Abstract
RNA exosomes are large multisubunit assemblies involved in controlled RNA processing. The archaeal exosome possesses a heterohexameric processing chamber with three RNase-PH-like active sites, capped by Rrp4- or Csl4-type subunits containing RNA-binding domains. RNA degradation by RNA exosomes has not been studied in a quantitative manner because of the complex kinetics involved, and exosome features contributing to efficient RNA degradation remain unclear. Here we derive a quantitative kinetic model for degradation of a model substrate by the archaeal exosome. Markov Chain Monte Carlo methods for parameter estimation allow for the comparison of reaction kinetics between different exosome variants and substrates. We show that long substrates are degraded in a processive and short RNA in a more distributive manner and that the cap proteins influence degradation speed. Our results, supported by small angle X-ray scattering, suggest that the Rrp4-type cap efficiently recruits RNA but prevents fast RNA degradation of longer RNAs by molecular friction, likely by RNA contacts to its unique KH-domain. We also show that formation of the RNase-PH like ring with entrapped RNA is not required for high catalytic efficiency, suggesting that the exosome chamber evolved for controlled processivity, rather than for catalytic chemistry in RNA decay.
INTRODUCTION
The eukaryotic and archaeal RNA exosomes and their distant relative, the bacterial degradosome, are large multiprotein assemblies that function as central cellular RNA processing and degradation machineries. The RNA exosome was originally found in yeast as an essential protein complex with 3′ → 5′ exonuclease activity. First, identified for the 3′-processing of the yeast 5.8S ribosomal RNA (1), the yeast RNA exosome subsequently turned out to be important for the trimming and degradation of the 3′-end of several nuclear RNA precursors (2). In addition, the exosome was shown to be also active in the cytoplasm by controlling mRNA turnover (3), and by its implication in various mRNA surveillance pathways like the non-sense-mediated and the non-stop decay pathways (4–7). Due to its involvement in all the different RNA processing and surveillance pathways the exosome is apparently one of the central exonucleases of a yeast cell [for reviews see for instance (8,9)].
Structural homologues of the yeast exosome were subsequently identified in humans, previously known as the PM-Scl (polymyositis–scleroderma overlap syndrome) complex, and in archaea (10–12). A variety of structural studies revealed a conserved architecture of exosome like complexes (13–18): exosomes consist of nine conserved core subunits, six RNase PH type subunits and three subunits with S1 and KH or zinc-ribbon domains. The six RNase-PH like domains form a ring, arranged as trimers of pseudo-dimers. In archaea, the ring is formed by three (archaeal)aRrp41:aRrp42 dimers, while human and yeast exosomes contain six different RNase PH type subunits.
The archaeal exosome possesses a central chamber within the RNase PH ring which contains three phosphorolytic active sites. The actual active site is located in the aRrp41 subunits, but the whole aRrp41:aRrp42 dimer is involved in positioning the RNA. These sites degrade single-stranded RNA (ssRNA) in a phosphate dependent manner in 3′ → 5′ direction. They also catalyse the reverse reaction of adding nucleoside diphosphates to the 3′-end of RNA (13), liberating inorganic phosphate. In archaea, this activity has been attributed to formation of poly-A-rich tails on RNA (19). A proposed RNA entry pore at one side of the chamber restricts entry to mostly unstructured ssRNA, providing an explanation for controlled RNA degradation. Furthermore, three Csl4 or Rrp4 type putative RNA recognition subunits are located on top of the (Rrp41:Rrp42)3 ring and frame the proposed RNA entry pore. Current models suggest that these domains recognize RNA substrates and help to funnel them into the processing chamber.
Although the human exosome is structurally related to the archaeal complex, including S1 and KH domain containing subunits (Csl4, Rrp4 and Rrp40), it has lost phosphorolytic activity (14). Instead, it gained additional ectopic subunits: the hydrolytic RNase Rrp44 (20,21) has both exonucleolytic and endonucleolytic activities, located in RNB and PIN domain subunits, respectively (14,20,22–24). A second hydrolytic RNase (Rrp6) was identified as transient part of the nuclear complex (10). Recent results indicate that despite the ectoptic placement of the nuclease active sites, RNA is still threaded through the nuclease deficient RNase PH type ring (25).
A variety of groups have biochemically observed processive RNA degradation, in particular for the archaeal exosome. From structural studies, it was proposed that RNA is channelled through an entry pore between the S1 domains of aCsl4 or aRrp4 trimers into the processing chamber, where the 3′-end of the RNA is positioned in one of the three phosphorolytic active sites, subsequently degrading RNA base-per-base (16,26,27). The presence of the cap proteins Csl4 and Rrp4 in general increases the degradation efficiency of the exosome, but it is unclear how they do so. For instance, if the cap proteins recruit RNA, one would expect an increase in the general binding affinity. However, once RNA has entered the processing chamber, high affinity binding to the ectopic domains should slow down processive degradation. Another mechanism that is not yet understood is why processivity depends on the length of the RNA molecules (28). To address these questions and to develop means to quantitatively analyse processive degradation, we performed quantitative high-resolution RNase degradation activity assays with different variants of the Archaeoglobus fulgidus exosome. We evaluated different kinetic models and developed a Markov Chain Monte Carlo (MCMC) analysis to fit the model to the data and derive appropriate rate constants of individual RNA degradation steps. Our data identify different structural contributors to processivity, suggesting that ectopic RNA-binding domains, the entry pore and the active site are different contributors to processive degradation. The methods should be easily applicable also to other processive enzymes, including the hydrolytic nucleases of the eukaryotic exosomes.
