Abstract
Recent studies showed that small interfering RNAs (siRNAs) and Piwi-interacting RNA (piRNA) in mammalian germ cells play important roles in retrotransposon silencing and gametogenesis. However, subsequent contribution of those small RNAs to early mammalian development remains poorly understood. We investigated the expression profiles of small RNAs in mouse metaphase II oocytes, 8–16-cell stage embryos, blastocysts and the pluripotent inner cell mass (ICM) using high-throughput pyrosequencing. Here, we show that during pre-implantation development a major small RNA class changes from retrotransposon-derived small RNAs containing siRNAs and piRNAs to zygotically synthesized microRNAs (miRNAs). Some siRNAs and piRNAs are transiently upregulated and directed against specific retrotransposon classes. We also identified miRNAs expression profiles characteristic of the ICM and trophectoderm (TE) cells. Taken together, our current study reveals a major reprogramming of functional small RNAs during early mouse development from oocyte to blastocyst.
INTRODUCTION
RNAs ranging from 19 to 32 nt contain functional small non-coding RNAs involved in gene regulation. Three major classes of functional small RNAs have been found: small interfering RNA (siRNA), microRNA (miRNA) and Piwi-interacting RNA (piRNA). siRNAs and miRNAs, with a typical length of 21–23 nt, are processed from longer transcripts forming double-strand and stem-loop structures, respectively, by digestion with an RNase III enzyme, Dicer, and then their single-strand elements are incorporated into the RNA-induced silencing complex (RISC) and function as mediators in gene silencing (1,2). piRNAs are ∼25–32 nt in length, specifically expressed in germ line cells and associated with the Piwi family proteins (3–6). Gene silencing involving such small non-coding RNAs appears to play essential roles in regulation of gene expression in development, differentiation and proliferation (7–18).
Recent studies have shown that siRNAs and piRNAs are expressed in mammalian germ cells and play important roles in retrotransposon-silencing and gametogenesis (13,14,18). Many siRNAs, like piRNAs, appear to be derived from repetitive sequences including retrotransposons. The contribution of such small RNAs (including miRNAs) to mammalian gametogenesis is further validated by failures of gametogenesis in mice carrying loss-of-function of the Piwi family genes and Dicer associated, respectively, with piRNAs (15–17) and siRNAs/miRNAs production (11,12).
In early development of pre-implantation mammalian embryos, the first transition from maternal to embryonic (zygotic) programs takes place as early as the two-cell stage (19), and qualitative and quantitative changes in gene expression occur over the subsequent development. At the blastocyst stage, when the embryo is composed of two distinct cell populations, the inner cell mass (ICM) and trophectoderm (TE), marked differences in gene expression between them can be detected (20,21). Although small non-coding RNAs play key roles in gametogenesis (13,14,18), little is known about their subsequent contribution to early mammalian development. In the present study we investigated the expression of small RNAs in mouse unfertilized (metaphase II: MII) oocytes, 8–16-cell stage embryos, blastocysts as well as pluripotent ICMs by high-throughput pyrosequencing. While the recent study presented the expression profile of known miRNAs in early mouse development (12), our current study has revealed comprehensive profiles of small RNAs containing uncharacterized small RNAs in pre-implantation embryos. The data thus demonstrate a drastic change in the expression of small RNAs associated with the transition from oocyte to embryo during mammalian development.
MATERIALS AND METHODS
Collection and culture of unfertilized eggs and embryos
Female ICR mice (5–8 weeks old) were superovulated via intraperitoneal injection of 7–10 i.u. of pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG) at 48 h intervals. The female mice were then mated with male ICR mice and inspected for vaginal plugs the next day. Unfertilized eggs (metaphase II eggs) were also collected from female mice without mating at 16–20 h post hCG, and subjected to treatment with hyaluronidase (300 U/ml in M2 medium). Fertilized embryos were collected from plug-positive female mice at the expected embryonic age as hours post-hCG: 8–16-cell stage embryo, 64–70 h; blastocyst, 88–94 h. Embryos were cultured in KSOM-AA medium containing 4 mg/ml BSA in a 5% CO2 humidified chamber (22).
Isolation of ICM and TE
Immunosurgery for isolation of ICM was carried out as described previously (23,24). Briefly, blastocysts were placed in acidic Tyrode’s solution (pH 2.5) to remove the zona pellucida and rinsed 3 times with M2 medium (Sigma). Zona-free embryos were incubated in anti-mouse antiserum (1:20 in M16 medium, Rockland) at 37°C for 10 min in a 5% CO2 humidified chamber. The embryos were then washed 3 times in M16 medium and incubated in guinea pig complement (1:20 with M16 medium, MP Biomedicals, LLC) for 30 min at 37°C in a 5% CO2 humidified chamber. After incubation and washing 3 times in M16 medium, ICM was isolated from the embryos by gentle pipetting with a glass micropipette. Microsurgery was carried out to isolate TE populations. Briefly, zona-free blastocysts were placed in a drop of M2 medium on a plastic Petri dish, and the drop covered with liquid paraffin. Excess medium was slowly removed using a glass micropipette so that the blastocyst could be fixed on the Petri dish in a position suitable for dissection (25). The blastocysts fixed onto the dishes were equatorially cleaved using a 30-G needle under a ZEISS stereomicroscope (Stemi 2000-C). Mural TE fragments were then collected via attachment to the tip of a 30-G needle.
