Abstract
Cell adaptation to the environment often involves induction of complex gene expression programs under the control of specific transcriptional activators. For instance, in response to cadmium, budding yeast induces transcription of the sulfur amino acid biosynthetic genes through the basic-leucine zipper activator Met4, and also launches a program of substitution of abundant glycolytic enzymes by isozymes with a lower content in sulfur. We demonstrate here that transcriptional induction of PDC6, which encodes a pyruvate decarboxylase isoform with low sulfur content, is directly controlled by Met4 and its DNA-binding cofactors the basic-helix–loop–helix protein Cbf1 and the two homologous zinc finger proteins Met31 and Met32. Study of Cbf1 and Met31/32 association with PDC6 allowed us to find a new mechanism of recruitment of Met4, which allows PDC6 being differentially regulated compared to sulfur amino acid biosynthetic genes. Our findings provide a new example of mechanism allowing transcriptional plasticity within a regulatory network thanks to a definite toolbox comprising a unique master activator and several dedicated DNA-binding cofactors. We also show evidence suggesting that integration of PDC6 to the Met4 regulon may have occurred recently in the evolution of the Saccharomyces cerevisiae lineage.
INTRODUCTION
The transcription activator Met4 in Saccharomyces cerevisiae is the master regulator of the MET gene network that coordinates synthesis of sulfur containing molecules, including the amino acids methionine and cysteine, the primary methyl donor S-adenosylmethionine, and the widespread thiol-containing antioxidant glutathione (GSH) (1–3). Met4 activity is tightly regulated according to the sulfur status of the cell. Under conditions of excess organosulfur compounds, Met4 is subjected to ubiquitylation by the SCFMet30 ubiquitin ligase (4,5), and ubiquitylation can, depending on the medium, either target Met4 for immediate degradation by the 26S proteasome, or prevent MET promoter binding (6,7). In contrast, under sulfur limiting conditions, Met4 is not modified by ubiquitin and it can activate transcription through recruitment of the SAGA histone acetyltransferase and the Mediator coactivator complex (6,8).
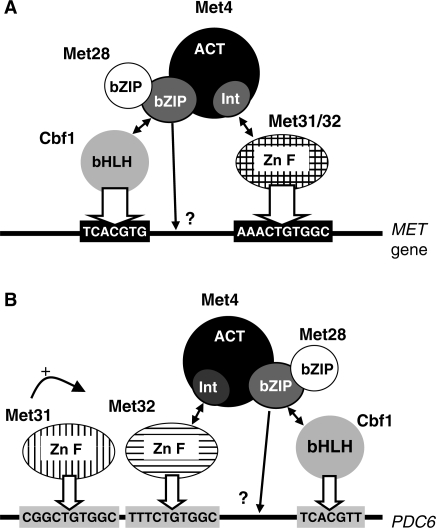
Met4 contains at its C-terminus a dimerization/DNA binding domain of the basic-leucine zipper (bZIP) family (2). Previous studies have shown that Met4 association with its target promoters depends not only on the bZIP domain, but also involves four DNA-binding cofactors, namely the bZIP protein Met28, the basic helix–loop–helix (bHLH) protein Cbf1 and the two related zinc finger proteins Met31 and Met32 (9,10). Contrary to Met4, these cofactors possess no intrinsic capacity to activate transcription and appear mainly dedicated to the recruitment of Met4 to its target genes (2,10,11). The reason for this complexity is not well understood, but is believed to reflect a requirement for the cell to fine-tune synthesis of the various sulfur containing molecules (9). Interestingly, Cbf1 is a multifunctional factor also involved in the maintenance of centromere. Its loss causes defects in both sulfur amino acid biosynthesis and chromosome segregation (12–14). Accordingly, Cbf1 DNA-binding site, the sequence TCACRTG (R = A/G), is present in MET promoters and in centromeres where it constitutes the highly conserved centromere DNA element 1 (CDEI) (1,15). The DNA-binding site for Met31 and Met32 is the sequence AAACTGTGG which is conserved in MET promoters (1,11). In vitro experiments have shown that Met4 can associate with DNA only in complex with its cofactors, and it is believed that Cbf1 and Met31/32 association with their respective DNA elements in MET promoters forms anchoring platforms for Met4 (9,16). As expected, Met4 has been shown to possess protein interaction domains for both Cbf1 and Met31/32 (9,10). Met28 has no sequence specific DNA-binding capability either; however, it has been shown, first, to enhance the DNA-binding activity of Cbf1 by a mechanism which is still unknown (16), and second, to heterodimerize with Met4 (10).