MATERIALS AND METHODS
Protein expression and purification
The Archaeaoglobus fulgidus RNA exosome with different Rrp4 and Csl4 caps, derivatives and mutants were expressed and purified as described (29). Site directed mutations were introduced using the QuickChange® Site-directed Mutagenesis Kit (Stratagene) and verified by sequencing. Oligonucleotide sequences are provided in the Supplementary Table S2.
Crystallization and structure determination
An amount of 120 μM Csl4-exosome (Csl4:Rrp41:Rrp42)3 or its Y70ARrp42 mutant (= 27 g/l) were incubated with 400 μM RNA (3.3-fold excess, 6-mer CCCCUC) for 10 min on ice. Protein:RNA complexes were crystallized by sitting drop vapour diffusion technique by mixing 1 μl protein and 1 μl of reservoir solution (0.1 M NaAcetate, pH 4.6, 30% 3-Methyl-1,5-pentadiol (MPD), 100 mM NaCl) at 20°C. Datasets were recorded at the ID-14-2 beamline (ESRF, Grenoble, France) to 2.4 Å (wild-type exosome) and at the PX I beamline (SLS, Villigen, Switzerland) to 3.0 Å (Y70ARrp42 mutant) and processed with X-ray Detector Software (XDS) (30). A model of the apo-Csl4-exosome complex (29) was used as a search model for molecular replacement using PHASER (31). Refinement to 2.4 Å and 3.0 Å, respectively was performed with CNS (32) and PHENIX (33). In the additional electron density RNA nucleotides were positioned using COOT (34). Refinement of the complete complexes was followed by iterative cycles of manual model completion with COOT and positional and B-factor refinement with CNS (Supplementary Table S1).
Small angle X-ray scattering
For small angle X-ray scattering (SAXS) studies, the (Rrp41:Rrp42:Rrp4)3 complex was purified as described above. To purify the exosome with endogenously bound Escherichia coli RNA the protocol was modified as follows: RNA was not washed off with high salt, and in all buffers the salt concentration was 250 mM or lower. After the Ni-NTA affinity chromatography, the complex was loaded on an anion exchange column to remove unbound nucleic acids and the procedure was repeated to assure the total removal of free RNA. Not until only one distinct peak was eluted, the fractions were pooled, concentrated and flash frozen. The apo-complex was measured at 5, 10 and 15 mg/ml and the RNA complex was concentrated to an absorption at 280 nm of A280 = 55 and measured in a 1: 0, 1: 1 and 1: 2 dilution to evaluate the concentration dependency of scattering. Both complexes did not show concentration dependent aggregation and were not affected by long exposure to high-energy X-rays. SAXS data collection was performed in 20 mM Tris pH 7.4 and 200 mM NaCl buffer at the SIBYLS beamline (Advanced Light Source, Berkeley, CA, USA) (35). The radius of gyration was calculated using the Guinier plot in the linear region (constraint: s · Rg <1.3) and the calculation of the pair distribution function was done with GNOM within PRIMUS (36). Ab initio modelling of the solution structures was done with GASBORp (37) and more than 10 identically calculated models were aligned and averaged using DAMAVER and SUPCOMB (38). For analysis of the bound RNA, the protein was separated from the RNA by running the complex on a denaturing 6 M urea and 20% polyacrylamide gel and elution of the RNA from the gel. The pelleted RNA was sent to Vertis Biotechnologie AG, where the sample was poly(A)-tailed using poly(A) polymerase followed by ligation of an RNA adapter to the 5′-phosphate of the small RNAs. First-strand cDNA synthesis was then performed using an oligo(dT)-adapter primer and M-MLV H- reverse transcriptase. The resulting cDNAs were PCR-amplified to 20–30 ng/μl in 19 cycles using standard Taq DNA polymerase. We cloned the cDNA products with EcoRV into pET21 vectors, transformed and amplified the plasmids and isolated and sequenced single clones.
Cross-linking
Site-specific crosslinking of the K37CRrp41:D143CRrp42 mutant was performed with a HBVS (1,6-Hexane-bis-vinylsulfon) crosslinker. The crosslinking reaction was performed with a 100-fold excess of crosslinker under oxygen-free conditions in a glove-box. We removed crosslinked protein from non-crosslinked protein complexes using a Superose 6 size-exclusion column, equilibrated with a running buffer containing 4 M guanidinium chloride. Protein from the peak corresponding to a crosslinked Rrp41/Rrp42 dimer was refolded in 50 mM Tris (pH 7.4), 200 mM NaCl, 500 mM arginine and 5% glycerol by dilution. The refolded protein was again applied onto a Superose 6 column in 50 mM Tris (pH 7.4) and 200 mM NaCl. Only the correctly refolded protein, verified by the formation of a hexamer in size exclusion chromatography, was used for further experiments. As a control sample, the same complex without crosslinker was partly denatured, purified and refolded in the same way as the crosslinked protein.