Construction of small RNA libraries from pre-implantation mouse embryos
Small RNA libraries were constructed based on previous protocols (26,27). Total RNA was extracted from 1470 unfertilized eggs, 960 embryos at the 8–16-cell stage, 438 blastocysts and 405 ICMs using TRIzol reagent (Invitrogen), and subjected to size fractionation using flash PAGE (Ambion) according to the manufacturer’s instructions. Approximately 17–40-nt RNA fragments were collected and subjected to ligation with 5 μM of the 3′-adaptor RNA oligonucleotide (Linker-1), which is 5′-adenylated and 3′-blocked with a dideoxy-C base (IDT, Supplementary Table S1), by T4 RNA ligase (Amersham) without ATP at 37°C for 1 h. The RNAs were then purified by polyacrylamide gel electrophoresis, eluted from gels in elution buffer (0.5 M ammonium acetate, 1 mM EDTA and 0.1% SDS), collected by ethanol precipitation and dissolved in H2O. The collected RNAs were further ligated to the 5′-adaptor RNA oligonucleotide (Supplementary Table S1) by T4 RNA ligase in the presence of ATP at 37°C for 1 h, and used as templates to synthesize the first strand complementary DNAs (cDNAs) using SuperScript II reverse transcriptase (Invitrogen) with the 3′ PCR Oligo (Supplementary Table S1) complementary to the Linker-1 (3′-adaptor oligonucleotide) according to the manufacturer’s instructions. The resultant cDNAs were subjected to amplification by PCR using the ABI GeneAmp PCR system 9700 (Applied Biosystems). The thermal cycling was carried out as follows. In the first PCR, template and 5′ PCR Oligo and 3′ PCR Oligo primers were heat denaturation at 96°C for 1 min, followed by 20 cycles of amplification at 95°C for 10 s, 50°C for 1 min and 72°C for 20 s. The second PCR was carried out using the first PCR product and the 2nd and 3rd PCR-F and -R primers for 8–10 cycles of the thermal cycling profile in the first PCR. The third PCR was carried out using the second PCR product with eight cycles of the thermal cycling profile of the second PCR. The PCR products were purified by polyacrylamide gel electrophoresis, and collected and dissolved in TE (pH 8.0) as described above after every PCR amplification.
High-throughput sequencing analysis and annotation of small RNAs
Sequence determination of the cDNAs prepared from small RNAs was carried out using the 454 pyrosequencing technology (Roche). The obtained sequence data were mapped to the mouse genome using Blastn (ftp://ftp.ncbi.nih.gov/blast), and the sequences that perfectly matched the mouse genome were selected for further analyses. Annotation of the sequences was preformed as described previously (13). Briefly, to identify small RNAs corresponding to various repeats such as rRNA, tRNA, retrotransposon and DNA transposon, genomic positions of the repeats were retrieved from the University of California, Santa Cruz (UCSC) website (http://hgdownload.cse.ucsc.edu/downloads.html) and compared with the genomic positions of small RNAs. If the genomic position of a particular small RNA overlapped with any repeats by at least 15 nt, this small RNA was considered to be repeat-derived. Repeat names were retrieved for all positions to which a small RNA was mapped, and if multiple repeat names were retrieved, the class (such as LTR/MaLR or rRNA) and subclass (such as IAP), where applicable, were determined according to the majority of positions. If the top two repeats had the same number of positions, the class or subclass was not determined. To identify small RNAs corresponding to tRNAs, rRNAs, snRNAs, snoRNAs, scRNAs, miRNAs, piRNAs (previously identified in adult and neonate mouse testes and growing unfertilized eggs) and mRNAs based on sequence similarity, the sequences of these RNAs were extracted from the flat files (sequence and annotation files) of GenBank (ftp://ftp.ncbi.nih.gov/genbank/) and sequences downloaded from the following databases: tRNAs, Genomic tRNA Database (http://lowelab.ucsc.edu/GtRNAdb); snoRNAs, snoRNA database (http://www-snorna.biotoul.fr) and RNA database (http://jsm-research.imb.uq.edu.au/rnadb); piRNAs, RNA database (http://jsm-research.imb.uq.edu.au/rnadb) and the Gene Expression Omnibus (GEO) database (accession number: GSE7414); miRNAs, miRBase (http://microrna.sanger.ac.uk/sequences); mRNAs, Refseq Genes (ftp://ftp.ncbi.nih.gov/refseq) and Ensembl Genes (http://www.ensembl.org). Blastn searches were then performed using the small RNA sequences determined in this study as queries and the downloaded sequences as a database. After the annotation of small RNA clones, small RNA clusters were identified and characterized using in-house programs based on our previous study (13).
We also carried out sequence determination of small RNA cDNA libraries by means of conventional DNA sequencing. The cDNAs prepared from 1470 MII oocytes and 438 blastocysts as described above were subjected to 20 cycles of amplification followed by eight cycles of the second-amplification by PCR, and the resultant PCR products were cloned into the pCR4-TOPO vector (Invitrogen) according to the manufacturer’s instructions. Approximately 600 and 700 colonies derived from the MII oocyte and blastocyst libraries, respectively, were examined by the ABI 3730 xl DNA Analyzer (Applied Biosystems) followed by sequence annotation as described above.
Synthetic oligonucleotides
DNA oligonucleotides and siTrio siRNA duplexes were obtained from Invitrogen and B-Bridge, respectively. DNA and RNA oligonucleotide sequences synthesized in this study are presented in Supplementary Table S1. The siTrio negative-control (B-Bridige) was also used as a non-silencing siRNA.
