and that their relative orientation is not fixed (6,8). Here we examine the relation between KH2 and KH3 and show that the two domains interact and form the structural core of the KSRP protein. The structure of the KH23 protein reveals that the two domains assume a well-defined orientation with the linker playing an important role. We discuss the implication of this novel inter-domain arrangement for RNA binding and test our conclusions in the cellular environment.
MATERIALS AND METHODS
Protein and RNA oligonucleotides preparation
Recombinant KH domains shown in Figure 1 were constructed by PCR from a plasmid containing full-length KSRP (5). The PCR products were ligated into NcoI and HindIII sites of pETM-30 to be expressed with TEV-cleavable His-GST tags. Unlabelled and isotopically 15N- and 13C-labelled proteins were expressed in Escherichia coli BL21 (DE3) (Invitrogen) in LB or minimal (M9) medium supplemented with 15NH4Cl or 15NH4Cl and 13C-D-glucose. All His-GST-fusion proteins were initially purified by nickel affinity chromatography. The bulky His-GST-fusion tags were then cleaved with TEV protease and removed by using a second nickel affinity step. The constructs were further purified on a Superdex-75 (Pharmacia) gel filtration column. Samples were concentrated to 0.3–1.5 mM in 10 mM Tris-HCl buffer (pH 7.4) with 50 mM NaCl and 1 mM TCEP. Protein concentration was determined by a combination of spectrophotometry with predicted extinction coefficients and ninhydrin analysis of protein hydrolysates.
All RNA oligonucleotides were chemically synthesized (Curevac and Dharmacon).
Circular dichroism spectroscopy
All circular dichroism (CD) spectra were recorded on Jasco J-715 spectropolarimeter equipped with a PTC-348 Peltier temperature-control system. CD intensities are presented as the CD absorption coefficient calculated by using the molar concentrations of the proteins. Thermal unfolding was monitored between 10 or 20°C and 90 or 95°C, depending on the constructs. Temperature was increased at a rate of 1°C/min and unfolding was monitored by recording the signal at 220 nm. Reversibility of the unfolding was assessed by cooling to 10 or 20°C at the same rate. Protein concentrations were 1–2 µM in 10 mM Tris–HCl buffer (pH 7.4), 100 mM NaCl, 1 mM TCEP. The data were fit to a two-state native-denatured model, while two independent unfolding transitions where used for the KH23 didomain. Data fit was performed with in house software as described in (9).
RNA binding was monitored by adding increasing amounts of protein to 1–2 µM AU-12mer RNA in 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 1 mM TCEP. A temperature of 5°C was chosen to optimize the signal change upon protein binding. The average signal between 255 and 265 nm was fitted against the protein concentration using in house software (9).
Nuclear magnetic resonance spectroscopy
The nuclear magnetic resonance (NMR) samples of the different KH constructs were prepared in 90% H2O/10% D2O solutions of 10 mM Tris-HCl buffer (pH 7.4), 100 mM NaCl, 1 mM TCEP, 0.02% NaN3 at concentrations in the range 0.3–1.5 mM. NMR spectra were recorded at 300K on Varian Inova and Bruker Avance spectrometers operating at 800 and 600 MHz 1H frequencies. The spectra were processed with NMRPipe (10) and analysed with Sparky (11).
Standard 3D NMR experiments (HNCACB, HNCA and HNCO) were used to obtain sequence specific 1HN, 15N, 13Cα, 13Cβ and 13C’ backbone assignments (12). Side-chain aliphatic proton and carbon assignments were achieved using data from a combination of 3D 15N and 13C-edited TOCSY and NOESY-HSQC spectra (13) with 70 and 100 ms mixing time for TOCSY and NOESY experiments respectively, plus a HCCH–TOCSY experiment (14). Water suppression was achieved by the WATERGATE pulse sequence (15). 3JHN-Hα scalar couplings were measured from HNHA experiment as described previously (16).
15N relaxation parameters (T1, T2 and {1H}-15N NOE) were obtained from standard experiments (17) recorded at 600 MHz 1H frequency and 300K and analysed using NMRPipe routines (10). The program TENSOR (18) was used to determine amplitude and rhombicity for the diffusion tensor of KH2, KH3 and KH4 in isolation and in two-domain constructs. The same program was used to estimate the reported rotational correlation times (τc). Estimates of the rotational correlation times of wild-type (8.5 ns) and double mutant (7.3 ns) were also obtained using a fast method that relies on changes in the intensity of the downfield region of 1D proton spectrum (Kelly et al., manuscript in preparation).
15N-1H amide residual dipolar coupling (RDCs) were measured using a magnetically oriented binary mixture of ∼5% (v/v) alkyl-poly(ethylenglycol) C12E5 and 1% (v/v) hexanol and IPAP experiments as described in (6). The fit between the 15N–1H amide RDCs predicted from the KH23 structure and the ones measured experimentally was calculated using the program Module (19).
The thermal stability of the domains was monitored by recording 15N-HSQC spectra on 600 MHz (single domains) and 800 MHz (KH23) Varian Inova spectrometers on ∼0.3 mM protein samples in 10 mM Tris–HCl buffer (pH 7.4), 50 mM NaCl, 1 mM TCEP. Spectra were recorded at 3°C intervals between 27 and 69°C. In all cases, except for KH2 + linker (233–324), the protein unfolding was fully reversible.
The effects of RNA complex formation on KH constructs were followed by acquiring 15N-HSQC spectra during titrations of a 50 µM sample of 15N-labelled KH domain(s) in 10 mM Tris-HCl buffer (pH 7.4), 50 mM NaCl, 1 mM TCEP with the three (5′-UAUUUAUU-3′, 5′-UAUUUAUUAU-3′ and 5′-UAUUUAUUAUUU-3′) RNA oligos. Titration curves were obtained by plotting chemical shift perturbations (ΔδBind) against the molar ratio of RNA/protein. Non-linear least squares fits to a 1:1 binding model (20) were performed in Origin 7.5 (Microcal. Inc.). This model accounts for a dilution effect of both molecules during the titration, with the ratio RNA/protein and ΔδBind as the independent and dependent variables, respectively. The dissociation binding constant (Kd) and the maximum chemical shift change (Δδmax) were the fitted parameters. A global fit of the data was performed in which the curves from different amide resonances were fitted simultaneously to a single Kd value, whereas the Δδmax for each resonance was allowed to vary. Weighted average values of 15N and 1H chemical shift perturbations (Δδavg) of each amide was calculated as follows: Δδavg = (([ΔδH]2 + [ΔδN / 5]2)/2)1/2.