RNase activity assays
We carried out RNase activity assays using 32P-labelled poly(rA)-oligoribonucleotides with different lengths as substrate (26). RNA was incubated with [γ-32P] ATP (Hartmann Analytics) and T4 polynucleotide kinase (NEB) for 45 min at 37°C and purified by using MicroSpin G-25 columns (GE Healthcare). For each reaction, the protein (30 nM for the Csl4- and the Rrp4-capped wild-type exosome and the interface mutant; 60 nM for the cap-less exosome and the single site mutants R65ERrp41 and Y70ARrp42; 120 nM for the crosslinked cap-less exosome) was incubated with RNA in buffer containing 20 mM Tris (pH 7.8), 60 mM KCl, 10 mM MgCl2, 10% glycerol, 2 mM DTT, 0.1% PEG 8000, 10 mM NaH2PO4 (pH 7.8) and 0.8 U/ml RNasin (Promega) at 50°C. Different time points were taken and the reaction was stopped by adding one volume of loading dye [0.75 g/l bromphenol blue, 0.75 g/l xylene cyanol, 25% (v/v) glycerol, 50% formamide]. The reaction products were resolved on a 20% polyacrylamide/6 M urea sequencing gel running at 50°C and were analysed by phosphorimaging (GE Healthcare). The gel bands were quantified using the ImageQuant Software (GE Healthcare) and data analysis, simulation and fitting was done with MatLab (Mathworks).
Models and kinetic data analysis
Kinetic models are shown in Figure 3A. They are described by four parameters: association rate ka,i, dissociation rate kd,i, cleavage rate kc,i and polymerization rate kp,i, one for each RNA of length i = 4, 5, …, 30. The corresponding set of differential equations that quantitatively describe RNA degradation is shown in the supplement Data (Chapter 1). Since the reaction takes place in an excess of inorganic phosphate (10 mM phosphate compared to only 3.6 μM ADP at the time all RNA molecules are totally degraded), we may assume no polymerization takes place, i.e. kp,i = 0 for all i. Consistently, we saw no synthesis of longer RNAs in our reactions. To obtain empirical estimates of the posterior parameter distribution, we implemented a MCMC approach based on the Metropolis–Hastings algorithm. The key ingredients are the likelihood function, the prior, and the proposal distribution. The likelihood function penalizes the estimation error produced by a given model. More precisely, it penalizes the residuals, i.e. the deviation of the measured RNA amounts at each time point from the amounts that have been predicted from the current parameter set. We assume that the residuals are independent realizations of Gaussian distributions with zero mean. Since the variances of these Gaussians are not known a priori, we assume a two-parameter error model with an additive and a multiplicative error component which has been proposed (39) in the context of spot quantification on arrays. We initialize the error model very conservatively (presuming large measurement errors). During the MCMC run, the error model is updated continuously by replacing it with an empirical estimate derived from the residuals that occurred in the Markov chain so far.
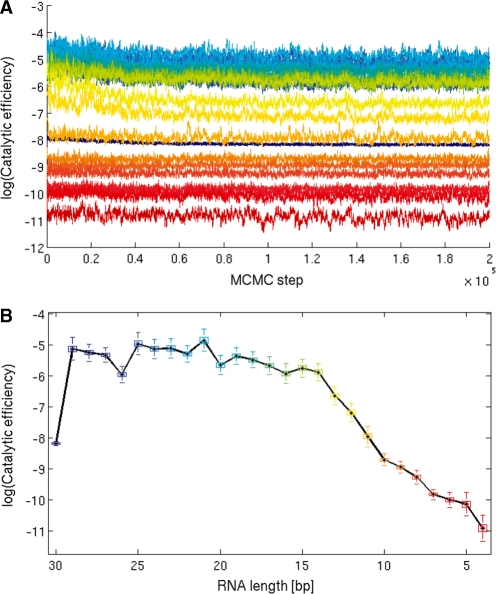
Figure 3.
Three different models to describe the kinetics of RNA degradation by the exosome were tested: (A) scheme for the general kinetic model, which includes cleavage and polymerization rates kc and kp as well as association and dissociation rates ka and kd for all RNAs from 30–4 nt. (B–D) Quantified concentrations of RNA intermediates from Figure 1A, along with least square fits to different kinetic models. (B) Strict processivity considers only 27 different cleavage rates kc,30 –kc,4. (C) cleavage-and-polymerization considers 27 different cleavage rates kc,30 –kc,4, 27 different polymerization rates kp,30–kp,4 and one initial association rate ka,30 (=55 rates). With models (C) and (B), no reasonable fit could be obtained. (D) By including association, dissociation and cleavage and making rational simplifications (see text) we can convincingly fit the data with a model containing 28 different rate constants.
The prior encodes prior knowledge/assumptions on the distribution of the parameters. It is sensible to require the kinetic parameters to vary smoothly with the RNA length i. This is made explicit by penalizing the difference of two successive kinetic parameters kx,i+1 and kx,i using a Gaussian prior on these differences. We emphasize that this does not impose any restrictions on the absolute level of the parameter values. The comparison of the parameter levels obtained by different experiments is virtually unaffected by our prior choice and therefore practically unbiased. The proposal distribution generates a new parameter set as a candidate for the next MCMC step that is based on the current parameter set. We simply use a multivariate log-normal distribution with fixed diagonal covariance matrix, which is centred at the current parameter set.
It turns out that the parameters of the model as stated above are not identifiable. We therefore fixed kd,i to one global constant kd, whereas the association parameters ka,i are sampled individually. The parameters kc,i are set equal to one length-independent parameter kc. The details of this approach and its justification through extensive simulations are given in the supplementary Data (Chapters 2–4).
RESULTS
RNA is not degraded with constant velocity
Despite intense structural and biochemical research on RNA exosomes, a kinetic model, quantitative analysis of processive RNA degradation and a biochemical identification of elements that contribute to processive degradation have not been studied, due to the complex kinetics involved. To address these issues, we performed RNase assays with 5′-radioactively labelled 30-mer oligo(A) RNAs and the A. fulgidus Csl4- (Csl4:Rr41:Rrp42)3 and Rrp4-exosomes (Rrp4:Rr41:Rrp42)3. The reaction products and their time evolution were resolved on a denaturing sequencing gel and quantified by phosphorimaging (Figure 1), controls are shown in the supplemental material (Supplementary Figures S4 and S5). Several characteristic features of substrate degradation by exosomes are revealed:
Figure 1.
