Abstract
In response to replication-blocking DNA lesions, proliferating cell nuclear antigen (PCNA) can be conjugated with a single ubiquitin (Ub) or Lys63-linked Ub chains at the Lys164 residue, leading to two modes of DNA damage tolerance (DDT), namely translesion synthesis (TLS) and error-free DDT, respectively. Several reports suggest a model whereby monoubiquitylated PCNA recruits TLS polymerases through an enhanced physical association. We sought to examine this model in Saccharomyces cerevisiae through artificial fusions of Ub to PCNA in vivo. We created N- and C- terminal gene fusions of Ub to PCNA-K164R (collectively called PCNA·Ub) and found that both conferred tolerance to DNA damage. The creation of viable PCNA·Ub strains lacking endogenous PCNA enabled a thorough analysis of roles for PCNA mono-Ub in DDT. As expected, the DNA damage resistance provided by PCNA·Ub is not dependent on RAD18 or UBC13. Surprisingly, inactivation of TLS polymerases did not abolish PCNA·Ub resistance to DNA damage, nor did PCNA·Ub cause elevated spontaneous mutagenesis, which is a defining characteristic of REV3-dependent TLS activity. Taken together, our data suggest that either the monoubiquitylation of PCNA does not promote TLS activity in all cases or PCNA·Ub reveals a currently undiscovered role for monoubiquitylated PCNA in DNA damage tolerance.
INTRODUCTION
With the surge in ubiquitin (Ub) research, a growing number of proteasome-independent roles are emerging for targets linked to single Ub molecules or atypical poly-Ub chains not linked via Lys48. Perhaps the most striking example is seen for proliferating cell nuclear antigen (PCNA), which was found to be a key target of Ub modification in the RAD6-dependent DNA damage tolerance (DDT) pathway (1).
In comparison to other DNA repair pathways in the budding yeast Saccharomyces cerevisiae, the mechanisms governing DDT are only now being elucidated. For many years DDT (also known as DNA post-replication repair, PRR) was best known in yeast as a genetic framework that was characterized through epistatic relationships based on DNA damage sensitivity. Genetic models of DDT elucidated the same underlying features; namely a branched pathway stemming from RAD6 and RAD18 leading to at least one error-free and one error-prone DDT sub-pathway (2–4). Null alleles of RAD6 and RAD18 are epistatic to all other DDT gene mutations, and these mutants exhibit severe sensitivity to a broad spectrum of DNA damaging agents sharing replication-blocking lesions as a common outcome of exposure/treatment. The discovery and characterization of the MMS2 gene established the error-free DDT pathway (5), into which RAD5 and UBC13 were later added (3,6,7). The error-prone branch consists of REV3, REV7 and REV1, which encode translesion synthesis (TLS) polymerases with a propensity for introducing mutations, although an exception lies in the relatively error-free ability of Rad30 (Polη) to bypass UV-induced DNA lesions (8). The relationship between the two branches of DDT is such that mutations of the error-free genes, such as MMS2, lead to dramatically increased mutagenesis due to channeling lesions into the error-prone TLS pathway (5). Recently, several studies began to shed light into the molecular activities of DDT gene products. It is now generally accepted that DDT is achieved through sequential Ub conjugation to PCNA by two Ub conjugating-Ub ligation (E2–E3) complexes: Rad6-Rad18 and Mms2-Ubc13-Rad5 (1,9–11). PCNA (encoded by POL30) forms an essential homotrimer that functions as a sliding clamp for the highly processive DNA polymerases in eukaryotic cells (12). PCNA is involved in numerous roles in DNA metabolism (13), and a pol30-46 allele was previously implicated in DDT (14). Hoege et al. (1) demonstrated that PCNA is modified in vivo at a single site (Lys164) by mono-Ub after DNA damage, via Rad6-Rad18, and by Lys63-linked Ub chains via Mms2-Ubc13-Rad5. The Lys164 residue and its modification by mono- and Lys63-linked poly-Ub is apparently conserved throughout eukaryotes (15–19). A mechanistic model controlling DDT was proposed whereby modification of PCNA by mono-Ub promotes TLS, whereas the further addition of Lys63-linked Ub chains promotes the error-free DDT pathway (1,20). How the error-free DDT pathway functions following the Lys63-linked poly-Ub signal remains unclear (21). Data supporting the model show that the pol30-K164R mutation prevents Ub conjugation and leads to DNA damage sensitive phenotypes that are epistatic to all DTT pathway mutations (1).
A molecular mechanism for mono-Ub-PCNA signaling in DDT has been proposed (22,23). Several reports support a model whereby the intrinsic affinity of PCNA for Y-family TLS polymerases containing the PCNA-binding (PIP) motif is enhanced by its monoubiquitylation (18,24,25). These polymerases, including Polη, Rev1, Polι and Polκ, are all found to contain specialized Ub-binding domains (UBM or UBZ) that aid their physical association with monoubiquitylated PCNA. While these studies were mainly performed with mammalian proteins, some supporting evidence has been found for Polη in S. cerevisiae (26).
In order to test the TLS signaling model with respect to DDT function in vivo, we created linear fusions of UB and POL30-164R in order to mimic the constitutive monoubiquitylation of PCNA (PCNA·Ub). We demonstrate that PCNA·Ub is able to confer S. cerevisiae strains with DDT. Our ability to delete the endogenous POL30 gene in strains harboring PCNA·Ub allowed us to critically test the function of PCNA·Ub in the context of DDT. Surprisingly, resistance to the alkylating agent methyl methanesulfonate (MMS) provided by PCNA·Ub was not dependent on the TLS polymerase ζ and PCNA·Ub did not increase mutagenesis levels associated with TLS activity. Taken together, our data adds to evidence challenging the current model of TLS signaling by monoubiquitylated PCNA and supports an alternative model in which monoubiquitylated PCNA may overcome replication blocks via a currently unknown mechanism (27).