Reverse transcription (real-time) PCR
Total RNA extracted from unfertilized eggs and embryos was subjected to concentration measurement by the RiboGreen assay (Molecular Probes) according to the manufacturer’s instructions. Reverse transcription (real-time) PCR (Q-PCR) for miRNA was carried out using the TaqMan MicroRNA assay and the ABI 7300 Real Time PCR system (Applied Biosystems) according to the manufacturer’s instructions. The TaqMan MicroRNA assays used (assay ID) were as follows: hsa-let-7b (000378); hsa-let-7c (000379); hsa-let-7g (002282); hsa-miR-16 (000391); hsa-miR-99b (000436); hsa-miR-200c (000505); hsa-miR-210 (000512); mmu-miR-290 (000187); mmu-miR-292-3p (001054); mmu-miR-294 (001056); mmu-miR-295 (000189); mmu-miR-467b (001671); mmu-miR-669a (001683); snoRNA135 (001230). In the case of Q-PCR for protein-coding genes, total RNA was subjected to first-round cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen) and oligo(dT) primer, and then Q-PCR was carried out using the AB 7300 Real Time PCR System (Applied Biosystems) with a TaqMan Universal PCR Master Mix together with Assays-on-Demand Gene Expression products (Applied Biosystems) or a SYBR Green PCR Master Mix (Applied Biosystems) and with Perfect Real Time Primers (TAKARA BIO) or designed PCR primers (Supplementary Table S1). The Assays-on-Demand Gene Expression products used and their assay ID were as follows: Cdx2, Mm00432449_m1; Pou5f1 (Oct3/4), Mm00658129_gH; Nanog, Mm02384862_gl; Gapdh, Mm99999915_g1. The Perfect Real Time Primers used were as follows (primer-set ID): Gapdh, MA050371; Dicer, MA043537. For examination of the MuERVL_Mm transcript, total RNA treated with Turbo DNase I (Ambion) was subjected to cDNA synthesis with oligo(dT) and semi-quantitative PCR was carried out.
Amanitin treatment
Alpha-amanitin (SIGMA-Aldrich) was used to inhibit RNA polymerase II. Thirty 1-cell stage embryos (28h post-hGC) were cultured in the presence or absence of 24 µg/ml alpha-amanitin (28), and collected after 16 h incubation: the resultant embryos appeared to normally develop to two-cell stage embryos. When total RNA was prepared from the embryos, in vitro synthesized DsRed mRNA was added as an external control for assessment of RNA preparation. The isolated total RNAs were subjected to reverse transcription followed by Q-PCR analysis.
In vitro transcription
To construct GFP fusion genes, the MuERVL-Mm retrotransposon and β-actin were amplified from cDNA prepared from two-cell stage embryos and blastocysts as a templates, respectively. The LINE-1 sequence in pd2EGFP-L1 (6) was also amplified by PCR. The resultant PCR products were digested with XbaΙ and NotΙ, and inserted into the phMGFP vector (Promega) treated with the same restriction enzymes. The PCR primer sets for amplification of the templates for synthesis of sense- and antisense-strand RNAs are presented in Supplementary Table S1. We also constructed a template plasmid encoding the DsRed gene via replacement of the SmaΙ–NotΙ fragment carrying GFP in the phMGFP vector with the SmaΙ–NotΙ fragment carrying DsRed, isolated from the pDsRed-Monome-N1 vector (Clontech). The constructed plasmids were digested with NotΙ and used as templates in RNA synthesis. In vitro transcription was carried out using a mMessage mMachine T7 ultra kit (Ambion) according to the manufacturer’s instructions.
Electroporation
Electroporation was carried out according to the previous reports (29,30). Approximately 20–30 fertilized eggs (one-cell stage) and 8–16-cell stage embryos were subjected to electroporation in 30 µl of HBS buffer [20 mM HEPES, pH 7.0–7.6 (SIGMA-ALDRICH), 150 mM NaCl] containing 2 µg of tetramethylrhodamine-labeled dextran (3000 MW) (Molecular Probes) or 2–4 µg of GFP-fusion gene mRNA together with 1 µg of the DsRed mRNA as a control. Three sets of four electric pulses (21 V for fertilized eggs and 28–30 V for 8–16-cell stage embryos; duration, 1 ms; interval, 99 ms) were delivered using an electronic pulse generator (model CUY-21, BEX, Tokyo) with electrodes having a gap of 1 mm (BEX, Tokyo) with 1-min intervals and polarity changes between the sets of pulses. Delivery of rhodamin-labeled dextran into the embryos was examined by a ZEISS fluorescent microscope (Axiovert 40 CFL). Embryos electroporated in the presence of the reporter mRNAs were incubated in KSOM-AA medium at 37°C in a 5% CO2 humidified chamber. Approximately 30 min after electroporation, 5–10 live embryos were collected for isolation of total RNA and the remaining samples were further incubated. At 18 h after electroporation (corresponding to the two-cell and early blastocyst stages), total RNA was isolated from 5 to 10 embryos. In the case of suppression of Dicer, two-cell stage embryos were electroporated in 30 µl of HBS buffer containing 10 µl of 100 µM siTrio siRNAs against Dicer (Supplementary Table S1) under the same conditions as for fertilized eggs. The electroporated embryos were then cultured in KSOM-AA medium as described above. The levels of suppression of Dicer were determined by Q-PCR.