Visualization of RNase activity of the archaeal exosome on denaturing polyacrylamide gels: the input (I) is a 30-mer polyA RNA radioactively labelled at the 5′-end that is degraded from the 3′-end to a final product (FP) of a 3-mer. Time points were taken in increasing intervals [in minutes: 0:10; 0:20; 0:30; 0:40; 0:50; 1:00; 1:10; 1:20; 1:40; 2:00; 2:20; 2:40; 3:00; 3:30; 4:00; 4:30; 5:00; 5:50; 6:00; 6:30; 7:00; 7:30; 8:00; 9:00; 10:00; 12:00; 14:00; 16:00; 18:00; 20:00; 25:00; 30:00; 35:00; 40:00; (B) ends at 8:00 min]. RNA degradation does clearly not occur with constant speed and the (Csl4:Rrp41:Rrp42)3 exosome (A) degrades RNA with a different time dependency than the (Rrp4:Rrp41:Rrp42)3 exosome (B).
First, RNA is not degraded at a constant speed, but the degradation of substrate has several phases and is distinct in different isoforms. In the Csl4 exosome (Figure 1A), after a slower first processing step, longer RNAs (>∼12–13 nt) are degraded very fast, seen by the low amount of intermediates in this range; shorter RNAs (<∼12–13 nt) are degraded slower and accumulate first before they are further degraded. On the contrary, the first processing step is faster in the Rrp4- than in the Csl4-exosome (Figure 1B). However, oligo-rA substrates >∼24 nt are degraded slower, intermediate substrates (∼24–13 nts) faster, and RNAs <13 nt slower again. This result is astonishing, considering homooligomeric sequences are used and the effect is consequently not sequence dependent. In addition, the unexpected slow-fast-slow kinetics of the Rrp4 isoform reveals a quite complex length dependency of RNA degradation speeds.
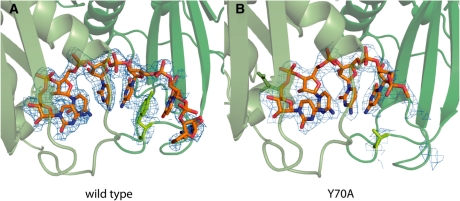
Second, the final degradation product is a 3-mer. Further degradation is extremely slow, comparable to spontaneous background hydrolytic cleavage under the present conditions. We hypothesized that features of the active site might interact specifically with the fourth base at the 3′-end. Previous structural analysis with the Sulfolobus solfataricus exosome has shown that at least 4 nt are stably bound in the phosphoropytic active sites (26), but in the case of the Pyrococcus furiosus exosome some nucleotides were recognized (16). To get direct structural information for the A. fulgidus exosome:RNA interaction, used in this study, we crystallized our Csl4-exosome with a 6-mer RNA molecule (Figure 2; Supplementary Table S1). Four nucleotides from the 3′-end are clearly visible in the unbiased Fo–Fc electron density, with weaker density for the two additional nucleotides. Interestingly, the side chain of Y70Rrp42 shows π-stacking with the fourth base (counting from the active site) and this seems to be a conserved feature among archaeal exosomes (16,26). This interaction specifically stabilizes the first 4 nt, while RNA positions +5 and +6 behind Y70Rrp42 appear not to be specifically recognized. To test the role of Y70Rrp42, we determined the co-crystal structure of the Csl4-exosome-Y70A mutant with a CCCCUC oligonucleotide. In fact, we only see clear electron density for 4 nt in the active site and the electron density at position +4 is weaker and less defined compared to the wild-type. Thus, the 3-mer as degradation end-product is likely the cause of inefficient recognition of RNA’s with <4 nt at the active site.
Figure 2.
Crystal structure of 6-mer RNA bound to the active site of the archaeal exosome. Rrp41 is shown in light and Rrp42 in dark green. The 2Fo–Fc electron density is contoured at 1.0σ and only shown for the RNA and the side chain of Y70Rrp41. (A) In the wild-type exosome Y70 is stacking with the fourth base of the bound RNA, and only weak density can be seen for the fifth and sixth base. (B) Electron density for the fourth base of the RNA is much weaker in the Y70ARrp41 mutant compared to the wild-type and no density can be detected at this contour level for additional nucleotides.
A kinetic model for processive RNA degradation by exosomes
To obtain a comprehensive picture of the exosomal RNA decay, we need to analyse the reaction speeds in a quantitative manner. The amount of RNA as function of time of intermediate i of an rA n-mer may be described by several rate constants (Figure 3A): an association rate constant ka,i of the 3′-end of RNA to the active site; a corresponding dissociation rate constant kd,i; a rate of formation of intermediate i by cleavage of intermediate i + 1, kc,i+1; and by adenylation (polymerization) of intermediate i−1, kp,i-1; a rate of disappearance of intermediate i by cleavage of i, kc,i and by adenylation of i, kp,i. The system kinetics is then given by a set of differential equations (Supplementary Data).