The gene network controlled by Met4 is induced upon exposure to cadmium (17,18), a non-essential toxic heavy metal detoxified in S. cerevisiae through conjugation with GSH (19). Cadmium interferes with Met4 regulation at two levels to enable rapid and maximal induction of the MET gene network needed for GSH production: first, cadmium induces dissociation of Met30 from SCFMet30, thereby preventing Met4 ubiquitylation; and second, cadmium activates an unknown deubiquitylating enzyme which removes inhibitory ubiquitin moieties from Met4, thereby restoring its activity (20,21). Saccharomyces cerevisiae response to cadmium is not limited to transcriptional reprogramming of the sulfur metabolism, but also involves a so-called ‘sulfur sparing’ program in which abundant enzymes involved in the carbohydrate metabolism are replaced by isozymes with a lower content in sulfur-containing amino acids, supposedly to maximize the cellular pool of cysteine available for glutathione synthesis (17). A striking illustration of this isozyme switching is provided by pyruvate decarboxylase: upon exposure to cadmium, the gene encoding the main isoform Pdc1 which contains 16 sulfur atoms is repressed, whereas the gene encoding the minor isoform Pdc6 which contains five sulfur atoms is strongly induced (17). Most interestingly, it was also observed that PDC6 transcription is no longer induced by cadmium in cells containing a null allele of MET4, which was unexpected considering that PDC6 promoter does not contain the DNA-binding sites for Cbf1 and Met31/32 found in MET genes.
The aim of this study was to examine PDC6 regulation in more details. We report here that (i) PDC6 is also activated in other conditions in which Met4 is active, such as in sulfur limitation or in the absence of its negative regulator Met30; (ii) PDC6 activation involve recruitment of Met4 and its DNA-binding partners Cbf1, Met28, Met31 and Met32; and (iii) recruitment of theses factors occurs through non-canonical DNA binding sites and involves an original mechanism. Moreover, we found that PDC6 is differentially regulated compared to MET genes, most likely because of the non-canonical DNA binding sites. Altogether, these findings shed new lights on the DNA-binding properties of zinc finger and bHLH factors, as well on the mechanism of assembly of the Met4/Cbf1/Met28/Met31/32 activation complex. They also provide a striking example of mechanism allowing transcriptional plasticity within a regulatory network thanks to a definite toolbox comprising a unique master activator and several dedicated DNA-binding cofactors. Finally, based on sequence comparison and transcriptional analysis in S. cerevisiae relatives, we suggest that addition of PDC6 to the Met4 regulon is a recent event in the evolution of the S. cerevisiae lineage.
MATERIALS AND METHODS
Yeast strains, plasmids and media
Saccharomyces cerevisiae strains used in this study (Table 1) were all derived from W303. Y444, Y645, Y646 and Y647 were generated using PCR-based, one-step integration strategies (22). The PDC6[Δ-518/-512]-KanMX6 and PDC6[Δ-458/-453]-KanMX6 fragments used to generate Y646 and Y647 were obtained in two steps. First, two fragments containing PDC6 open reading frame (ORF) preceded by the upstream region lacking sequences −518/−512 or sequences −458/−453 were generated by PCR. Then, the two fragments were independently mixed with a PCR fragment containing the kanamycine gene flanked by two short sequences corresponding to PDC6 3′-end, and the mixture was subjected to PCR to generate hybrid fragments. Y648, Y650, Y657, Y675 and Y677 were obtained by integrating at the ura3 locus of Y444 integrative URA3 plasmids containing wild-type (WT) or mutated PDC6 fragments spanning positions −550 to +1692 followed by ADH1 terminator. Mutations in PDC6 promoter were generated by in vitro site-directed mutagenesis. Y663 and Y664 were generated by disrupting MET32 in Y648 and Y650, respectively. Saccharomyces paradoxus (CLIB228), S. bayanus (CLIB181) and Candida glabrata (CLIB298) strains were obtained from the CIRM-Levures (AgroParisTech-Grignon).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| CC718-1A | MATa cbf1::TRP1 cbf1::TRP1 | (10) |
| CC767-6A | MATa met28::TRP1 | (10) |
| CC849-8A | MATalpha met4::TRP1 | (4) |
| CC932-8B | MATa met4::GAL1-MET4 met30::LEU2 | (25) |
| CD158 | MATa met31::HIS3 | (9) |
| CD159 | MATalpha met32::HIS3 | (9) |
| CD163 | MATa met31::TRP1 met32::HIS3 | (9) |
| CY202-4B | MATalpha met4::HA3MET4Δ[590-612]-CBF1[210-351] cbf1::URA3 | (7,30) |
| W303-1A | MATa ade2 ura3 his3 leu2 trp1 | R. Rothstein |
| Y444 | MATa pdc6Δ::His3MX6 | This study |
| Y461 | MATa MET4::18myc-TRP1 | (45) |
| Y464 | MATa MET32::9myc-TRP1 | (45) |
| Y465 | MATa CBF1::9myc-TRP1 | (45) |
| Y645 | MATa PDC6::PDC6-KanMX6 | This study |
| Y646 | MATa PDC6::PDC6[Δ-518/-512]-KanMX6 | This study |
| Y647 | MATa PDC6::PDC6[Δ-458/-453]-KanMX6 | This study |
| Y648 | MATa pdc6::His3MX6 ura3::PDC6-tADH1 | This study |
| Y650 | MATa pdc6::His3MX6 ura3::PDC6[AAA]-tADH1 | This study |
| Y657 | MATa pdc6::His3MX6 ura3::PDC6[AAA,CACGTG]-tADH1 | This study |
| Y663 | MATa met32::KanMX6 pdc6::His3MX6 ura3::PDC6-tADH1 | This study |
| Y664 | MATa met32::KanMX6 pdc6::His3MX6 ura3::PDC6[AAA]-tADH1 | This study |
| Y675 | MATa ura3::PDC6[ΔCACGTT]-tADH1-URA3 pdc6::HisMX6 | This study |
| Y677 | MATa ura3::PDC6[CACGTG]-tADH1 pdc6::His3MX6 | This study |
| CY302-3C | Mata his3 leu2 ade2 ura3 trp1Δ2 | Y. Surdin-Kerjan |
Plasmid p6hisCbf1ΔN, p6hisMet31 and p6hisMet32, containing the C-terminal part of Cbf1 (codons 210–351), and full-length Met31 or Met32 inserted in pET28a were described in our previous studies (11,16).