Gene expression analyses under Dicer knockdown
More than 100 two-cell stage embryos were subjected to electroporation with the siRNAs against Dicer as described above. Three days after electroporation, total RNA was extracted from 71 electroporated embryos, which developed to blastocysts, and examined by the Affymetrix GeneChip® Mouse Genome 430 2.0 Array (Affymetrix) followed by analyses using the GeneChip Operating Software Program ver1.4 (Affymetrix) with default parameters (considered significance: P < 0.005) according to the manufacturer’s instructions. Genes presenting with possibly significant changes in their expressions were further examined by Q-PCR. Total RNA was extracted from ∼15–20 Dicer-knockdown embryos and subjected to cDNA synthesis as described above, and Q-PCR using primers indicated in Supplementary Table S1 was carried out.
RESULTS
Small RNAs present in pre-implantation mouse embryos
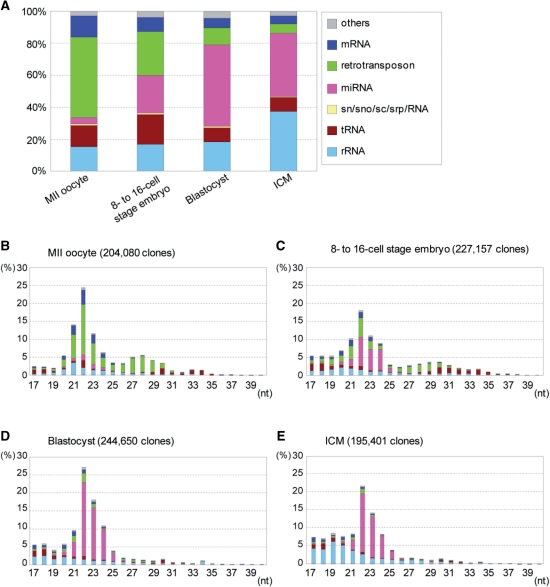
To investigate small RNAs in pre-implantation mouse embryos, we collected 960 embryos at the 8–16-cell stage [2.5 days postcoitum (dpc)], 438 blastocysts (3.5 dpc) and also 1470 unfertilized eggs (MII oocytes). Small RNAs were extracted from the samples and examined by 454 pyrosequencing followed by sequence annotation. We obtained 204080, 227157 and 244650 small RNA sequences completely matched the mouse genome from the MII oocytes, 8–16-cell embryos and blastocysts, respectively. The annotated small RNA sequences revealed a marked increase in the known miRNA population and a marked decrease in small RNAs corresponding to retrotransposons sequences over the course of development (Figure 1A). The size distribution of the small RNAs revealed two peaks, one at 22 nt and the other at 27–30 nt (Figure 1B–D). The most prominent peak at 22 nt was consistently present at all stages, but its major constituent changed from retrotransposon- and mRNA-derived small RNAs (MII oocyte and 8–16-cell embryo) to miRNAs (blastocyst). The other peak at 27–30 nt was observed in MII oocytes and 8–16-cell embryos, but hardly in blastocysts; and it enriched retrotransposon sequences. The data described above appear to be reproducible, because similar results have been obtained from independent experiments by means of conventional DNA sequencing procedures (Supplementary Figure S1).
Figure 1.
Small RNA profiles in stages of early development of mouse embryos and in ICM. (A) Small RNA libraries from each stage were examined by 454 pyrosequencing followed by assignment to RNA class as described in ‘Materials and Methods’ section. The distribution of the RNA classes for each developmental stage is given as a percentage of the total annotated small RNAs. (B–E) Distribution of nucleotide lengths of annotated small RNAs as in A. The number of clones matched the mouse genome for each library is indicated in parentheses.
Many of the 22-nt peak small RNAs other than miRNAs and many of the 27–30-nt small RNAs were mapped to the previously identified oocyte siRNA and piRNA clusters, respectively (13,14) (Supplementary Table S2); and new small RNA clusters were also detected. But, the number of the small RNA clusters progressively decreased over the course of development (Supplementary Table S2).
Small RNAs derived from retrotransposons
The cloning frequency of the retrotransposon-derived small RNAs was decreased as a whole over the course of development: the LINE-1 (L1)-derived small RNAs, which constitute a major fraction in MII oocytes and contain both putative siRNAs and piRNAs, obey this rule (Supplementary Figure S2A and B). However, the small RNAs derived from some retrotransposons exhibited unique expression patterns. For example, the frequency of small RNAs derived from a member of the LTR/ERVL family, MERVL-Mm, showed a transient increase at the 8–16-cell stage (Supplementary Figure S2A and C) and the increased small RNAs were putative siRNAs.
The frequency of small RNA derived from SINE/B1 was also initially low but increased at the 8–16-cell stage and stayed at a similar level thereafter (Supplementary Figure S2A). Interestingly, the SINE/B1 small RNAs in MII oocytes comprise only putative siRNAs, but the small RNAs increased at the 8–16-cell and blastocyst stages appear to contain both putative siRNAs and piRNAs (Supplementary Figure S2B). The expression profiles of the Piwi family genes indicated that Mili, whose protein can be associated with piRNAs ranging from 25 to 27 nt (3), was transiently expressed at the eight-cell stage (Supplementary Figure S3), suggesting the possibility that the SINE/B1 piRNAs may be associated with the transiently expressed Mili in the embryos.