MATERIALS AND METHODS
Plasmids and plasmid construction
To create a pol30Δ disruption cassette, a 0.28-kb StyI-EcoRI fragment within the POL30 ORF was deleted from pBL230 (28) (YCp, TRP1, POL30, from Dr P. Burgers, University of Washington, St Louis) and replaced with a 1.15-kB HIS3 gene from YDp-H (29) to form ppol30Δ::HIS3. The pol30Δ::HIS3 disruption cassette was released by MluI-XbaI digestion prior to transformation. Plasmid pubc13ΔBg (7) was used to clone a 0.8-kb TRP1 gene from YDp-W (29) to form pubc13Δ::TRP1. The ubc13::TRP1 disruption cassette was released by SphI-MluI digestion prior to transformation.
Plasmid YCpT-Pol30-Ub was created by first cloning the synthetic UBI4 gene without the C-terminal two Gly residues as a BamHI-KpnI fragment into YCplac22 (30). A 0.55-kb POL30 C-terminal untranslated region was then PCR amplified and cloned as a KpnI-SacI fragment into the 3′-end of UBI4 to form YCpT-Pol30-UbT. The pol30-164R gene and its own 0.5-kb upstream untranslated region plus a 7xHis coding sequence at the 5′-end of pol30-164R was PCR amplified as a 1.3-kb BamHI fragment and cloned into YCpT-Pol30-UbT to form YCpT-Pol30-Ub. The entire pol30-UB cassette was released as a 2.3-kb SacI-PstI fragment and cloned into YCplac111 to form YCpL-Pol30-Ub.
To create a construct expressing the N-terminal Ub-Pol30 fusion protein, the modified UBI4 gene without the stop codon was PCR amplified as a NotI fragment and cloned into YCpL-Pol30-164R at the NotI site, which is located immediately upstream of the 7xHis-pol30-164R coding sequences, to form plasmid YCpL-Ub-Pol30. All cloned PCR-amplified fragments were verified by sequencing the entire insert.
Saccharomyces cerevisiae strains, cell culture and transformation
The haploid S. cerevisiae strains DBY747, HK578-10A and HK578-10D were used as the wild-type source for the creation of all strains listed in Supplementary Table S1.
Yeast cell cultures and plasmid transformations were performed as previously described (31). For disruption of genomic POL30, yeast cells were transformed with the plasmid pBL211 (28) (YCp, URA3, POL30, from Dr P. Burgers) by a modified lithium acetate method (32). One-step targeted gene disruption was then performed (33) by the pol30Δ::HIS3 disruption cassette released from plasmid ppol30Δ::HIS3 with restriction enzymes MluI-XbaI prior to transformation. Deletion of the genomic copy of POL30 was confirmed by the reliance of cells on pBL211 for survival, as indicated by its inability to grow on a 5-fluoroorotic acid (FOA) plate (34). Plasmids carrying the POL30 gene derivatives were transformed into the strain, and subsequent loss of the pBL211 plasmid was confirmed by an ability to grow on media containing FOA.
The complete MMS2 open reading frame (ORF) was deleted by an mms2Δ::TRP1 cassette (4). The rev3 null mutant was made using a rev3Δ::hisG-URA3-hisG cassette obtained by KpnI digestion of plasmid pDG347 (35). The ubc13 deletion mutant was made with a ubc13Δ::TRP1 cassette. The RAD18 ORF was deleted by using a rad18Δ::TRP1 cassette as previously described (36).
DNA damage sensitivity assays
Gradient and dilution plate assays were performed to measure the relative sensitivity of S. cerevisiae cells to MMS and UV radiation, respectively. Saccharomyces cerevisiae cells were transformed with YCpL-Pol30 derivatives and at least four individual transformants were examined but a single representative clone for each strain is shown. The gradient plate assay was performed as previously described (36). Briefly, the gradient was made by pouring 30 ml of molten YPD agar containing the predetermined MMS concentration into a tilted square petri dish. After solidification for 1 h, the dish was returned flat and 30 ml of the same molten agar without MMS was poured to form the top layer. Overnight cell cultures grown in selective media were printed across the gradient and the plate was incubated at 30°C for 48 h or otherwise as indicated before taking the photograph. For the serial dilution plates, 10 μl of overnight cell culture grown in selective minimal media was spotted as 10-fold dilutions in water on YPD agar plates. The spots were allowed to dry and the plates were subsequently exposed to UV doses as indicated. Plates were incubated at 30°C for 48 h before taking the photograph. For each of the above assays, multiple doses were performed but only the results from a single dose are presented.
Spontaneous mutagenesis
Spontaneous mutagenesis experiments were performed as previously described (37).
Protein purification and analysis
Saccharomyces cerevisiae cells for generating cell lysates were subcultured 1:5 in YPD broth from overnight cell cultures and grown to an OD600 nm between 0.8 and 1.0 prior to the following preparations. Cells for whole-cell lysate analysis were pelleted and resuspended in YeastBuster Protein Extraction Reagent (Novagen, #71186) as per the manufacturer’s instructions. Samples were combined with 2× sample buffer and separated by 12% SDS–PAGE. Cells for the purification of His7-tagged PCNA were resuspended in 1.5 vol. of binding buffer (20 mM sodium phosphate, 0.5 mM NaCl, 40 mM imidazole, pH 7.4) to which 1 volume of glass beads was added. Cells were then bead-beat in 30-s pulses for a total of 2 min, keeping the samples on ice between pulses. Cellular debris was removed with centrifugation at 15 000g for 30 min and the supernatant, typically 15 ml, was added to a Bio-Rad prep-column containing a 150-μl bed-volume of His-binding resin (GE Healthcare, #17-5318-01). Columns were washed five times with binding buffer and eluted with 200 μl elution buffer (20 mM sodium phosphate, 0.5 mM NaCl, 500 mM imidazole, pH 7.4). Samples were combined with 6× sample buffer and separated by SDS–PAGE. Steps involving the purification of His7-tagged PCNA were performed at 4°C.