Structure calculation and analysis
Structure calculations for KH2 (221–305) and KH23 (233–418) were performed with ARIA 1.2 (21) using distance and dihedral angles restraints. Dihedral restraints were obtained from experimentally measured scalar couplings (ϕ; (22)) or from the chemical shift-based TALOS database (ϕ/φ; (23)). Experimental distance restraints (Table 1) were derived from the integration (XEASY; 24) of Sparky (11)-derived NOE peak lists (6). H-bond constraints were added when unambiguously identified by the structural analysis of preliminary structures.
Table 1.
NMR and water refinement statistics
| KH2 (221–305) | KH23 (233–418) | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance restraints | 2149 | 3229 |
| Intra-residue | 886 | 1373 |
| Inter-residue | 1263 | 1856 |
| Sequential (|i – j| = 1) | 428 | 551 |
| Medium range (1 < |i – j| < 4) | 321 | 401 |
| Long-range (|i – j| > 4) | 514 | 904 |
| Experimental ϕ constraints | 20 | 66 |
| TALOS ϕ and φ constraints | 52 | 107 |
| Hydrogen bond constraints | 28 | 51 |
| Structure statistics | (25 conformers) | (20 conformers) |
| Mean total energy (kcal/mol) | −3498 ± 53 | −6857 ± 132 |
| NOE violations >0.3 Å | 0.2 ± 0.4 | 0.9 ± 0.2 |
| Mean NOE energy (kcal/mol) | 33 ± 7 | 81 ± 9 |
| RMSD from idealized covalent geometry | ||
| Bond lengths (Å) | 0.0036 ± 0.0001 | 0.0039 ± 0.0009 |
| Bond angles (°) | 0.483 ± 0.019 | 0.542 ± 0.015 |
| RMSD from the mean structure (Å) | ||
| Whole structured domaina,b | ||
| Backbone atoms | 0.79 ± 0.18 | 1.43 ± 0.33 |
| Heavy atoms | 1.45 ± 0.25 | 2.08 ± 0.37 |
| Secondary structurec,d | ||
| Backbone atoms | 0.49 ± 0.10 | 0.95 ± 0.26 |
| Heavy atoms | 1.19 ± 0.22 | 1.52 ± 0.29 |
| Ramachandran plot analysis | ||
| Most favoured regions (%) | 87.9 | 85.5 |
| Additional allowed regions (%) | 11.1 | 12.0 |
| Generously allowed regions (%) | 0.1 | 1.0 |
| Disallowed regions (%) | 0.9 | 1.5 |
aKH2 whole structured domain: 234–304.
bKH23 whole structured domain: 234–392.
cKH2 secondary structure: 234–249, 254–270, 281–304.
dKH23 secondary structure: 234–249, 254–270, 281–304, 325–338, 344–359, 368–392.
Two hundred randomized conformers underwent simulated annealing with a standard CNS protocol as described in (25) and the 25/20 lowest energy structures for KH2/KH23 were refined in water (21). Structural statistics are shown in Table 1, structure quality was evaluated by using the program PROCHECK-NMR (26). The program MOLMOL (27) was used to visualize the conformers, and to create figures. Multiple sequence alignment was executed within the CLUSTALX program (28).
Cell transfections and ribonucleoprotein complexes immunoprecipitation
HEK293 cells were transfected (using Lipofectamine Plus, Invitrogen) with empty pCMV-TAG2B vector (mock) or the same vector expressing either wild-type KSRP (KSRP) or KSRPF311A/Y317A double mutant (KSRPF311A/Y317A). ribonucleoprotein complexes immunoprecipitation (RIP) assays were performed as previously described by (29) with some modifications. Briefly, cells lysates were immunoprecipitated with either Protein A- or ProteinA/ProteinG-Sepharose-coupled antibodies at 4°C overnight. Pellets were sequentially washed with the following buffers: Buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 150 mM NaCl); Buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 500 mM NaCl) and Buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1). Total RNA was prepared using Trizol (Invitrogen, Carlsbad, CA, USA) retro-transcribed using random primers and amplified by PCR.
The primer sequences are:
hsa.PP2ACA forward 5′— GAT GGA GGG ATA TAA CTG GTG CC —3′
hsa.PP2ACA reverse 5′— ACG AGG TGC TGG GTC AAA CT —3′
hsa.CTNNB1 forward 5′— TTG GAA CCT TGT TTT GGA CA —3′
hsa.CTNNB1 reverse 5′— ACC GCA TTT TCT CTT GAA GC —3′
RESULTS
Characterization of the KH2–KH3 protein
We have used NMR data on chemical shift perturbation (CSP), hydrodynamic behaviour and stability to define the relation between the KH2 and KH3 domains. Our data indicate that an interaction between KH2 and KH3 is taking place in which the linker plays a pivotal role.
The superposition of the 15N-HSQC spectra (Supplementary Figure S1) of the isolated KH2 (233–305) and KH3 (317–418) domains on that of the two-domain KH23 construct (233–418) shows significant chemical shift differences (Δδavg > 0.02 ppm) for resonances of both domains which are mapped in Figure 2A. The largest chemical shift changes are observed for residues of KH2’s α1/α3 and KH3’s β1 (see the structure of KH2 in the next paragraph and ref. 6), indicating that the linker is likely to be sandwiched between the two domains. Indeed, the addition of the linker to the KH2 domain (KH2+linker construct) results in very substantial chemical shift changes in the KH2 resonances (Supplementary Figure S1), while inclusion of KH3 causes only small additional chemical shift perturbations. Furthermore, resonances of the last seven amino acids of the linker (317–324), which are not visible when this short stretch is attached to KH3 in the (317–418) construct, become clearly visible in the KH23 construct, indicating that further contacts are taking place in the full two-domain construct. Importantly, no significant chemical shift differences are observed between the isolated KH23 and the two domains within the KH1234 construct (Figure 2B), which confirms that the features of the KH23 interaction are conserved in the longer protein.