Small RNAs derived from retrotransposons mediate gene silencing
We investigated whether the retrotransposon-derived small RNAs play an active role in gene silencing. A previous study demonstrated that exogenously introduced target RNAs containing the L1 sequence were specifically degraded in oocytes by an RNAi-dependent mechanism (6). To examine whether a similar degradation of L1-containing RNAs occur in pre-implantation embryos, in vitro synthesized GFP RNAs carrying the L1 sequence were introduced into fertilized one-cell and 8–16-cell embryos by means of electroporation (Supplementary Figure S4A), and degradation of the target RNAs was monitored by real-time PCR (Q-PCR). As a result, the RNAs carrying the L1 sequence were specifically degraded at both the one-cell and 8–16-cell stages (Supplementary Figure S4B), suggesting that a silencing mechanism similar to that in oocytes may also operate in pre-implantation embryos.
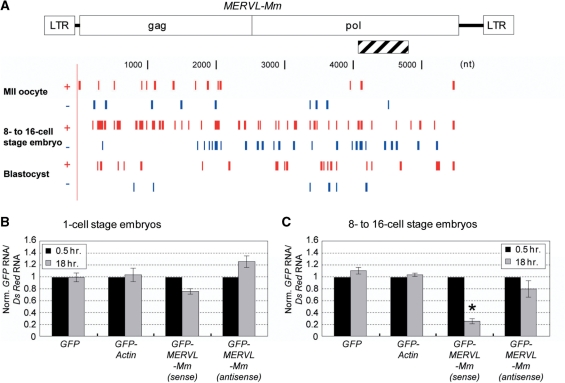
When GFP RNAs carrying the MERVL-Mm sequences (Figure 2A) were introduced into one-cell and 8–16-cell embryos, the level of the GFP RNAs carrying the MERVL-Mm sense-strand sequence was markedly decreased in the 8–16-cell embryos, and slightly reduced in the one-cell embryos (Figure 2B and C). The GFP RNAs with the MERVL-Mm antisense-strand sequence was unaffected and slightly affected in one-cell and 8–16-cell stage embryos, respectively. Thus, our data indicate a stage-specific silencing of MERVL-Mm in early developing mouse embryos, and the silencing is likely dependent upon the level of the siRNAs derived from the MERVL-Mm retrotransposon itself (Figure 2A and Supplementary Figure S2A and C). In addition, Dicer-knockdown embryos (Figure 3A) showed a marked increase in the level of the MERVL-Mm transcript, consistently suggesting the involvement of RNAi in the MERVL-Mm retrotransposon silencing [ref. (31), Figure 3B].
Figure 2.
Active small RNAs derived from retrotransposons. (A) Small RNAs (19–24 nt) of the plus strand (red) and minus strand (blue) matched the MERVL-Mm retrotransposon in mouse MII oocytes and pre-implantation embryos. Hatched bar represents an insertion into the GFP 3′ UTR. (B and C) Degradation of GFP fusion mRNA carrying MERVL-Mm. The GFP, GFP-Actin, GFP-MERVL-Mm (sense), or GFP-MERVL-Mm (antisense) mRNAs was introduced together with the DsRed mRNA as a control into 1-cell and 8–16-cell stage embryos by electroporation. After 0.5 and 18 h of electroporation, total RNA was extracted from 5 to 10 embryos and the introduced mRNAs were quantified by Q-PCR. The levels of GFP and its fusion mRNAs were normalized to those of DsRed mRNAs, and then further normalized to those obtained at 0.5 h in each case [n = 3; error bars represent SEM; *P < 0.05 (t-test)].
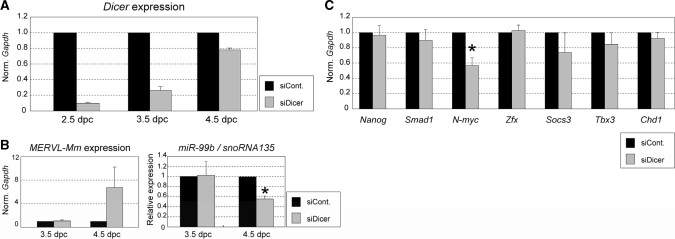
Figure 3.
Effect of Dicer-knockdown on gene expressions in pre-implantation embryo (A) Suppression of Dicer expression by siRNA. The siRNAs against Dicer (siDicer) or non-silencing control siRNAs (siCont.) were introduced into two-cell stage embryos (1.5 dpc) by electroporation, and total RNA was extracted from the electroporated embryos (2.5, 3.5 and 4.5 dpc). The expression level of Dicer was examined by Q-PCR and normalized to that of Gapdh examined as a control. The resultant level of Dicer in the presence of siDicer was further normalized to that in the presence of siControl (siCont.) as 1 (n = 3; error bars represent SEM). The data indicate that the level of Dicer mRNA in the presence of siDicer remains low until 3.5 dpc and recovers toward normal level thereafter, suggesting that Dicer-knockdown at the RNA level appears to last until 3.5 dpc. (B) MERVL-Mm and miR-99b expressions under Dicer-knockdown. The expression of MERVL-Mm, which appears to be regulated by endogenous siRNAs, and miR-99b, which appears to be a zygotic miRNA, were examined by Q-PCR using the same samples as in A. Marked increase and decrease in the levels of MERVL-Mm and miR-99b, respectively, were detected at 4.5 dpc, i.e. the effect of Dicer-knockdown on the expression of MERVL-Mm and miRNA became evident on the third day after the introduction of siDicer when the Dicer mRNA level recovered from RNAi suppression. These suggest that there is a time lag in changes in Dicer mRNA and protein levels, and we then examined the effect of Dicer-knockdown on the stability of transcripts of interest on the third day after siDicer was introduced (4.5 dpc). (C) Influence of Dicer-knockdown on gene expression. Electroporation with the siRNAs against Dicer (siDicer) and siControl (siCont.) and also preparation of total RNA were carried out as in B. The expression levels of indicated genes were examined by Q-PCR and normalized to those of Gapdh as a control. The resultant expression levels in the presence of siDicer were further normalized to those in siControl as 1. Data are averages of at least three independent experiments [error bars represent SEM; *P < 0.05 (t-test)].