Western blots were performed using the following antibodies. Mouse polyclonal anti-yeast PCNA antibody was raised in-house using purified Pol30 from bacterial cells as an antigen, and was used at a 1:2500 dilution. Anti-Ub (Upstate, #07-375) and anti-His (Santa Cruz, #SC-803) antibodies were used at a 1:5000 dilution. Horseradish peroxidase-conjugated secondary anti-mouse (Upstate, #12-349) and anti-rabbit (Upstate, #12-348) antibodies were used at a 1:5000 dilution. A Western Lightning Chemiluminescence kit (Perkin Elmer Life Sciences, #NEL104) was used for signal development.
Structural imaging
Molecular images were generated using the protein data bank files indicated and PyMOL version 0.96 by DeLano Scientific (http://www.pymol.sourceforge.net).
RESULTS
Design and structural model of the Ub and PCNA fusions
Several considerations were taken into account in designing the fusion of Ub to PCNA. Since PCNA levels have been shown to fluctuate in the cell (1), we created each of the gene fusions in yeast centromeric plasmid (YCp)-based single-copy plasmids under the control of the native POL30 promoter, in order to achieve physiologically relevant genetic regulation. The POL30 ORF used for Ub fusion contains a K164R substitution (pol30-164R) in order to prevent the natural Ub modification of PCNA following DNA damage (1), which would complicate experimental interpretation. We created both N- and C-terminal genetic fusions of UBI4 to pol30-164R (Figure 1A) in order to take into account different structural implications in each case (Figure 1B and C). Each construct also contains an N-terminal 7xHis tag to facilitate purification and analysis. Not depicted in the N-terminal Ub construct is the deletion of the C-terminal Gly–Gly residues of Ub in order to prevent Ub protease cleavage. As seen in Figure 1B, the PCNA C-terminal and Lys164 residues reside on the same ridge, albeit on separate faces of the molecule. The PCNA N-terminus is situated on a separate but similar ridge to Lys164 and is in a medial location between the faces. Hence, the N- and C-terminal Ub fusions may represent two types of structures resembling the natural aspects of PCNA ubiquitination at K164. However, we note that the orientation of the C-terminal fusion causes an atypical ‘inverted’ attachment of Ub to PCNA when compared with a model showing a natural Lys164 modification (Figure 1C).
Figure 1.
Structures of the Ub and PCNA fusions. (A) Schematic diagrams of the Ub fusion constructs to the N-terminus (top) or C-terminus (bottom) of PCNA-164R. (B) Molecular structure of the PCNA homotrimer from both front and side views shown at left and right, respectively. (C) Predicted PCNA-Ub structures for the N- and C-terminal fusions as well as the in vivo PCNA-Lys164 monoubiquitylation, The PDB file for PCNA (1AXC) (52) was used to generate the molecular structure. Each subunit of the PCNA homotrimer is colored differently and the natural Ub modification site at Lys164 is in yellow. The N- and C-terminal residues of PCNA are colored blue and red, respectively.
The fusion of Ub to PCNA partially and specifically rescues the rad18 mutant
The PCNA·Ub fusions were first analyzed in RAD18 deletion strains. When S. cerevisiae cells are inflicted with replication-blocking damage caused by agents such as MMS or UV, the Rad6-Rad18 E2-E3 heterodimer promotes DDT through conjugation of a single Ub to PCNA (1). Since Rad6 is involved in several other S. cerevisiae pathways, whereas Rad18 functions solely in DDT (2), the RAD18 deletion mutant serves best for abolishing the DDT pathway. Thus, even in the presence of wild-type PCNA, the rad18Δ strain ensures that the PCNA·Ub gene fusions are the only source of ubiquitylated PCNA.
The phenotype of the rad18Δ mutant carrying PCNA·Ub fusion constructs is presented in Figure 2, which shows that rad18Δ cells harboring plasmids containing either POL30 or pol30-164R were extremely sensitive to MMS. We also note that the rad18Δ strain containing pol30-164R was slightly more resistant to MMS than the one harboring wild-type POL30. This is consistent with a previous report (1) and is likely due to its inability to be modified by SUMO (20). Nevertheless, the pol30-164R sensitivity to MMS is partially alleviated when rad18Δ strains contain either of the plasmids expressing the PCNA·Ub fusions. The function of pol30-UB and UB-pol30 is not limited to DNA damage caused by MMS (Figure 2A) because we observed the same effect in cells that were treated with UV (Figure 2B). Interestingly, the N-terminal Ub-PCNA fusion appeared to provide more resistance than the C-terminal PCNA-Ub fusion (Figure 2). Taken together, the data indicate that the DNA damage sensitivity of rad18Δ can be partially alleviated by the fusion of Ub to either PCNA terminus, despite the presence of endogenous wild-type PCNA.
Figure 2.