Figure 2.
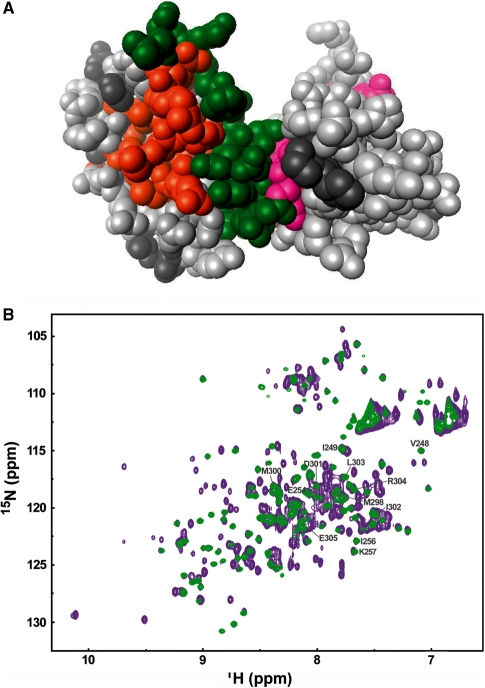
Interaction between KH2 and KH3 in the two- and four- (KH1–KH4) domain constructs. (A) Mapping of the residues of KH2 and KH3 whose 15N and 1H amide chemical shift changes in the two domain construct on a space filling representation of KH23. Residues of KH2 (233–305) and KH3 (317–418) whose amide resonances Δδavg (ppm) is <0.2 are in light grey, while residues whose Δδavg ≥ 0.2 are in orange for KH2 and magenta for KH3. Residues in the linker are in green, unassigned residues in dark grey. (B) The KH2–KH3 arrangement is conserved in KH1234 Superimposition of 15N-HSQC spectra of KH23 (233–418), green, and KSRP KH1234, purple. Grey labels correspond to KH2 signals which shift upon addition of the KH2–KH3 linker and/or KH3 binding. The absence of significant chemical shift changes in the resonances of KH2, KH3 and the linker indicate that the relative orientation of the KH2 and KH3 domains and their contacts with the linker are conserved in the longer protein.
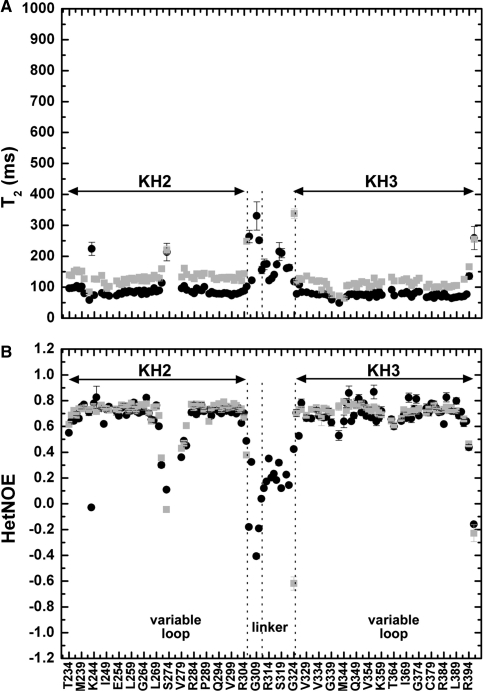
15N T1, T2, and heteronuclear NOE values recorded on the KH2, KH3 and KH23 proteins indicate that KH23 encompasses two relatively rigid regions (corresponding to the two KH domains) joined by a more mobile 18-residue long linker (Figure 3 and Supplementary Figure S2). The intra-domain trends of 15N T1, T2 and heteronuclear NOE values for KH2 and KH3 are consistent with what is observed for other KH domains and do not change from the single to the di-domain constructs (Figure 3). The linker comprises two distinct regions corresponding to amino acids 306–310 and 311–324. The trend and values of the relaxation parameters of amino acids 306–310 are consistent with a fully flexible protein chain (30), while the ones of residues 311–324 are in between those of the structured KH domains and those of the flexible 306–310 residues (Figure 3). This second part of the linker is much more flexible in the KH2+linker construct (Supplementary Figure S2) in agreement with the linker being sandwiched between the two KH domains. The KH23 construct has a rotational correlation time (τc) of 8.92 ns, in the range of τc/MW reported by Dayie and co-workers (31) and significantly larger than the one of the isolated KH2 and KH3 domains (5.27 and 6.36 ns in the isolated domains), consistently with the two domains interacting.
Figure 3.
Internal motions in KH2, KH3 and KH23. T2 (A) and Heteronuclear NOE (B) values of KH2 (233–305) and KH3 (317–418) amide resonances, in grey (as reported in 8 and 6, respectively), are compared with equivalent data on KH23 (233–418) in black. The positions of the variable loop(s) and inter-domain linker in the protein sequence are indicated at the bottom of the figure. The arrows span the KH2 and KH3 domains and two vertical lines define the boundaries of the more and less flexible parts of the linker. Residues in the invariant loops of KH2 and KH3 are characterized by T2 and heteronuclear values shorter than the average, suggesting conformational exchanges with rates near µs to ms time scale (30) while residues in the variable loops displays a behaviour consistent with motions in the nanosecond range.
The rhombicity (r) values of KH2 and KH3 in solution change very significantly between isolated domains and domains within the two-domain construct (Supplementary Table S1). This contrasts with what we observe for the KH34 construct, where the non-interacting KH3 and KH4 (6) have similar r values to the isolated KH3 and KH4 domains (Supplementary Table S1).