Expression of miRNAs in mouse MII oocytes and pre-implantation embryos
As a basis for analyzing the expression of miRNAs during early mouse development, an appropriate control(s) is necessary in order to normalize the levels of miRNAs. We thus examined the expression levels of snoRNA135 and the miR-16 and -200c miRNAs, whose cloning frequencies were essentially unchanged in our small RNA libraries, by means of Q-PCR with equal amount (∼11 ng) of total RNAs prepared from MII oocytes, 8–16-cell embryos and blastocysts. As a result, similar levels of expression in each of the examined small RNAs among the samples were detected, suggesting that miR-16, miR-200c and snoRNA135 are suitable for the control (Supplementary Figure S5A).
The expression of the miRNAs belonging to the let-7 family and miR-290 and -467 clusters during early mouse development was investigated by Q-PCR and normalized to the level of the controls (miR-16, miR-200c or snoRNA135). As shown in Supplementary Figure S5B, similar expression profiles of the miRNAs normalized to any of the controls were detected and the profiles were compatible with the data of the cloning frequencies.
To investigate miRNAs during transition from maternal to embryonic (zygotic) programs in early development, we treated zygotes (one-cell stage embryos) with alpha-amanitin, a RNA polymerase II inhibitor, for 16 h and examined by Q-PCR the expression of the miRNAs belonging to the let-7 family (let-7b and -7g), which were present in MII oocytes and markedly decreased during pre-implantation development, and the miR-290 cluster (miR-292 and -294), which were barely present in MII oocytes and significantly increases toward 8-cell and blastocyst stages (see Supplementary Table S3). The alpha-amanitin treated embryos, developed during the 16 h culture into the two-cell stage, showed lower levels of the miR-292 and -294 expressions compared with the control embryos untreated with alpha-amanitin, in contrast to the let-7s and miR-16, a ubiquitous miRNA, with little difference in their levels (Supplementary Figure S6). The data suggest that, while maternal miRNAs reportedly decrease in two-cell stage embryos (12), the zygotic expression of miRNAs most likely initiates as early as the two-cell stage.
Small RNAs present in ICM
The blastocyst is composed of two distinct cell populations: the ICM and TE. The ICM cells exhibit pluripotency and generate the embryo proper, and the TE cells participate in the formation of the placenta after implantation. To investigate small RNAs in the ICM, we collected 405 ICMs from blastocysts by immunosurgery (Supplementary Figure S5C) (23) and prepared small RNAs. High-throughput sequencing analysis as described above was carried out and we obtained 195401 small RNA sequences that matched the mouse genome (Figure 1A and E). As shown in Figure 1E, miRNAs were the most predominant small RNA class of the ICM as in the blastocyst profile and the members of the miR-290 cluster appeared to be expressed abundantly. The piRNA peak was hardly detected (Figure 1E) and 22-nt putative siRNAs corresponding to retrotransposons and other sequences occupied a small fraction. Numerous rRNA derivatives were detected, but they were likely to be degradation products resulting from the immunosurgery. This is because they showed a broad distribution in the small size range (<21 nt) (Figure 1E) and were not so abundant in the blastocyst profile (Figure 1A and D).
Asymmetries in miRNA expressions between ICM and TE
Based on the data of each miRNA in the ICM and blastocyst, we estimated miRNAs exhibiting asymmetrical expression between the ICM and TE (Supplementary Table S3). To verify such an asymmetric distribution of miRNAs, we collected TE cells from blastocysts by microsurgery (Supplementary Figure S5D), and their total RNAs were examined by Q-PCR for the expression of Oct3/4, Nanog (ICM marker) and Cdx2 (TE marker) (20,24,32,33), in comparison with those from the blastocyst as a control. The ratio of the expression level of either Oct3/4 or Nanog to that of Cdx2 is substantially smaller in the TE sample than in the blastocyst (BL) sample (Supplementary Figure S5E), thus validating our method for collection of the TE sample. Using those RNA samples, we investigated the levels of miR-99b and miR-210 by Q-PCR followed by normalization to the level of miR-200c as a control. The results indicated that the relative amount of either miR-99b or miR-210 in the TE was significantly larger than that in the BL, suggesting predominant expression of the miRNAs in the TE cells (Supplementary Figure S5F).