Relative DNA damage sensitivity assays to evaluate the in vivo function of the POL30-UB gene fusions. The provision of DNA damage resistance by UB-pol30 and pol30-UB is tested by their ability to protect a given mutant strain from killing by MMS or UV treatment. (A) A 0.005% MMS gradient plate assay of the rad18Δ mutant (WXY971) carrying different plasmids. (B) A serial dilution plate assay of the rad18Δ mutant (WXY971) carrying different plasmids exposed to 10 J/m2. (C) A serial dilution plate assay of the rad10Δ mutant (WXY9537) carrying different plasmids exposed to 1.5 J/m2. (D) A 0.002% MMS gradient plate assay of the rad52Δ mutant (WXY9560) carrying different plasmids. All plates were incubated at 30°C for 48 h before photography. For gradient plates, arrows point to increasing MMS concentration. For serial dilution plates, cells were diluted 10-fold from left to right.
In order to determine if the function of PCNA·Ub is specific to DNA repair via DDT (i.e. RAD18) or whether it simply provides DNA damage resistance in general, we tested the function of PCNA·Ub constructs in yeast strains inactive in the nucleotide excision or homologous recombination DNA repair processes. RAD10 is a member of the RAD3 epistasis group and its deletion disrupts nucleotide excision repair (38). As a result, rad10Δ cells are severely sensitive to UV irradiation. RAD52 represents the DNA repair epistasis group functioning in homologous recombination repair and its mutation causes strong sensitivity to DNA damage caused by MMS (38). In contrast to the effect seen in the rad18Δ strain, PCNA·Ub fusions did not provide DNA damage resistance in either rad10Δ (Figure 2C) or rad52Δ (Figure 2D). We noted that the relative DNA damage sensitivity of rad10Δ and rad52Δ strains to UV and MMS, respectively, is comparable to rad18Δ and as such provide a good reference for the specificity of PCNA·Ub function. In addition, we did not observe an enhanced DNA damage resistance phenotype in wild-type yeast strains harboring PCNA·Ub constructs (data not shown). Taken together, these results demonstrate that the function of PCNA·Ub is confined to the RAD6 DDT pathway.
In this study, we also tested to see whether other factors related to PCNA modifications affect the function of PCNA·Ub fusions. We previously showed that the checkpoint protein Rad17 is another target of mono-Ub modification via Rad18 following DNA damage (39) and questioned this modification in the context of PCNA·Ub function. We found that DNA damage resistance by PCNA·Ub was still present in a rad17Δ rad18Δ strain (Supplementary Figure S1A and B). Since PCNA is also modified by the small Ub-like modifier (SUMO) via the Siz1 SUMO ligase (20), we tested to see if PCNA·Ub function was dependent on SIZ1, and found that SIZ1 deletion had no effect on PCNA·Ub-mediated damage tolerance to either MMS or UV (Supplementary Figure S1C and D).
POL30 deletion strains are viable when complemented with POL30·UB gene fusions
Although we observed functionality of the PCNA·Ub fusions, we sought an experimental system whereby the fusions could be directly tested in the absence of endogenous wild-type PCNA. This approach would offer several important advantages, such as ensuring that wild-type PCNA does not compete with the PCNA·Ub fusion and that each of the trimeric subunits is constitutively monoubiquitylated. Using a plasmid shuffling approach we were able to create S. cerevisiae strains in which viable pol30Δ is made possible by expressing UB-pol30 or pol30-UB. Equivalent pol30Δ strains harboring the corresponding POL30 or pol30-164R plasmids were also created for proper experimental controls.
Figure 3A shows that these strains grew equally well in the absence of DNA damage by a colony-size assay and we observed no apparent difference in growth between the strains in subsequent experiments. Anti-PCNA western blots of whole-cell extracts prepared from pol30Δ strains harboring POL30 and either UB-pol30 or pol30-UB indicate equal protein levels for the PCNA·Ub fusions as compared with wild-type PCNA (Figure 3B, lanes 1 and 2). The successful loss of the wild-type POL30 plasmid in these strains was also confirmed (Figure 3B, lanes 3 and 4); however, a minor band corresponding to unmodified PCNA was observed in the pol30Δ strain containing the UB-pol30 construct (Figure 3B, lane 4 and Figure 3C). This unmodified PCNA band observed in the UB-pol30 fusion strain is unlikely from cells harboring wild-type POL30 plasmid, since the single colony (i.e. clone) has been through several passages of FOA selection. Additional bands are seen for proteins that cross-react with the α-PCNA polyclonal antibodies and provide a comparison for total protein loaded in each sample (Figure 3B). In order to better analyze the PCNA·Ub fusion protein in the pol30Δ strains, immobilized metal affinity chromatography was carried out to purify the PCNA derivatives. Western blot analysis of these samples reveals that PCNA-Ub is a stable fusion, whereas a small proportion of the Ub-PCNA fusion (but not the PCNA-Ub fusion) is cleaved such that the 7xHis-Ub portion is removed, leaving a small amount of PCNA(K164R). In summary, this analysis indicates that cells are viable and can grow well when all PCNA subunits are constitutively fused to Ub.
Figure 3.
Analysis of cell growth and proteins produced by yeast cells harboring the UB-pol30 and pol30-UB gene fusions. (A) Cell growth by a colony size assay. Equal inoculations of HK578-10D pol30Δ::HIS3 cells harboring different plasmids as indicated were streaked onto YPD agar and incubated at 30°C for 48 h. Differences in growth rates are observed if the colony sizes vary noticeably between strains. (B) Analysis of PCNA protein from whole-cell lysates. POL30 deletion strain HK578-10D pol30Δ::HIS3 containing the plasmids as indicated was used to create whole-cell lysates. The anti-PCNA western blot reveals the PCNA derivatives contained within each strain. Non-specific cross-reacting bands indicated with asterisks demonstrate equal protein loading throughout the samples. (C) Analysis of affinity purified PCNA protein. Lysates made from HK578-10D pol30Δ::HIS3 derivatives that produce 7xHis-tagged PCNA were subjected to His-affinity chromatography. Western blots of the samples using anti-PCNA (left), anti-Ub (center), and anti-His6 (right) antibodies are shown. The non-specific co-purifying bands detected (marked with asterisks) in the anti-His blot indicate that similar amounts of total protein were used for each sample.