However, it is possible that this more general model can be further simplified. For instance, we likely can neglect adenylation (kp,i = 0) because our reaction conditions contain 10 mM phosphate compared to only 3.6 μM ADP at the time all RNA molecules are totally degraded, strongly shifting the reversible reaction towards degradation. In addition, the exosome might be strictly processive, i.e. association and dissociation rates of RNA intermediates are negligible compared to the cleavage rates (ka,i = kd,i = 0). Furthermore, all cleavage rates may be independent of the length of RNA, because they could be a local active site property (kc,i = kc,j). Hence we analysed three simplified models (Figure 3B–D). Once initial values for the rate constants, enzyme concentration and RNA substrate concentration (rA 30-mer) are provided, this corresponding set of differential equations can be used to calculate the concentrations of all RNA intermediates over time. We then minimized the resulting least square deviations between the calculated and experimental concentrations of reaction intermediates by optimizing the rate constants using the ‘fminsearch’ parameter optimization procedure as implemented in Matlab.
Using the ‘strict processivity’ model with 27 independent variables (ka,i = kd,i = kp,i = 0) (Figure 3B), we obtained no reasonable fit of the experimental data. A second model including the adenylation reaction (55 independent variables, ka,i = kd,i = 0) could also not properly interpret the data (Figure 3C). Thus, simply adding more parameter does not automatically lead to reasonable fits and the RNA degradation activity cannot be convincingly explained by strict processivity. Consequently, we added association and dissociation of RNA intermediates to the equations and used the following alternative simplifications: (i) adenylation is omitted (kp,i = 0; see above); (ii) the same cleavage rate is used for all RNA molecules (kc,i = kc,j), i.e. cleavage rate is a local active site property and not dependent on RNA length. We estimated starting values for kc and validated this simplification from analysis of the initial exponential decay of RNAs substrates with different initial lengths (data not shown). (iii) Due to our experimental approach, we cannot experimentally distinguish between bound and free RNA since the gel bands represent the sum of free and exosome-bound RNA intermediates of length i. For that reason, we cannot reconstruct dissociation-, association- and cleavage rate constants independently of each other. Consequently, we do not treat the association and the dissociation rate constants independently, but analyse the ratio of ka,i/kd,i by setting kd,i’s to a constant low value, leaving ka,i free to vary. Variation of the value for kd did not result in significant changes in the analysis (Supplementary Data). These three reasonable simplifications leave essentially one free parameter per intermediate plus one overall cleavage rate constant. Although this model has less degrees of freedom than the second model (55 versus 28), it can convincingly interpret the experimental data for the both Csl4 and Rrp4 exosome variants and most mutants (Figure 3D).
MCMC analysis of degradation
To address the problem of multidimensional parameter fitting and to assess the variance in parameter estimation, we established MCMC simulations. Because of the difficulty in determination of separate values for the single rate constants, we defined an RNA length-dependent quantity vi
| (1) |
with Km,i the Michaelis–Menten constant
| (2) |
vi is called ‘catalytic efficiency’ or ‘specificity constant’, as it is a measure of the velocity of RNA intermediate i degradation by the exosome. We are now in a position to test exosome features important for vi. We observe that for the Csl4 exosome, vi is highly dependent on the RNA length: the initial RNA processing step, likely determined by the initial association of RNA with the exosome, is generally slow. Once RNA is bound, vi is large and relatively constant for RNA lengths >13 nt. vi then progressively decreases for RNA lengths <13 nt until the final 3-mer appears (Figures 4B and 5A). This length dependency may be explained by the exosome structure: RNA molecules longer than 13 nt might still reach through the ‘neck’, and this topological interaction will induce a higher ‘local concentration’ of RNA at the active site with increased vi. Short RNAs, on the contrary, will lose this contact and due to their smaller size more easily diffuse out of the processing chamber, therefore decreasing vi.
Figure 4.
Catalytic efficiency vi for all RNA intermediates present during the degradation of a 30-mer RNA by the Csl4-Rrp41-Rrp42 exosome was determined with MCMC simulations. (A) shows the traceplot and (B) the final parameter set (burnin = 150 000). It can be seen that the MCMC chains vary in convergence speed as well as in variability. The boxplots in (B) illustrate the main advantage of the MCMC approach: it not only offers a set of parameters that best describe the measured data, but it also yields a posterior distribution for each catalytic efficiency parameter and thus provides a more comprehensive summary of the data.
Figure 5.
Comparisons of the catalytic efficiency vi of different exosome variants versus RNA lengths: (A) differences in the cap proteins influence catalytic activity. This is shown by comparison of vi from the cap-less exosome (Rrp41:Rrp42)3 in magenta, the Csl4 capped exosome (Csl4:Rrp41:Rrp42)3 in red and the Rrp4 capped exosome (Rrp4:Rrp41:Rrp42)3 in blue. (B) Tyr70Rrp42 close to the active site is especially important to efficiently degrade small RNAs. The wild-typ Csl4 exosome is shown in red and the Y70ARrp42 mutant in green. (C) The role of the ring architecture and dynamics for catalytic activity is shown by comparing wild-type cap-less exosome (Rrp41:Rrp42)3 in magenta with the dimeric and open interface mutant (Rrp41:Rrp42)1 and a rigidified crosslinked variant that is less dynamic in yellow. A total of 1000 parameter sets have been randomly drawn from the stationary phase of the Markov chain. Thus for each RNA length and each timepoint, we obtained 1000 estimates whose distribution is displayed by boxplots.