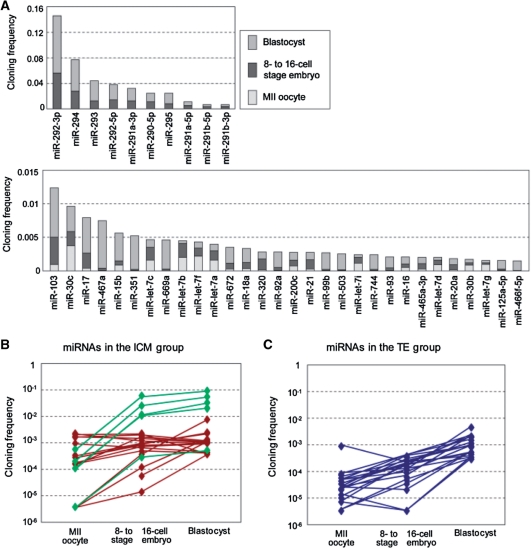
We further selected the top twenty miRNAs in each lineage (the ICM and TE groups) and examined their cloning frequencies in MII oocytes, 8–16-cell embryos and blastocysts (Figure 4B and C). As a result, while most of the miRNAs in the TE group exhibited a gradual increase in expression over the course of development, many of the miRNAs in the ICM group, which included five miR-290 cluster members, showed an increase at the 8–16-cell stage followed by a smaller change between the 8–16-cell and blastocyst stages. Accordingly, the data suggest distinct differential expressions of miRNAs between the ICM and TE cell lineages.
Figure 4.
miRNA profiles in pre-implantation mouse embryos. (A) Major miRNAs present in MII oocytes and pre-implantation embryos for the 40 most highly expressed miRNAs based on cloning frequencies. (B and C) Profiles of miRNAs abundant in ICM or TE. The 20 miRNAs in each of ICM (ICM group) (B) and TE (TE group) (C) were selected with the ratios of miRNAs (ICM/Blastocyst) (see Supplementary Table S3) and plotted for three stages from MII oocyte to blastocyst. The members belonging to the miR-290 cluster are indicated in green.
Dicer-knockdown in pre-implantation development
We wished to examine potential involvement of small RNAs in pre-implantation development, and performed suppression of Dicer, a key enzyme for production of both siRNAs and miRNAs in gene silencing, by means of electroporation using siRNAs against Dicer (Figure 3). Expression profiles in pre-implantation mouse embryos with Dicer knockdown indicated marked increase in the level of MERVL-Mm and also the reduction of the levels of several genes including miR-99b (Figure 3B). Interestingly, Q-PCR confirmed the significant reduction of the expression of N-myc, which appears to be associated with pluripotency, in Dicer-knockdown embryos (Figure 3C).
DISCUSSION
Maternal and zygotic siRNAs and piRNAs in pre-implantation mouse embryos
Endogenous siRNAs and piRNAs in mammalian germ cells play essential roles in retrotransposon silencing and gametogenesis (13,14,18). Our current study has investigated subsequent contribution of such small RNAs to early development of pre-implantation mouse embryos. Our data revealed that, some siRNAs and piRNAs derived from retrotransposons are transiently (zygotically) upregulated and likely directed against specific retrotransposons, although siRNAs and piRNAs as a whole are markedly decreased over the course of development (Figure 1, Supplementary Figure S2 and Table S2).
The transient increase in the MERVL-Mm small RNAs represents a new production, and most of the small RNAs are putative siRNAs. Since previous studies indicated that sense and antisense MERVL-Mm transcripts were abundantly expressed at the two-cell stage, but not at other stages (34,35), and since the MERVL-Mm transcript was significantly increased in Dicer-knockdown mouse embryos (Figure 3B) (31), our present evidence strongly suggests that endogenous siRNAs are zygotically produced from the MERVL-Mm transcripts following fertilization and work as mediators in RNAi-dependent silencing directed against the MERVL-Mm transcript itself. As a result, the level of MERVL-Mm is reduced after the two-cell stage. Thus, an autonomous suppression of MERVL-Mm via RNAi appears to occur.
The transiently increased piRNAs and siRNAs, which are derived from SINE/B1, together with the increased Mili at the same stage might also participate in autonomous suppression of the SINE/B1 retrotransposon.
In addition to zygotically produced siRNAs and piRNAs, maternally-derived small RNAs may also contribute to the early development. For example, it is noteworthy that maternally derived L1 small RNAs appear to work to at least the 8–16-cell stage, although the small RNAs are markedly decreased following fertilization (Supplementary Figure S2A and B). Given that genome-wide reprogramming including protamine/histone exchange and DNA demethylation takes place in early mammalian development (36–38), maternally and zygotically produced siRNAs and piRNAs presumably participate in the defense against harmful retrotransposons activated during the reprogramming, which are presumably silenced by chromatin modifications such as DNA methylation in somatic cells.
Expression of miRNAs in pre-implantation mouse embryos
MiRNAs play essential roles in gene regulation during early development (7–9). Various mouse miRNAs are synthesized after fertilization and the miRNA complexity rapidly increases (Figures 1 and 4A, Supplementary Figure S5B, S6 and Table S3). An increase in miRNAs of the miR-290 cluster, which are specifically expressed in embryonic stem (ES) cells (39–41), is particularly remarkable (Figure 4A), as previously described (12). In the ICM, miRNA is also the most predominant small RNA class as in the blastocyst profile and the members of the miR-290 cluster are expressed abundantly. Based on the expression ratio of each miRNA in the ICM and blastocyst, we estimated miRNAs exhibiting asymmetrical distribution between the ICM and TE (Supplementary Table S3), and the biased presence of some of the miRNAs was verified using dissected TE samples (Supplementary Figure S5F). The data suggest that difference in miRNA expression occurs between the ICM and TE cell lineages, which may contribute to specialization of cells in the two cell lineages, and maintenance of stemness specifically in the ICM cell lineage. To elucidate such contributions, more extensive studies need to be carried out.