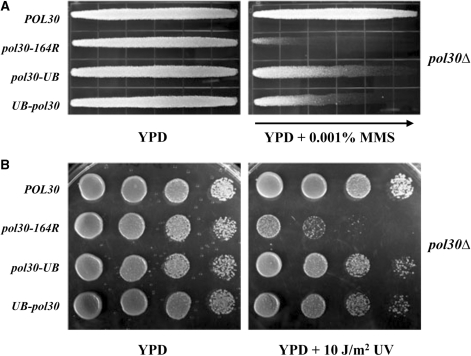
The PCNA·Ub fusions confer DNA damage resistance in a pol30Δ strain
The successful generation of pol30Δ strains maintained by PCNA derivatives enabled us to directly analyze the DDT function of the PCNA·Ub fusions. As seen in Figure 4, the pol30Δ strain carrying pol30-164R exhibited an enhanced sensitivity to DNA damage by both MMS (Figure 4A) and UV (Figure 4B), and this sensitivity was alleviated by fusion of a single Ub in each of the pol30-UB and UB-pol30 constructs. The partial level of resistance provided by the PCNA·Ub fusions, as compared with wild-type PCNA, was expected because mono-Ub is proposed to promote only a single branch of DDT, namely TLS (1,20). Furthermore, we have been unable to observe PCNA polyubiquitylation in these strains (Figure 3 and data not shown). The finding that pol30-UB provided stronger resistance to DNA damage than UB-pol30 is possibly attributed to the cleavage of some of the N-terminal Ub-PCNA(K164R) fusion protein (Figure 3), although we cannot rule out a contribution from structural differences (Figure 1C). We also note that the PCNA·Ub fusion phenotypes observed in this experiment were significantly stronger than rad18Δ cells containing endogenous wild-type POL30 (Figure 2).
Figure 4.
Relative sensitivity to DNA damaging agents of HK578-10D pol30Δ::HIS3 cells harboring the pol30·UB gene fusions constructs. The provision of DNA damage resistance by UB-pol30 and pol30-UB is tested through their ability to complement a pol30Δ yeast strain with respect to killing by MMS or UV treatment by comparing with isogenic cells containing the PCNA-164R mutation. (A) 0.001% MMS gradient plate assay. (B) Serial dilution plate assay with 10 J/m2 UV irradiation. Experimental conditions were as described in Figure 2.
The DNA damage resistance provided by PCNA·Ub fusions is independent of RAD18, error-free DDT and TLS
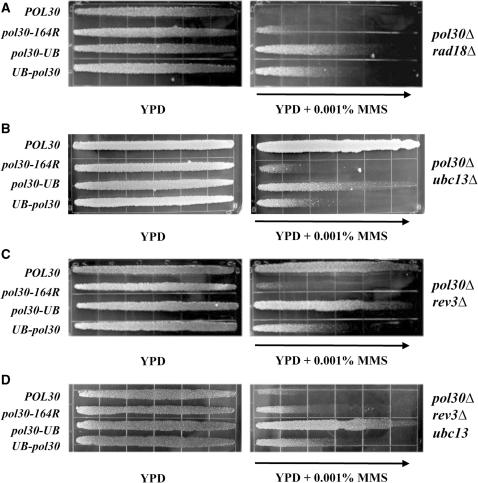
We wanted to determine the nature of the MMS resistance provided by PCNA·Ub fusions within the DDT pathway. To help confirm that the PCNA·Ub fusions are indeed functional within DDT, we first tested their function in a pol30Δ rad18Δ double-deletion strain. As discussed above, RAD18 and RAD6 govern the entire DDT pathway and as such, deletion of either gene causes a complete loss in DNA damage resistance via DDT. As seen in Figure 5A, each of the fusions provided MMS resistance in a pol30Δ rad18Δ strain, suggesting that Ub-PCNA and PCNA-Ub can at least partially replace Rad18′s role for modifying PCNA with mono-Ub.
Figure 5.
MMS gradient plate assays to assess the effects of deleting DDT pathway genes on HK578-10D pol30Δ::HIS3 cells harboring pol30·UB gene fusions. (A) In the pol30Δ rad18Δ strain background; (B) in the pol30Δ ubc13Δ background; (C) in the pol30Δ rev3Δ background; and (D) in the pol30Δ ubc13Δ rev3Δ background. Experimental conditions were as described in Figure 2.
We next determined whether the activity of the PCNA·Ub fusions was attributed to elongation of the single Ub to a poly-Ub chain. Since the Ub-conjugating enzyme Ubc13, along with the accessory protein Mms2, is required for the synthesis of Lys63-linked poly-Ub chains on PCNA to promote the error-free DDT pathway (1,5,7), we tested the MMS resistance phenotype of the fusion constructs in a pol30Δ ubc13Δ strain. As seen in Figure 5B, the PCNA·Ub fusions provided more MMS resistance in the pol30Δ ubc13Δ strain as compared with the non-fused pol30-164R control. We conducted the same experiment with a pol30Δ mms2Δ strain and observed a similar phenotype (Supplementary Figure S2A). These data allowed us to conclude that the Lys63-linked Ub conjugation activity of Ubc13-Mms2 is not required for the function of the PCNA·Ub fusions in providing MMS resistance in DDT.