To test this idea, we analysed the Y70A mutant of the Csl4-exosome. The length profile of the catalytic efficiency has a similar shape than for the wild-type, although the catalytic efficiency is lower for all RNA intermediates (Figure 5B). For RNAs >13 nt, the difference in vi is ∼2- to 3-fold (about one log unit). However, the drop in vi for RNAs <13 nt is progressively more pronounced compared to the wild-type and towards short RNAs (<8 nt), the mutant is ∼20- to 150-fold (three to five log units) slower than the wild-type. This is consistent with the idea that for long RNAs the neck provides additional interaction and thus overcomes in part the destabilizing effect of Y70A. For shorter RNAs, the active site becomes the sole attachment, leading to a rapid drop of catalytic efficiency in the Y70A mutant.
We also analysed the ‘neck’ mutant R65E, which has been shown to severely reduce exosome activity (16,26,27). This mutant exhibited a substantially delayed onset of degradation, presumably because RNA is unable to efficiently enter the active site (data not shown). A likely reason is the formation of non-productive RNA:protein complexes with RNA trapped on the outside of the exosome (29). At present, our model cannot deal with this scenario and we could not convincingly include—as only variant—the R65E mutant in the analysis. However, the data of the analysis of R65E are provided in the supplementary Figure S15 and following.
Role of exosome ring formation and ring dynamics for RNA binding
Although the initial binding of RNA appears slow, it seems unlikely that the 3′-end directly finds its way through the small hole in the neck. It is perhaps more likely that the ring structure ‘breathes’—as observed e.g. in hexameric helicases—and allows some lateral entry at the neck. To explore this idea we analysed a crosslinked exosome, where the ring is rigidified by three site specific crosslinks, and a mutant that disrupts the ring structure into Rrp41:Rrp42 pairs. We compared these isoforms with the corresponding wild-type, the cap-less hexameric (Rrp41:Rrp42)3 ring (Figure 5C). From the structural analyses, it was observed that the Rrp41 and Rrp42 subunits possess two interfaces. One interface is larger, and characterized by contiguous β-sheets between Rrp41 and Rrp42 (40). The other interface is smaller, presumably more dynamic, and was chosen for the crosslinking and mutagenesis analysis (Supplementary Figure S2). (Rrp41:Rrp42)3 exhibit a biphasic length dependence of vi, similar to the Csl4-exosome. However, the catalytic efficiency of (Rrp41:Rrp42)3 is ∼5- to 10-fold higher across the RNA spectrum than that of the Csl4-exosome, indicating that the Csl4 cap subunits do not substantially promote degradation of this model substrate. In addition, vi drops for RNAs <13 nt even for the (Rrp41:Rrp42)3 particle, indicating that the neck not e.g. the S1 domains of caps are responsible for the higher catalytic efficiency on longer RNAs.
To explore the effect of the hexameric ring formation, we mutated Lys51 to Glu, located in the ‘smaller’ interface between alternating Rrp41 and Rrp42 pairs. This resulted in stable Rrp41:Rrp42 dimers that do not assemble into hexamers anymore (Supplementary Figure S3). The Rrp41:Rrp42 dimers exhibit catalytic efficiencies that are only slightly lower than the (Rrp41:Rrp42)3 particle for RNAs >13 nt, and almost identical to the corresponding hexamers for RNAs <13 nt. As a result, the drop around 13 nt from a faster to a slower degradation is much less pronounced in the Rrp41:Rrp42 dimer, further supporting the idea that encapsulation in the neck is responsible for higher degradation speeds. The relatively high activity of the dimers is possibly also a result of the effective ‘triplication’ of active sites, i.e. only one RNA molecule can be degraded by a (Rrp41:Rrp42)3, while three RNA molecules can be degraded by three Rrp41:Rrp42 dimers. In addition, while RNA probably dissociates faster from the dimers, this effect could be compensated by a faster association of RNA to the readily accessible active sites in the open dimers.
The opposite is observed, when the ring structure is crosslinked. We introduced cysteines on the outside of the RNase-PH ring and crosslinked the three Rrp41:Rrp42 dimers via a thiol specific bifunctional crosslinker. This procedure resulted in a hexameric RNase PH ring with wild-type-like size and shape according to gel filtration (Supplementary Figure S3). Comparison of νi between the crosslinked isoform with the (Rrp41:Rrp42)3 hexamer, revealed a dramatically reduced νi (∼500- to 2000-fold) indicating that rigidifying the exosome by the crosslink severely affects catalytic efficiency. We cannot formally rule out that the crosslinking affects activity by other means, but considering that the hexamer disrupting mutation at the same interface does not severely reduce activity, a plausible scenario is that the rigidified exosome does not allow efficient association with RNA anymore.
Thus, taken together, the self-compartmentalization of exosomes is probably not an evolution for high activity, but rather for controlled RNA degradation.
Effect of the cap structures
To learn about the role of the cap proteins in exosome activity, we compared the rate constants of the cap-less exosome with the Csl4 and the Rrp4 exosome. The initial rates for degradation of the 30-mer rA are similar for the Csl4-exosome and the cap-less version, but considerably faster for the Rrp4-exosome (Figure 5A). This indicates that cap proteins can influence recognition and recruitment of RNA substrates and that this step is more efficient for the Rrp4 exosome. However, while RNA degradation for medium and short RNAs is quite comparable between the Csl4 and Rrp4 exosomes, there is an interesting difference for long RNA molecules (>24 nt). The Rrp4 exosome is quite slower for RNAs >24 nt, faster for RNA between 24 and 13 nt, and then progressively slower for RNAs <13 nt. This remarkable length dependency is clearly evident in degradation profiles (Figure 1). The most likely explanation is that long RNAs might still have contacts with Rrp4, where a more specific binding site holds them partially back from rapid degradation. In principle, this could be viewed as molecular friction. When RNAs are shorter, they loose contact to Rrp4 and degradation speed is increased. The Csl4 protein and the Rrp4 protein differ in their domain structures. While Csl4 contains a Zn-ribbon domain, Rrp4 possesses a KH-domain, which is a typical RNA-binding domain and could recognize the oligo-rA. Such a binding could be responsible for the faster first degradation step, because it more efficiently sequesters RNAs on the exosome surface, but may subsequently slow down degradation until RNAs are too short to maintain simultaneous contacts at the KH domain and active site.