Potential involvement of small RNAs in pre-implantation development
It should be noted that the expression of N-myc is significantly decreased in Dicer-knockdown embryos (Figure 3C), although the N-myc gene regulation involving functional small RNAs produced by Dicer remains to be investigated. N-myc is reported to be involved in maintenance of stemness in ES cells (42,43), and its expression is reduced after differentiation of the cells (44,45). N-myc is also predominantly expressed in the ICM cells at the blastocyst stage (46). As for the association of Dicer with pluripotency, Dicer-deficient mice are lethal at embryonic day 7.5, lacking pluripotent cells (47). Since our Dicer knockdown was carried out in pre-implantation embryos, the gene silencing involving Dicer may contribute to the maintenance of stemness even before implantation, and N-myc might play a key role in such maintenance of stemness. Taken together, the data suggest the possibility that the gene silencing may participate in the maintenance and differentiation of pluripotent cells in addition to the suppression of retrotransposons over the course of pre- and post-implantation development.
Transition of a major small RNA from siRNA/piRNA to miRNA during pre-implantation development
Together with the findings of recent studies (3,5,6,12–14), our current study has revealed that the transition of a major small RNA class from siRNA/piRNA to miRNA takes place during pre-implantation development (Figure 5). The data further suggest that the zygotic expression of miRNAs as the beginning of the transition presumably starts as early as the two-cell stage. Given that miRNA is a major mediator of gene regulation, and predominantly present in mammalian somatic cells including pluripotent ES cells (40,48,49), it is conceivable that early mammalian embryos shape the somatic type small RNA modality and gene regulation involving miRNAs prior to their implantation.
Figure 5.
Proposed transition of functional small RNAs during mammalian oogenesis and early embryogenesis. Based on the present and previous study (13), possible changes in the levels of functional small RNAs from growing oocytes to pre-implantation embryos are represented schematically. Putative expression levels of zygotic siRNAs and piRNAs are represented by dotted lines and are not drawn in scale.
ACCESSION NUMBERS
The GeneBank accession numbers of all the small RNAs derived from 454 sequencing in this study are ALAAA0000001-ALAAA0130942 (MII oocyte), ALAAB0000001-ALAAB0116883 (8- to 16-cell stage embryo), ALAAC0000001-ALAAC0092019 (Blastocyst), ALAAD0000001-ALAAD0057749 (ICM).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Health, Labour and Welfare of Japan, Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and National Institute of Genetics (NIG) Cooperative Research Program (2008-A60). Funding for open access charge: Ministry of Health, Labour and Welfare of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank all the technical staff of the Sequence Technology Team at RIKEN Genomic Sciences Center (GSC) for their assistance, and also K. Hanaoka, D. Watanabe and Y. Mikami for their helpful advices.
REFERENCES
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 5.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 8.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 9.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 10.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 11.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 14.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 16.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 17.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 20.Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 21.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 22.Biggers JD, McGinnis LK, Raffin M. Amino acids and preimplantation development of the mouse in protein-free potassium simplex optimized medium. Biol. Reprod. 2000;63:281–293. doi: 10.1095/biolreprod63.1.281. [DOI] [PubMed] [Google Scholar]
- 23.Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc. Natl Acad. Sci. USA. 1975;72:5099–5102. doi: 10.1073/pnas.72.12.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 25.Hohjoh H. M.S. Thesis. Tokyo Metropolitan University; 1987. Analysis of the proteins in the preimplantation mouse embryos. [Google Scholar]
- 26.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 27.Pfeffer S, Lagos-Quintana M, Tuschl T. Cloning of small RNA molecules. In: Ausubel FM, et al., editors. Current Protocols in Molecular Biology. New York: Wiley InterScience; 2005. pp. 26.4.1–26.4.18. [DOI] [PubMed] [Google Scholar]
- 28.Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 2005;283:40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 29.Grabarek JB, Plusa B, Glover DM, Zernicka-Goetz M. Efficient delivery of dsRNA into zona-enclosed mouse oocytes and preimplantation embryos by electroporation. Genesis. 2002;32:269–276. doi: 10.1002/gene.10076. [DOI] [PubMed] [Google Scholar]
- 30.Soares ML, Haraguchi S, Torres-Padilla ME, Kalmar T, Carpenter L, Bell G, Morrison A, Ring CJ, Clarke NJ, Glover DM, et al. Functional studies of signaling pathways in peri-implantation development of the mouse embryo by RNAi. BMC Dev. Biol. 2005;5:28. doi: 10.1186/1471-213X-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 32.Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 33.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 34.Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D, et al. Systems biology of the 2-cell mouse embryo. Cytogenet. Genome Res. 2004;105:240–250. doi: 10.1159/000078195. [DOI] [PubMed] [Google Scholar]
- 35.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 37.Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127:643–651. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- 38.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 39.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 40.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 43.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 44.Kelly DL, Rizzino A. DNA microarray analyses of genes regulated during the differentiation of embryonic stem cells. Mol. Reprod. Dev. 2000;56:113–123. doi: 10.1002/(SICI)1098-2795(200006)56:2<113::AID-MRD1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 45.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 46.Yoshikawa T, Piao Y, Zhong J, Matoba R, Carter MG, Wang Y, Goldberg I, Ko MS. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr. Patterns. 2006;6:213–224. doi: 10.1016/j.modgep.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 48.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 49.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.