The mono-Ub modification of PCNA after DNA damage treatment has been proposed to promote the TLS pathway of DDT through signaling to one of the TLS polymerases. Of the TLS polymerases in S. cerevisiae, Polζ, which consists of two subunits encoded by REV3 and REV7, is primarily responsible for the bypass of MMS-induced damage (40). REV3 encodes the catalytic subunit of Polζ and is absolutely required for this activity. Therefore, we sought to test the function of the PCNA·Ub fusions in a pol30Δ rev3Δ strain. To our surprise, each of the pol30-UB and UB-pol30 constructs was still able to alleviate MMS sensitivity in the pol30Δ rev3Δ strain (Figure 5C). To examine the phenotype further and to rule out that the error-free DDT pathway is providing MMS resistance in the rev3Δ background, we tested PCNA·Ub fusions in a pol30Δ ubc13Δ rev3Δ triple mutant. As seen in Figure 5D, each of the fusions still provided resistance to MMS. These results indicate that neither REV3 nor UBC13 is required for the MMS resistance attributed to pol30-UB and UB-pol30, and raises the possibility that the PCNA·Ub fusions do not promote DDT through Polζ.
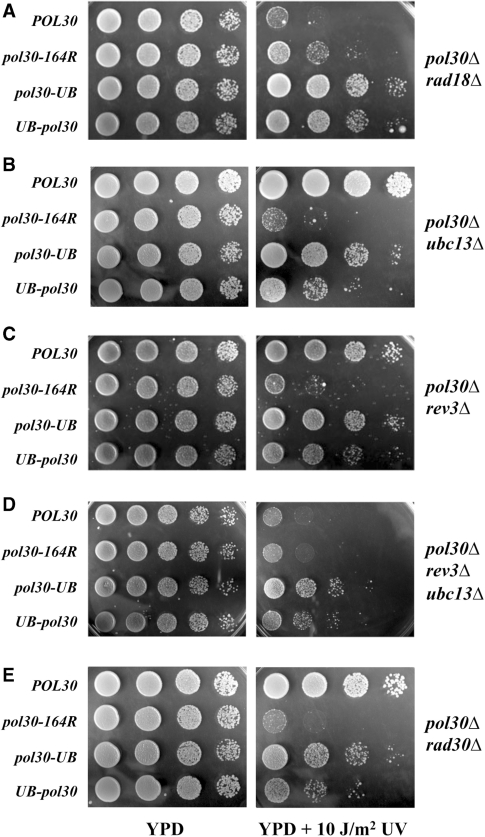
To ask whether the above phenotypes are unique to MMS-induced DNA damage, we examined the phenotypes of various pol30Δ ddtΔ mutants to UV-induced killing and found once again that DNA damage resistance conferred by PCNA·Ub fusions is independent of RAD18 (Figure 6A), UBC13 (Figure 6B), REV3 (Figure 6C), and REV3 UBC13 (Figure 6D).
Figure 6.
Relative UV sensitivity by a serial dilution plate assay to determine the function of the pol30·UB gene fusions in the context of DDT pathway mutants. (A) pol30Δ rad18Δ; (B) pol30Δ ubc13Δ; (C) pol30Δ rev3Δ; (D) pol30Δ ubc13Δ rev3Δ; and (E) pol30Δ rad30Δ. Experimental conditions were as described in Figure 2.
Since RAD30-encoded Polη is a TLS polymerase that efficiently bypasses UV-induced lesions (8,41), we created a pol30Δ rad30Δ strain to critically test TLS dependency of PCNA·Ub resistance to UV treatment. As seen in Figure 6E, the UV resistance imparted by PCNA·Ub does not require RAD30, since pol30Δ rad30Δ cells carrying PCNA-Ub fusions are clearly more resistant to UV treatment compared with the non-fused PCNA-K164R control. Resistance to MMS-induced DNA damage by PCNA·Ub was also seen in the pol30Δ rad30Δ strain (Supplementary Figure S2B).
To thoroughly rule out the possibility that other known TLS components are responsible for PCNA·Ub-mediated DNA damage resistance, we examined other known genes involved in TLS. In a rad18Δ strain harboring PCNA·Ub fusion constructs, deletion of REV1 encoding the third TLS polymerase (42) still retains PCNA·Ub-mediated UV resistance (Supplementary Figure S2C). Similarly, deletion of POL32, encoding a non-essential subunit of Polδ that plays a role in TLS (37), does not abolish the UV resistance conferred by PCNA·Ub fusions (Supplementary Figure S2D). Taken together, we conclude that constitutively monoubiquitylated PCNA appears to tolerate different replication-blocking lesions by a mechanism independent of the RAD6-RAD18-mediated DDT pathway.
PCNA·Ub fusions do not cause an increase in spontaneous mutations
A hallmark of the TLS DDT pathway in yeast is an increased level of mutagenesis that is primarily attributed to Polζ activity (43). For example, mutations in MMS2 result in a dramatically increased rate of spontaneous mutagenesis due to the sequestering of DNA lesions to the TLS branch that is absolutely dependent on REV3 (4,5). Since the mono-Ub modification of PCNA is proposed to promote the TLS pathway, we reasoned that if our PCNA·Ub fusions were to promote TLS in a similar manner, we would observe increased rates of spontaneous mutagenesis. However, we found no significant elevation in spontaneous mutagenesis rates for either of the PCNA·Ub fusions, whether in the presence (data not shown) or absence of endogenous wild-type POL30 (Table 1). In contrast, a small decrease in spontaneous mutagenesis was observed when PCNA was constitutively monoubiquitylated. The significance of this decrease is currently unclear.
Table 1.
Spontaneous mutagenesis in the DBY747 pol30Δ strain
| POL30-UB gene fusion carried | Mutation rate ± SD (×10−7); (fold over wild type) |
|---|---|
| POL30 | 0.64 ± 0.20 (1) |
| pol30-164R | 0.43 ± 0.41 (0.7) |
| UB-pol30 | 0.28 ± 0.23 (0.4) |
Results are the average of four independent experiments with standard deviations.