However, the Rrp4 isoform is more efficient for smaller RNA species than the Csl4 and capless isoforms. Since these shorter RNAs cannot form dual contacts with the active site and outside the caps, the Rrp4 could also influence the dynamics or other properties of the RNase-PH ring, for instance to help in loading of RNA into the ring structure.
SAXS structure of the Rrp4 exosome with endogenously purified bacterial RNA
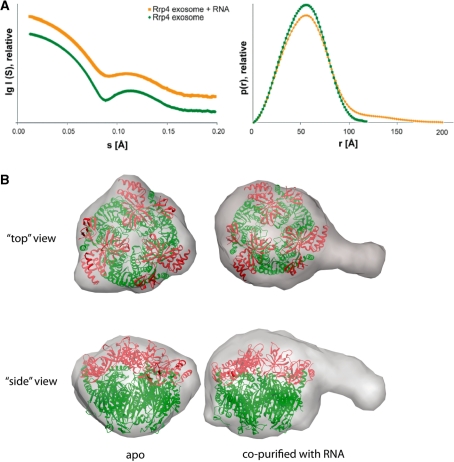
To explore the role of the Rrp4 cap in efficiently recruiting RNAs further, we performed SAXS studies with a nuclease deficient nine-subunit Rrp4 exosome bound to RNA: we had noticed that this nuclease deficient Rrp4-exosome (D180A in Rrp41) very efficiently co-purifies with E. coli RNA. To determine the kind of RNA that binds to the exosome we run it on a denaturing gel together with RNAs with known sizes and could estimate the size of the RNA to be between 55 and 65 nt (Supplementary Figure S1). Cloning and sequencing of bound RNA molecules revealed a set of much shorter inhomogeneous mixed sequences (Supplementary Table S3). It is possible that the bound RNAs are a mixture of various mRNAs from E. coli, although the isolated RNA is larger than the identified sequences and it is possible that highly structured RNAs such as tRNAs are underrepresented due to inefficient amplification and cloning. Comparison of the SAXS structure of apo-Rrp4-exosome with the RNA bound complex shows an increase in the radius of gyration from 39.6 Å to 46.8 Å when RNA is bound and the corresponding pair distribution functions contains longer vectors (Figure 6A), likely because additional scattering elements from RNA protrude from the compact protein core. The resulting ab initio model of the complex overlaid with the crystal structure of the apo-complex clearly indicates additional mass from the bound RNA (Figure 6B and C). This clear additional mass is distributed in the centre of the cap structure on top of the neck region but also protrudes away from the complex. When looking at the overlay with the crystal structure it appears that the RNA is bound at the KH and the S1-domains. The SAXS analysis supports the model that RNA binds near the KH-domain on the outside of the caps and reveals a low-resolution image of trapped exosome–RNA complexes.
Figure 6.
SAXS structure of the Rrp4 exosome with endogenously purified bacterial RNA. (A) SAXS data of the Rrp4 exosome (green) and the Rrp4 exosome with RNA (orange) (curves show the scattering intensity I(q) as a function of the scattering angle 2θ and X-ray wavelength λ, where q = (4sin/θ)) and the pair-distribution function describing intramolecular distances; in the presence of RNA longer distances occur and the radius of gyration increases. (B) Average of 10 independent ab initio models for the apo exosome and the RNA-bound complex superimposed with the crystal structure. The additional density for the RNA is clearly visible.
DISCUSSION
RNA exosomes are large, self-compartmentalized nucleases, implicated in processive, controlled degradation of a large variety of RNAs. While the archaeal exosome possesses three phosphorolytic active sites within the compartment, the eukaryotic exosomes apparently have lost this activity but adopted hydrolytic RNase subunits that are bound at the outside of the evolutionary conserved core. Nevertheless, recent data suggest that RNA is still threaded through the eukaryotic core exosome before it is degraded in ectopic hydrolytic active sites, suggesting that the core particle retained critical ‘structural’ functions regarding RNA degradation such as increased processivity or controlled RNA degradation (25).
To be able to quantitatively address RNA exosome activities, we derived a kinetic model for the complex RNA degradation of the archaeal RNA exosome using Markov Chain Monte Carlo analysis. The kinetic model gives a realistic assessment of the velocity of the exosome and mutant variants during processive degradation of a rA 30-mer oligonucleotide. The considerable effort we had to put into the MCMC simulation pays off eventually. We are now able to derive a realistic joint posterior distribution of kinetic parameters, enabling us to quantify the relation of different parameters in either the same or in distinct exosome mutants. This would have been impossible with a conventional least squares fit of the data, which produces very unstable parameter estimates (see Supplementary Data for a comparison), although the obtained fits are very good (Figure 3D).
With this in hand, we find several interesting and unexpected features of RNA degradation activities. First, kinetic evaluation of RNA degradation of exosomes needs to include association and dissociation rate constants. Thus the kinetics cannot be treated as strictly processive, at least for RNA species in the assessed length range. This does not necessarily imply that RNA dissociates and rebinds completely from the exosome. Longer RNAs may be retained within the neck as well as cap domains, while binding and dissociating from the active site in the processing chamber. The association and dissociation constants can thus be understood as ‘ratcheting’ constants that influence the rate of translation along the RNA to and from the active site. For short substrates that are unable to simultaneously bind neck and active site this connection is lost, the dissociation increases, degradation speeds drop and the exosome changes from being fast and processive to a slower distributive enzyme.