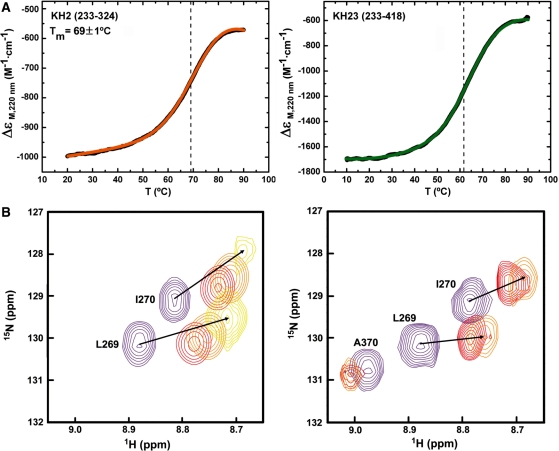
Finally, significant changes in the stability of one of the two domains confirm that an inter-domain interaction is taking place (32). CD spectroscopy showed that the isolated KH2 and KH3 domains have Tm of 69.1 (Figure 4A) and 62°C (6), respectively. The unfolding curve for KH23 (Figure 4A) cannot be reproduced by summation of the curves for the individual domains, which should be the case if no interaction between domains was present. It is worth noting that addition of the KH2-KH3 linker to KH2 does not change its Tm. A change in KH2 stability in the KH23 construct is confirmed by the NMR spectra recorded during the thermal unfolding of KH2 and KH23 (3°C intervals over the temperature range of 27–69°C), where unfolding of KH2 and KH3 domains can be followed separately. Comparison of the KH2 resonances in the two experiments shows that the domain is ∼10°C more stable when in isolation (Figure 4B). Fittingly, no such change is observed in the unfolding of KH1, KH2, KH3 and KH4 in isolation and in the KH12 and KH34 constructs (6,8)—where no inter-domain contact is present.
Figure 4.
Thermal denaturation of KH2 and KH23. (A) 220 nm CD signal of KH2 (233–324) (left) and KH23 (233–418) (right). Smoothed data are in black, fitted curves in colour and the transition midpoints are indicated by dashed lines. (B) NMR superimposition of four 15N-1H HSQC NMR spectra recorded during the thermal unfolding of the KH2 (233–305) (left) and KH23 (233–418) (right) proteins. The spectra are recorded at 27°C (purple), 48°C (red), 51°C (orange) and 63°C (yellow). KH2 resonances shift towards the random-coil region of the spectrum and disappear at temperatures above 51°C in the KH23 construct, but they are still visible at 63°C in the isolated domain.
Structure of KH2 and KH23 and role of the linker in inter-domain arrangement
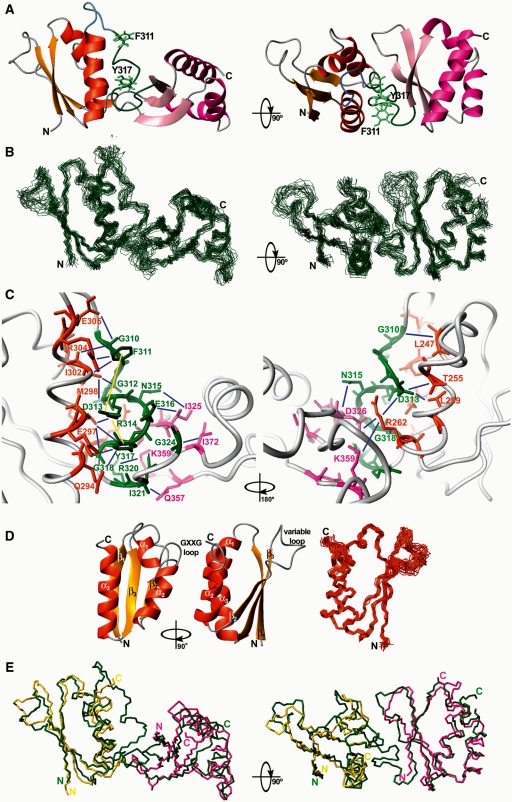
Next, we solved the structures of the KSRP KH2 (221–305) and KH23 (233–418) proteins using NMR spectroscopy. In KH23, KH2 and KH3 are positioned at a ∼ 90° angle to form an L-shaped structure, in a KH–KH orientation never observed before (Figure 5A–C). The KH2–KH3 linker starts with a short four-amino acid flexible turn followed by a partially structured stretch that runs anti-parallel to KH2 α3 and lies between by KH2 α1, α2 and α3. Then the linker turns and makes contact with KH3. The conformation of the section of the linker interacting with KH2 is not extended; the polypeptide backbone forms one helical turn and a broad double turn (Figure 5A). Subsequently, the backbone is arranged in two kinks that form the joint of the KH23 L-shaped structure and define domain–domain orientation. The surface buried by the contacts between KH2 and the linker is ∼1500 Å2 and the one between KH3 and the linker is ∼800 Å2. This difference explains the more extensive chemical shift changes for KH2 described in the previous paragraph. The relative orientation of KH2 and KH3 is defined mainly by domain(s)–linker contacts but also by a few domain–domain ones that bury 285 Å2 of surface. Domain–domain and domain–linker interactions are mediated by side chain–side chain interactions and do not rely on the formation of stable H-bonds or any extension of the secondary structure of the protein. In particular, two hydrophobic amino acids from the linker (F311 and Y317) make extensive contacts with KH2 (Figure 5C). The two aromatic side chains sandwich the side chain of R314 creating two cation–π interactions and stabilizing the relative position of the three residues. The inter-domain arrangement is defined by 151 linker-domain (KH2 α1, α2 and α3 and KH3 β1 and β3), 83 linker–linker and 5 domain–domain NOEs (Figure 5C, Supplementary Table S2). The orientation of the two domains is also confirmed by residual dipolar coupling (RDC) analysis. RDCs recorded on a sample of KH23 fit to the values predicted from the KH23 structure (Supplementary Figure S4). It is interesting to note that the relaxation measurements reported above indicate that the backbone of residues in the linker shows a significant flexibility. This flexibility could be related to the observed lack of H-bonds involving backbone moieties of the linker or/and could indicate the existence of minor conformers with a less structured linker.
Figure 5.