DISCUSSION
TLS polymerases have long garnered attention for their involvement in critical cellular roles, the most prominent being the prevention of cell death through replicating past DNA lesions and the subsequent effect on mutagenesis (2,43). The significance of TLS activity is underscored in the human disease xeroderma pigmentosum variant (XPV) caused by defective Polη activity (44,45). The direct physical association between Y-family TLS polymerases and the essential DNA replication clamp protein PCNA through a PIP motif apparently provides the foundation for linking high-fidelity processive DNA replication with TLS (46,47). The discovery that PCNA is monoubiquitylated by Rad6-Rad18 (1) and subsequent genetic and biochemical studies have led to a model whereby TLS activity is regulated through enhanced physical associations mediated by several newly identified Ub binding domains found in Y-family TLS polymerases and the Ub moiety of monoubiquitylated PCNA (17,24,48). In this study, we addressed this model by creating linear fusions of the Ub and PCNA genes to mimic monoubiquitylated PCNA in vivo.
We show that PCNA·Ub partially restores DNA damage resistance in rad18Δ cells that are incapable of ubiquitylating PCNA but not in cells defective in other DNA repair pathways. The partial rescuing effect was expected because PCNA conjugated to mono-Ub does not promote the error-free branch of DDT (1). We also reasoned that the function of PCNA·Ub could be competed for by endogenous wild-type PCNA. To overcome this problem, we successfully generated viable pol30 null mutant strains and showed that the essential replication function of PCNA can be maintained by PCNA·Ub, a constitutively monoubiquitylated form of PCNA. Not only were the fusion proteins able to replace PCNA, but no apparent growth differences were observed when comparing the PCNA·Ub strains with isogenic strains carrying wild-type POL30 or pol30-164R. During these studies, another group reported that similar linear fusions of Ub and PCNA could not rescue PCNA null mutant lethality; however, their observed lethal phenotype likely resulted as a consequence of the several additional point mutations introduced in their constructs (26).
Our strains lacking wild-type POL30 represent a significant improvement for testing PCNA·Ub, which contains the K164R mutation to prevent natural Ub modification. Indeed, we observed much stronger PCNA·Ub phenotypes in the pol30Δ rad18Δ strain as compared to the rad18Δ strain, indicating that unmodified PCNA does in fact impede the DNA damage resistance function of PCNA·Ub. Additional inferences are also made possible when using these strains that lack endogenous PCNA. For example, our experimental system reflects a scenario whereby every subunit of the PCNA homotrimer is linked by mono-Ub. This phenomenon has been suggested as the true modification state of PCNA after DNA damage (17), and may explain the stronger DNA damage resistance phenotype as compared with the strains containing a mixture of Ub-modified and unmodified PCNA. In addition, our strains yield a constitutive PCNA-Ub fusion in the absence of DNA damage that is apparently not detrimental to cell growth. This finding is remarkable because it implies that the numerous roles and physical interactions of PCNA (13) are not affected by Ub modification. It also suggests that the removal of Ub from PCNA may not be immediately required following DNA damage.
With numerous Ub-fusion studies emerging in the literature, our thorough analysis of PCNA·Ub fusions raises important considerations for such studies. We note the possible susceptibility of N-terminal Ub fusions to Ub proteases because they form natural polypeptide bonds with the Ub C-terminus that are likely recognized by Ub C-terminal hydrolases, and possibly other Ub-specific proteases. Despite our mutation of the Ub C-terminal Gly–Gly motif, we still detected small fraction of cleavage of the Ub-PCNA fusion. Selection pressure for unmodified targets might specifically enhance the removal of N-terminal Ub fusions from particular proteins. In contrast, fusion of Ub to the C-terminus of target proteins is likely stable because it results in an inverted attachment through the Ub N-terminus that would not be recognized by Ub hydrolases. This highly unnatural Ub modification can nonetheless impart some portion of the natural Ub modification function(s) as evidenced by this study.
The fact that each of the N- and C-terminal PCNA·Ub constructs is functional suggests some structural tolerance for the signaling imparted by monoubiquitylated PCNA. However, when compared to the N-terminal fusion, we did note that the C-terminal PCNA-Ub fusion caused stronger DNA damage resistance in every strain tested lacking genomic POL30. We infer that the C-terminal fusion is preferred because it leads to the attachment of Ub in closer proximity to PCNA-Lys164, and suggests that the location of Ub attachment is more important than the unconventional inverted Ub orientation (Figure 1C). Interestingly, the N-terminal fusion conveyed a slightly stronger DNA damage resistance phenotype than the C-terminal fusion in strains carrying wild-type POL30. We suspect that the opposite effects are due to endogenous Pol30 that forms mixed PCNA trimers with PCNA·Ub fusions.
When testing strains lacking endogenous PCNA in the context of the DDT pathway, we found that the DDT attributed to PCNA·Ub was not dependent on RAD18 or MMS2/UBC13/RAD5. Since Rad6-Rad18 and Mms2-Ubc13-Rad5 are required for the mono- and poly-Ub modifications of PCNA, respectively, this result eliminated the possibility that PCNA·Ub function was dependent on subsequent ubiquitylation. In accordance, we failed to detect additional Ub modifications in western blots of PCNA·Ub protein (data not shown). These findings also indicate that events downstream of PCNA·Ub do not require any other characterized activities or protein interactions mediated by Rad18 or Mms2-Ubc13-Rad5.