Our results also show how neck region and active site features contribute to exosome activity. Although we could not quantitatively address the importance of Arg65 in the neck with the simplified model in hand, this residue appears to be important for loading RNA into the processing chamber, but not for efficient degradation once RNA is bound. This conclusion is derived from the observation that while the initial degradation is substantially delayed, the appearance of smaller RNA species is qualitatively similar to the wild-type Csl4 exosome. Taken together with the observation that crosslinking severely reduces processing and the RNase PH ring needs to breath or display some conformational dynamics, it is unlikely that RNA is simply threaded into the processing chamber like a yarn through the eye of a needle. Rather, we propose that initial RNA binding includes some lateral entry near the neck.
We are also in the position now to address the influence of the cap proteins Rrp4 and Csl4. These proteins possess a variety of domains with unclear function in exosome activity. While eukaryotic exosomes have defined heterotrimeric caps, the stoichiometry of cap proteins in archaeal exosomes is not defined in vitro and perhaps variable in vivo. For the archaeal exosome, the Csl4 capped isoform displays similar degradation kinetics to that of the capless variant, and the function of this type of cap remains to be shown. However, the Rrp4 isoform substantially differs from the other two variants and our analysis suggests that Rrp4 more efficiently recruits RNA to the exosome. In fact, RNA from the heterologous expression in E. coli is very tightly bound to the Rrp4 exosome. It must be noted that the gene coding for Rrp4 is in the same operon as genes for Rrp41 and Rrp42, indicating that this cap is perhaps a ‘default’ isoform of the exosome, while the Csl4 cap, located elsewhere in the genome, might be differentially regulated.
The cap structures, however, also influence the degradation of short RNAs. This is to some extent surprising, since short RNAs (<13 nt) cannot bind to the caps and the active site at the same time. However, the Rrp4 subunit more intimately interacts with the RNase PH ring than the Csl4 protein and might influence also the dynamics of the RNase PH type ring. Likewise, binding of RNA to the KH domains, consistent with the lateral density of RNA in the SAXS models, may position it better for loading into the processing chamber.
In sum, we present here a robust method to analyse the complex degradation kinetics of a partially processive degradation enzyme in a quantitative manner, with estimates of the posterior distribution of the model parameters. We applied this analysis to degradation of RNA by several isoforms and variants of the archaeal exosome and could reveal a variety of features that are important for catalytic efficiency. The objective of this manuscript is to derive a general method that can now be used to unravel the biochemistry of exosomes in a more quantitative manner. The method can now form a basis for comprehensive analysis of different substrates, other RNA sequences, as well as mutants of this system or related systems such as the eukaryotic exosome.
ACCESSION NUMBERS
3M7N, 3M85.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (HO2489/3 and SFB646); Center for Integrated Protein Science Munich. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Christian Luginsland for help in protein purification, Katharina Büttner for exosome constructs and Katja Lammens and Gregor Witte for helpful discussions. The authors thank the staff of the European Synchrotron Radiation Facility (beamline 14–2) and the Swiss Light Source (beamline PX I) for help with diffraction data collection and Michal Hammel from the Advanced Light Source (SIBYLS beamline) for help with scattering data collection.
REFERENCES
- 1.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–>5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 2.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 5.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol. Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 7.Vasudevan S, Peltz SW, Wilusz CJ. Non-stop decay–a new mRNA surveillance pathway. Bioessays. 2002;24:785–788. doi: 10.1002/bies.10153. [DOI] [PubMed] [Google Scholar]
- 8.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 9.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ –> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Büttner K, Wenig K, Hopfner KP. The exosome: a macromolecular cage for controlled RNA degradation. Mol. Microbiol. 2006;61:1372–1379. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 15.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Navarro MV, Oliveira CC, Zanchin NI, Guimaraes BG. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J. Biol. Chem. 2008;283:14120–14131. doi: 10.1074/jbc.M801005200. [DOI] [PubMed] [Google Scholar]
- 17.Ramos CR, Oliveira CL, Torriani IL, Oliveira CC. The Pyrococcus exosome complex: structural and functional characterization. J. Biol. Chem. 2006;281:6751–6759. doi: 10.1074/jbc.M512495200. [DOI] [PubMed] [Google Scholar]
- 18.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc. Natl Acad. Sci. USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, Schuster G. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus. EMBO Rep. 2005;6:1188–1193. doi: 10.1038/sj.embor.7400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 21.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, Greimann JC, Lima CD. Reconstruction, activities, and structure of the eukaryotic RNA exosome. Erratum. Cell. 2007;131:188–189. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 23.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 24.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 26.Lorentzen E, Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter P, Klein F, Lorentzen E, Ilchmann A, Klug G, Evguenieva-Hackenberg E. Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2006;62:1076–1089. doi: 10.1111/j.1365-2958.2006.05393.x. [DOI] [PubMed] [Google Scholar]
- 29.Büttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. Automatic data processing of rotation diffraction data from crystals of initially unkonwn symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 31.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. D Biol. Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 37.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocke DM, Durbin B. A model for measurement error for gene expression arrays. J. Comput. Biol. 2001;8:557–569. doi: 10.1089/106652701753307485. [DOI] [PubMed] [Google Scholar]
- 40.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.