Structure of KH2 and KH23. (A) 90° rotated ribbon representations of the solution structure of the KSRP KH23 (233–418) di-domain. KH2 is displayed in orange, KH3 in pink. The backbone of the KH2–KH3 linker is coloured in light blue for the more flexible (306–310) residues and in dark green for the less flexible (311–324) ones. The side-chains of F311 and Y317 are shown in light green. Domain–linker and domain–domain contacts define the inter-domain orientation. (B) 90° rotated superpositions of the KH23 backbone (Cα trace, in dark green) for the 20 lowest energy conformers on the average structure. (C) 180° rotated close ups of the domain–linker and domain–domain interactions within the KH23 structure. KH2 residues interacting with the linker and with KH3 are in orange, while linker residues are in green and KH3 residues interacting with the linker and KH2 are in pink. A selection of long range NOEs between the linker and the two domains and between domains are represented as blue lines, while yellow lines represent the intra linker NOEs connecting the side chains of the two F311 and Y317 aromatic residues with the positively charged R314. (D) Left and middle—90° rotated ribbon representations of the solution structure of KH2 (221–305). Right—superposition of the KH2 backbone (Cα trace, in orange) for the 25 lowest energy conformers and the average structure. The KH2 structure is well defined and shows that the domain assumes a typical KH fold. (E) 90° rotated superimpositions of the backbone traces of KH23 di-domain (green), KH2 (yellow) and KH3 (magenta) (lowest energy conformers). N- and C-termini are labelled.
The comparison of the structures of the isolated KH2 (this article, Figure 5D) and KH3 (6) with the one of the two domains in the KH23 construct shows no major changes in the arrangements of the secondary structure elements, with the most significant differences observed at the C-end of KH2 α3 and the N-end of KH3 β1 (Figure 5E). Differences were also observed in the average conformation of the flexible (Figure 3) β2–β3 loops of both domains, most likely reflecting the scarce structural information available for these regions. The structures of the isolated KH2 and KH3 domains (Figure 5D) are superimposable to the ones of the domains within the KH23 construct with an RMSD of 0.95 Å (KH23–KH2, backbone atoms) and 1.05 Å (KH23–KH3, backbone atoms). Both structures are well defined and the Ramachandran plots indicate that the very few residues in the disallowed regions (1.5% for KH23 and 0.9% for KH2) (Table 1) belong to loops that are only partially assigned. Importantly the orientation of the two domains is also well defined (Figure 5A and B).
The structure presented here provides the first example of an intra-molecular KH–KH interaction mediated by the inter-domain linker. The high level of conservation of the linker amino acids in the KSRP (and FBP) family (Figure 1B)—especially of the two aromatic residues F311 and Y317—strongly suggests that the orientation of the KH2 and KH3 domains is conserved in man, rat and chicken.
Inter-domain arrangement and RNA binding
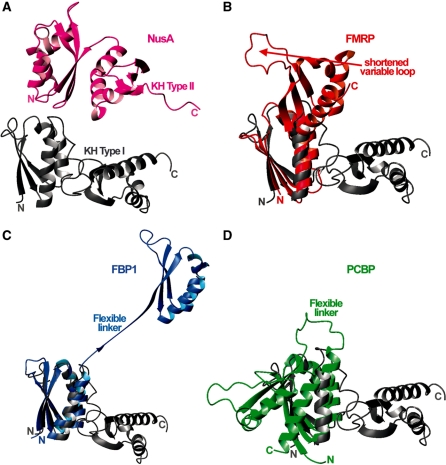
The structure of the KH23 construct shows that the nucleic acid binding grooves of KH2 and KH3 are ∼90° rotated and non-contiguous, adopting an orientation very different from the ones reported so far in KH–KH di-domains (33–36) (Figure 6). To understand how the KH2–KH3 orientation relates to RNA binding we tested the interaction of KH23 with a 12mer RNA (UAUUUAUUAUUU) that recapitulates the sequence of the TNFα ARE in vitro. The same RNA had been previously used to test the RNA binding of the KH3–KH4 domain pair, while shorter oligos spanning the 12mer sequence had been used to test the four isolated domains (6–8).
Figure 6.
Comparison between the structure of KH23 and the ones of the other two-KH domain constructs available in the PDB. The ribbon representation of the KH23 structure (grey) is compared with similar representations of the (A) NusA KH12 in magenta; (B) FMRP KH12 in red; (C) FBP1 KH34 in blue and (D) PCPB KH12 in green, structures. As the topologies of the E.coli NusA KH domains are different from the eukaryotic ones, this structure was displayed above KH23, while the amino terminal domains of FMRP KH12, FBP1 KH34 and PCBP KH12 were superimposed to KH2 within KH23.
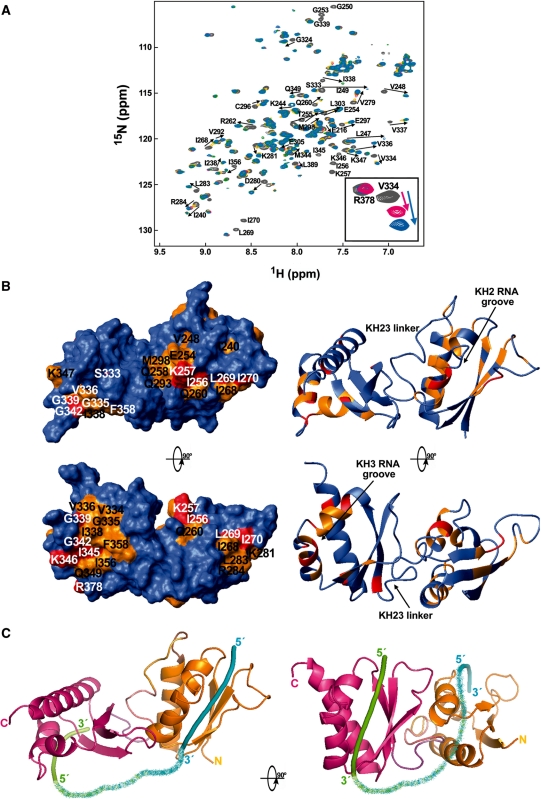
NMR CSP data obtained titrating 15N-labelled KH23 with the TNFα 12mer (Figure 7A) revealed not only that both KH2 and KH3 bind RNA using the ‘classical’ nucleic acid-binding grooves (Figure 7B) (37), but also that KH2- and KH3-binding surfaces do not coalesce. The lack of significant chemical shift changes in the resonances of the KH2–KH3 linker indicate that the linker is not directly involved in RNA binding and that no significant structural rearrangement is taking place in the domains interface. Smaller but clearly detectable chemical shift perturbations involve residues V276, C280, E281, M282, M284, L286 and E288 on KH2 α3. These small changes are also observed when short UAUUUA and UAUUAU oligos bind to KH2 and KH3 (data not shown) as well as in Nova-1 KH3-RNA binding (38) and they are probably caused by minor conformational effects linked to RNA interaction.