Commensurate with the TLS signaling model, we expected that the DDT contributed by PCNA·Ub would depend on the TLS activity by Polζ, Polη or Rev1. Surprisingly, none of these TLS polymerases was required in our experimental setting. Furthermore, PCNA·Ub did not lead to the increased rate of spontaneous mutagenesis that is a hallmark of Polζ function. The above observations and conclusion differ from a recent report (26), in which simultaneous inactivation of REV1, REV3 and RAD30 abolished DNA damage resistance to UV conferred by PCNA-Ub fusions. This discrepancy is likely due to the presence of endogenous unmodified PCNA in the other study and/or the additional mutations introduced into their PCNA-Ub fusion constructs, such as PCNA-K127R, which may interfere with sister chromatid cohesion (49). Strain differences are also possible, since we have not observed the reported increase in sensitivity to DNA damaging agents between rad18 single and rad18 tlsΔ mutants (data not shown). We do note, however, that a recent report (50) has challenged the significance of the yeast Polη UBZ motif and its related in vitro and in vivo TLS functions.
In summary, we were able to demonstrate that both N- and C-terminal fusions of Ub to PCNA provide yeast cells with enhanced DDT that is specific for the RAD6-RAD18 pathway, but could not prove that it was due to enhanced TLS activities. Our findings leave at least two possible explanations: (i) the constitutive fusions employed in the study created an artifact that functions in the context of the RAD6/RAD18 epistasis group and does not accurately represent endogenous PCNA monoubiquitylation, or (ii) monoubiquitylated PCNA promotes TLS through a mechanism more complex than the current prevailing model (24). Evidence in support of the latter possibility is emerging. For example, studies have shown that unmodified PCNA is sufficient to stimulate DNA synthesis by Polκ, primarily by reducing the Km for nucleotide incorporation (51). Also, in a reconstituted DNA synthesis reaction, PCNA that is monoubiquitylated on all three subunits did not enhance TLS activity by Y-family polymerases (27). Thus, the results presented in the present study add countervailing evidence that should further raise awareness and call for alternative models for DDT signaling.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research operating grant MOP-93612 to WX. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Dr P. Burgers for the POL30 plasmids, Barry Ziola and Lindsay Ball for technical assistance.
REFERENCES
- 1.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 2.Broomfield S, Hryciw T, Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 3.Xiao W, Chow BL, Broomfield S, Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao W, Chow BL, Fontanie T, Ma L, Bacchetti S, Hryciw T, Broomfield S. Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. 1999;435:1–11. doi: 10.1016/s0921-8777(99)00034-8. [DOI] [PubMed] [Google Scholar]
- 5.Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl Acad. Sci. USA. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 7.Brusky J, Zhu Y, Xiao W. UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 2000;37:168–174. doi: 10.1007/s002940050515. [DOI] [PubMed] [Google Scholar]
- 8.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 10.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 12.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 13.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Torres-Ramos CA, Yoder BL, Burgers PM, Prakash S, Prakash L. Requirement of proliferating cell nuclear antigen in RAD6-dependent postreplicational DNA repair. Proc. Natl Acad. Sci. USA. 1996;93:9676–9681. doi: 10.1073/pnas.93.18.9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastushok L, Xiao W. DNA postreplication repair modulated by ubiquitination and sumoylation. Adv. Protein Chem. 2004;69:279–306. doi: 10.1016/S0065-3233(04)69010-3. [DOI] [PubMed] [Google Scholar]
- 16.Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 21.Ball LG, Zhang K, Cobb JA, Boone C, Xiao W. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol. Microbiol. 2009;73:89–102. doi: 10.1111/j.1365-2958.2009.06748.x. [DOI] [PubMed] [Google Scholar]
- 22.Kannouche P, Lehmann A. Localization of Y-family polymerases and the DNA polymerase switch in mammalian cells. Methods Enzymol. 2006;408:407–415. doi: 10.1016/S0076-6879(06)08025-6. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 25.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 30.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 31.Pastushok L, Moraes TF, Ellison MJ, Xiao W. A single Mms2 “key” residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. J. Biol. Chem. 2005;280:17891–17900. doi: 10.1074/jbc.M410469200. [DOI] [PubMed] [Google Scholar]
- 32.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 34.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 35.Roche H, Gietz RD, Kunz BA. Specificity of the yeast rev3 delta antimutator and REV3 dependency of the mutator resulting from a defect (rad1 delta) in nucleotide excision repair. Genetics. 1994;137:637–646. doi: 10.1093/genetics/137.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao W, Chow BL, Rathgeber L. The repair of DNA methylation damage in Saccharomyces cerevisiae. Curr. Genet. 1996;30:461–468. doi: 10.1007/s002940050157. [DOI] [PubMed] [Google Scholar]
- 37.Hanna M, Ball LG, Tong AH, Boone C, Xiao W. Pol32 is required for Pol zeta-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat. Res. 2007;625:164–176. doi: 10.1016/j.mrfmmm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Friedberg EC. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 1988;52:70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Zhu Y, Zhang K, Yeung M, Durocher D, Xiao W. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133:601–611. doi: 10.1016/j.cell.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 40.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 1999;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 42.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 43.Lawrence CW. Mutagenesis in Saccharomyces cerevisiae. Adv. Genet. 1982;21:173–254. doi: 10.1016/s0065-2660(08)60299-0. [DOI] [PubMed] [Google Scholar]
- 44.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 45.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 46.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol. Cell. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 48.Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- 49.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Acharya N, Brahma A, Haracska L, Prakash L, Prakash S. Mutations in the ubiquitin binding UBZ motif of DNA polymerase eta do not impair its function in translesion synthesis during replication. Mol. Cell. Biol. 2007;27:7266–7272. doi: 10.1128/MCB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol. Cell. Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.