Figure 7.
Binding of KH23 to the 5′ UAUUUAUUAUUU 3′ RNA. (A) Superposition of a series of 15N-HSQC spectra recorded during a titration of KH23 (233–418) with increasing amounts of UAUUUAUUAUUU RNA. The spectra are coloured according to RNA:protein ratios: grey for free protein, yellow for 0.5:1 RNA:protein ratio, magenta for 0.75:1, green for 1:1 and cyan for 1.5:1. Resonances that shift most during the titration (Δδavg ≥ 0.05 ppm) are labelled in black. (B) Left, top—KH23 (233–418) protein surface, coloured according to the chemical shift changes undergone by the NH resonances upon RNA binding. Residues with Δδavg (ppm) <0.05 are in blue, while residues with Δδavg (ppm) ≥0.05 are in orange. Red indicates residues whose resonances are broadened beyond detection. Left, bottom—90° rotated surface. Right—Molmol ribbon representation of the KH23 (233–418) structure with an equivalent colour coding and orientation. The RNA binds in the classical nucleic acid-binding grooves of KH2 and KH3 defined by helices α1 and α2 and the GXXG loop on one side and the β2 strand on the other. Resonances of the linker are not affected upon binding. (C) Pymol ribbon representations of the KH23 (233–418) structure. KH2 is represented in orange, KH3 in magenta and the linker follows a colour gradient between the domain colours. In these representations, two short nucleic acid molecules (pentamers) have been placed in the KH2 and KH3 nucleic acid-binding grooves (cyan and green) based on existing structures (FBP KH34–DNA complex) and connected by a ribbon that follows a colour gradient between the colours of the two RNAs. On the left, the shortest route for a connection allows a RNA 12mer to bind both domains simultaneously and follows the route mapped by our CSP data. The alternative route, on the right panel, is significantly longer. In both cases the RNA must undergo a change of direction.
During the titration, signals in fast exchange regime on the NMR timescale shifted linearly with concentration, reaching saturation at a ∼ 1 : 1 stoichiometry (Figure 7A). This suggests tight binding with a Kd in the low micromolar range for the complex and is consistent with the Kd obtained for the same complex by monitoring a reverse titration (RNA into protein) by CD spectroscopy (1.0 ± 0.1 µM, Supplementary Figure S3a). As a comparison, the isolated KH2 and KH3 bind to the two UAUUAU and UAUUUA 6mers with Kd value ranging from ∼100 to ∼400 µM (6,7), while the Kd value of the complex between KH3–KH4 and the UAUUUAUUAUUU RNA is 0.2 µM (6). These in vitro results indicated that the RNA-binding surfaces of KH2 and KH3 are both accessible and involved in RNA binding but they do not form a continuous surface so that the RNA chain must bend to interact with both binding grooves, as shown in Figure 7C. However, the relation between KH2–KH3 orientation and the accessibility of single-stranded regions within the RNA structure could be important in defining the target in the context of the structured 3′UTR.
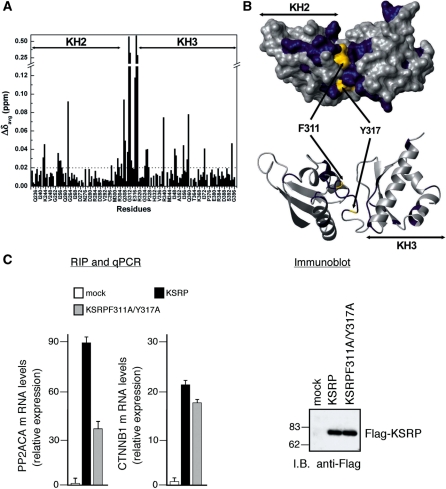
In order to assess if the KH2–KH3 orientation plays a role in the discrimination of the ARE targets within the large protein–RNA complexes assembled on the mRNA 3′UTR we have tested the effect of interfering with the inter-domain arrangement on the capability of KSRP to interact with its ARE targets in the HEK293 cell line. Based on the structure of KH23, we designed a double mutant (F311A/Y317A)—where removal of the two aromatic side chains in the linker perturbs KH2–KH3 interaction. The chemical shift perturbations in the 15N-HSQC spectrum of the wild type upon mutating F311 and Y317 indicate not only that substantial changes have taken place in the interface, but also that the domains maintain the correct fold (Figure 8A and B). Furthermore, we have measured the effect of the double mutation on the hydrodynamic properties of the molecule and we showed that the correlation time of the alanine double mutant is 1.2 ns lower than the one of the wild type, confirming a change in the inter-domain arrangement.
Figure 8.
KH2–KH3 orientation regulates RNA binding in the cell. (A) Averaged chemical shift differences (ΔδAvg) between the wild-type KH23 and the KH23F311A/Y317A double mutant plotted against the protein sequence. The dashed line marks the ΔδAvg = 0.02 cut-off used for the surface representation in Figure 8b. (B) Top—surface representation of the KH23 (233–418) structure displaying the chemical shift changes of Figure 8a. In blue are residues with ΔδAvg ≥ 0.02, in grey are residues with ΔδAvg < 0.02. F311 and Y317 are in yellow. Bottom – Ribbon representation of KH23 with the same orientation and colour coding. (C) HEK293 cells were transiently transfected with a control empty pCMV–TAG2B vector (mock) or a vector expressing either wild-type KSRP (KSRP) or KSRPF311A/Y317A double mutant (KSRPF311A/Y317A). Cells were lysed, total cell extracts were immunoprecipitated with anti-FLAG antibody, RNA was purified from immunocomplexes and analysed by qPCR to detect PP2ACA and beta-catenin (CTNNB1) mRNA. The values shown are averages (±SEM) of three independent experiments performed in triplicate (left panel). Western blot analysis of FLAG-tagged proteins in total extracts of transfected HEK293 cells (right panel).
We then assessed the capability of the full length KSRPF311A/Y317A double mutant to (i) bind the UAUUUAUUAUUU RNA in vitro and (ii) bind to two different (Supplementary Figure S6) and well characterized KSRP mRNA targets. Interestingly, the double mutation in the linker affected the interaction of KSRP with the UAUUUAUUAUUU RNA only to a very small degree (Supplementary Figure S7) with the binding to the double mutant being marginally higher (0.4 ± 0.05 versus 1.0 ± 0.2 µM) than the wild type one. Instead, the double mutation affected in vivo binding by two different target mRNAs to a very different degree. The interaction of the KSRPF311A/Y317A double mutant to PP2ACA mRNA was more than halved when compared to wild-type KSRP, while the interaction with the β-catenin mRNA decreased only 15% (Figure 8C). This indicates that the role of the inter-domain orientation is mRNA target-dependent, and presumably relates to the 3′UTR context enclosing the AREs.
DISCUSSION
The structure of KH23 reveals that the two central domains of the KSRP protein assume a novel inter-domain arrangement (Figure 6). Key to the relative positioning of the KH2 and KH3 domains is the 18-amino acid-long inter-domain linker that is sandwiched between domains. Although the two domains make a few direct contacts via KH2 α2 and KH3 β1/variable loop, the inter-domain interface is very small compared with the one between the domains and the linker. The linker acts as a pivot between the two domains and, indirectly, provides a novel structural solution to the recognition of low-sequence complexity RNA targets.
The role played by the linker in KH23 is different from what is observed for the structures of other KH domain pairs that have either very short linkers that constrain the inter-domain orientation by simple tethering or very flexible linkers that do not interact with the domains. In NusA (33), the short 6-nt linker is used to position the two domains and creates a continuous RNA-binding surface (Figure 6A). Similarly, in FMRP KH1–KH2 (34) only a few residues separate the two domains (Figure 6B). In contrast, in FBP1 (Figure 6C), the KH3–KH4 linker is a long and flexible 30-residue connector and this flexibility allows KH4 to bind a DNA sequence 6 nt 5′ of the sequence bound by KH3 (35). In PCBP (36) the long KH1–KH2 linker allows the domains to assume an anti-parallel orientation and to form a six-stranded inter-domain β-sheet (Figure 6D).
The simultaneous engagement of several RNA-binding domains increases the RNA-binding affinity of a protein whether or not the two domains make contact. Tethering of two RNA-binding domains implies that binding of the first domain increases the local concentration of the second, facilitating RNA binding. A simple model, in which the linker does not contact the RNA, was developed to estimate the effect of the linker length on RNA binding affinity (39) and demonstrates how a short linker results in a stronger cooperative effect. However, a fixed inter-domain orientation has further implications. By using two domains in a well-defined orientation it is possible to fix the distance between binding sites on the RNA, elongating the binding sequence and increasing the specificity. For example, NusA KH1–KH2 makes contacts with 11 RNA nucleotides (33). Furthermore, although a direct participation of the linker in the recognition of the RNA has not been reported for KH domains this mechanism has been observed in RNA recognition by RRM domains and zinc-fingers (40). There, functional groups of the protein linker take part in the interaction and the linker itself can become structured upon binding, boosting the binding affinity. In KSRP KH23 instead the KH2 and KH3, RNA-binding surfaces do not coalesce in a continuous area and the linker is not directly involved in the interaction.
Chemical shift perturbation data confirm that RNA binding is mediated by the canonical RNA-binding grooves of KH2 and KH3. The two RNA stretches bound by these grooves could be connected on either side of KH23 (Figure 7C), but these two putative connections would have very different length. Considering that at least 4 nt are bound by each KH domain, the length of the RNA target able to span two sites (∼12 nt) argues that the RNA is binding KH2 and KH3 in a 5′–3′ orientation, bridging the gap between binding surfaces by the shortest possible route (Figure 7C). This model is consistent with the lack of chemical shift changes on the linker side of the molecule when both sites are bound. It is interesting to consider that a 5′–3′ direction of protein–RNA binding would be similar to the one observed for oriented the KH1 and KH2 domains of NusA and is the reverse than the one observed for the KH3 and KH4 domains of FBP, that do not assume a fixed inter-domain orientation.
The affinity of KSRP KH23 for the KSRP target UAUUUAUUAUUU RNA is comparable (slightly lower in fact) to the affinity of KH34 for the same RNA despite the fact KH3 and KH4 do not contact each other. Here, tethering of the KH2 and KH3 domains does not create a protein–RNA high-affinity binding unit. Indeed, perturbing the inter-domain orientation by mutating two aromatic residues engaged in linker–KH2 interactions does result in only a very small increase in the capability of the protein to bind the UAUUUAUUAUUU RNA—that could be due to the two domain assuming a better orientation—or to spurious contributions by the positively charged residues present in the linker. Previous analysis of the sequence preference of the KH domains of KSRP (7) shows that the protein does not have specificity for AU-rich sequences per se. Indeed, the ARE targets of KSRP cover a broad range of sequences indicating that recognition of a long sequence in a very specific fashion is unlikely to be a major factor in target discrimination—a likely scenario is a direct role of the KH23 architecture in selecting KSRP ARE targets. The data reported in this manuscript support this hypothesis. In our system, binding of the two domains implies a bend in the backbone of the RNA polymer (Figure 7C)—similarly to what observed for the PTB protein (41)—regardless in fact of their orientation. The AU-rich targets of KSRP are located in highly structured 3′UTRs and the change in the direction of the RNA chain could contribute to the general architecture of large KSRP–RNA–protein complexes by stabilizing a suitable conformation. The results of our in cell assays, indicate a target-dependent role of the KH2–KH3 interaction confirming that KSRP–RNA recognition is a multi-factorial process that cannot be explained with a simple zip code recognition model as we proposed earlier (7).
In this manuscript we show that the central KH domains of KSRP interact, forming a single structural unit. Purpose of this interaction is not to create a high-affinity binding unit for AU-rich targets or to elongate the sequence recognized by KSRP but rather to relate the context of the 3′UTR to the domain arrangement of the protein.
ACCESSION NUMBER
The coordinates of the KH2 (221–305) and KH23 (233–418) domains from KSRP have been deposited in the PDB with the accession identifiers 2OPV and 2JVZ, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust Grant WT082088MA and the MRC Grant-in-Aid U117574558. I.D.M. was supported by European Molecular Biology Organization (EMBO), fellowship number 240-2005. The work of P.B. and R.G. has been partly supported by grants from Italian ISS (526D/39) to P.B., ISS (527B/2B/6), Associazione Italiana per la Ricerca sul Cancro (AIRC), and CIPE 2007 (Regione Liguria) to R.G. Funding for open access charge: MRC grant-in-aid (U117574558).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr C. De Chiara for advice on the ARIA protocols, Dr T. A. Frenkiel and Dr A. Oreggioni for their help in recording spectra, Dr A. Pandini for help with Bioinformatics tools and Dr L. Masino for general advice on data fitting. The plasmid of the original KSRP clone was provided by Dr CY Chen. All NMR spectra were recorded at the Medical Research Council Biomedical NMR centre.
REFERENCES
- 1.Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol. Cell. 2004;96:479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Kontoyannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 3.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2006;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 5.Gherzi R, Lee KY, Briata P, Wegmüller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.García-Mayoral MF, Hollingworth D, Masino L, Díaz-Moreno I, Kelly G, Gherzi R, Chou CF, Chen CY, Ramos A. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–498. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 7.García-Mayoral MF, Díaz-Moreno I, Hollingworth D, Ramos A. The sequence selectivity of KSRP explains the recognition of its RNA targets. Nucleic Acids Res. 2008;36:5290–5296. doi: 10.1093/nar/gkn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Díaz-Moreno I, Hollingworth D, Frenkiel TA, Kelly G, Martin S, Howell S, García-Mayoral MF, Gherzi R, Briata P, Ramos A. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localisation and mRNA decay. Nat. Struct. Mol. Biol. 2009;16:238–246. doi: 10.1038/nsmb.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin SR, Biekofsky RR, Skinner MA, Guerrini R, Salvadori S, Feeney J, Bayley PM. Interaction of calmodulin with the phosphofructokinase target sequence. FEBS Lett. 2004;577:284–288. doi: 10.1016/j.febslet.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Delaglio F, Grzesiek S, Vuister GV, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 11.Goddard TD, Kneller DG. SPARKY. San Francisco: University of California; 2004. [Google Scholar]
- 12.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc. Chem. Res. 1993;26:131–138. [Google Scholar]
- 13.Fesik SW, Zuiderweg ERP. Heteronuclear 3-dimensional NMR spectroscopy. A strategy for the simplification of homonuclear two-dimensional NMR spectra. J. Magn. Reson. 1988;78:588–593. [Google Scholar]
- 14.Kay LE, Xu G, Singer AU, Muhandiram DR, Forman-Kay JD. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlation in H2O samples of proteins. J. Magn. Reson. 1993;101:333–337. [Google Scholar]
- 15.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 16.Vuister GV, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 1993;115:7772–7777. [Google Scholar]
- 17.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 18.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 19.Dosset P, Hus JC, Blackledge M, Marion D. A novel interactive tool for rigid-body modeling of multi-domain macomolecules using residual dipolar couplings. J. Biomol. NMR. 2001;20:223–231. doi: 10.1023/a:1011206132740. [DOI] [PubMed] [Google Scholar]
- 20.Kannt A, Young S, Bendall DS. The role of acidic residues of plastocyanin in its interaction with cytochrome f. Biochim. Biophys. Acta. 1996;1277:115–126. [PubMed] [Google Scholar]
- 21.Linge JP, O'Donoghue SI, Nilges M. Automated asignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 22.Ye JQ, Mayer KL, Mayer MR, Stone MJ. NMR solution structure and backbone dynamics of the CC chemokine eotaxin-3. Biochemistry. 2001;40:7820–7831. doi: 10.1021/bi010252s. [DOI] [PubMed] [Google Scholar]
- 23.Cornilescu G, Delaglio F, Bax A. A protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 24.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 25.DeChiara C, Menon RP, Adinolfi S, DeBoer J, Ktistaki E, Kelly G, Calder L, Kioussis D, Pastore A. The AXH domain adopts alternative folds: the solution structure of HBP1 AXH. Structure. 2005;13:743–753. doi: 10.1016/j.str.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, Rullman JA, MacArthur MW, Kaptein R, Thorton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 27.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 29.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jürchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 30.Schwalbe H, Fiebig KM, Buck M, Jones JA, Grimshaw SB, Spencer A, Glaser SJ, Smith LJ, Dobson CM. Structural and dynamical properties of a denatured protein. Heteronuclear 3D NMR experiments and theoretical simulations of lysozime in 8M urea. Biochemistry. 1997;36:8977–8991. doi: 10.1021/bi970049q. [DOI] [PubMed] [Google Scholar]
- 31.Dayie KT, Wagner G, Lefevre JF. Theory and practice of nuclear relaxation in proteins. Ann. Rev. Phys. Chem. 1996;47:243–282. doi: 10.1146/annurev.physchem.47.1.243. [DOI] [PubMed] [Google Scholar]
- 32.Masino L, Martin SR, Bayley PM. Ligand binding and thermodynamic stability of a multidomain protein, calmodulin. Protein Sci. 2000;9:1519–1529. doi: 10.1110/ps.9.8.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–3587. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valverde R, Pozdnyakova I, Kajander T, Venkatraman J, Regan L. Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure. 2007;15:1090–1098. doi: 10.1016/j.str.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Braddock DT, Louis JM, Baber JL, Levens D, Clore GM. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;15:1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 36.Du Z, Fenn S, Tjhen R, James TL. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J. Biol. Chem. 2008;283:28757–28766. doi: 10.1074/jbc.M803046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 38.Ramos A, Hollingworth D, Major SA, Adinolfi S, Kelly G, Muskett FW, Pastore A. Role of dimerization in KH/RNA complexes: the example of Nova KH3. Biochemistry. 2002;41:4193–4201. doi: 10.1021/bi011994o. [DOI] [PubMed] [Google Scholar]
- 39.Shamoo Y, Abdul-Manan N, Williams KR. Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res. 1995;23:725–728. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell. Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vitali F, Henning A, Oberstrass FC, Hargous Y, Auweter SD, Erat M, Allain FH. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labelling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.