YPD medium contains 1% yeast extract, 2% bacto-peptone and 2% glucose. YNB medium contains 0.7% yeast nitrogen base, 0.5% ammonium sulfate and 2% glucose. B medium is a synthetic medium lacking organic and inorganic sulfur sources (23).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously (8). The antibodies used in this study include mouse monoclonal antibodies to the HA and Myc epitopes (F7 and 9E10, Santa Cruz Biotechnology), and rabbit polyclonal antibodies to Met28 and Met32 (kindly provided by Mike Tyers, Samuel Lunenfeld Research Institute). DNA was quantified by real time PCR using the LightCycler 480 instrument, and reaction kits containing SYBR Green I (Roche). A typical run included duplicates of each IP and input DNA, and serial dilutions of one input DNA to create a standard curve and determine the efficiency of the amplification. Data was analyzed with the LightCycler 480 software using the ‘second derivative maximum’ method for quantification. Occupancy at a genomic location was calculated by dividing the immunoprecipitated (IP) over input DNA ratio for a fragment encompassing the location by the IP over input DNA ratio of a fragment of reference (typically, a sequence within the open reading frame of the meiotic gene IME2).
RNA extraction and analyses
Total RNA was isolated by extraction with hot acidic phenol (24). Reverse transcription quantitative polymerase chain reaction (RT–PCR) was conducted using a two-step procedure. First, cDNA were synthesized with SuperScript II (Invitrogen) or RevertAid H Minus M-MuLV (Fermentas) reverse transcriptases according to manufacturers’ protocol. A mixture of random hexamers (Roche), anchored oligo(dT)23 (Sigma) and 25S rDNA oligonucleotide was used for priming. cDNA levels were then quantified by real time PCR using the LightCycler 480 system, and reaction kits containing SYBR Green I (Roche). Data was analyzed with the LightCycler 480 software using the ‘second derivative maximum’ method for quantification. Amplification efficiency was determined for each experiment based on a standard curve established using serial dilutions of one of the cDNA samples.
Gel shift assays
His-tagged recombinant proteins were expressed in BL21(DE3). In the case of 6hisCbf1ΔN, cells were induced at OD650 = 0.6 in the presence of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C, and his-tagged proteins were purified using Novagen BugBuster protein extraction reagent and Novagen His•bind purification kit. The protein preparation was dialyzed against 20 mM Tris–HCl (pH 8.0), 120 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol and 20% glycerol. In the case of 6hisMet31 and 6hisMet32, cells were induced at OD650 = 0.6 in the presence of 1 mM IPTG for 2 h, collected by centrifugation, resuspended in three volumes of buffer containing 20 mM Tris–HCl (pH 8.0), 0.5 mM NaCl, 10% glycerol and 1 mM of the protease inhibitor Pefabloc SC (Roche), and broken in an Eaton press. Extracts were subsequently sonicated to fragment DNA and reduce viscosity, and debris was eliminated by centrifugation.
Gel shift assays were performed in a final volume of 20 µl containing 25 mM HEPES pH 7.6, 60 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 5 mM MgCl2, 0.5 mg/ml acetylated-bovine serum albumin, 7.5% glycerol and 0.8 µg poly(dIdC)–poly(dIdC). DNA probes were 5′-labeled by using T4 DNA kinase and [γ-32P]ATP (3000 Ci/mmol). Samples were incubated for 40 min in ice and subjected to electrophoresis in a 5% polyacrylamide gel in 0.25× TBE for 3–4 h at 9 V/cm and at 7°C. The gel was dried, exposed to a storage phosphor screen for several hours, and imaged using Storm 820 (Molecular dynamics). Signals were quantified using ImageQuant software.
RESULTS
Transcriptional activation of PDC6 requires Met4 and its cofactors
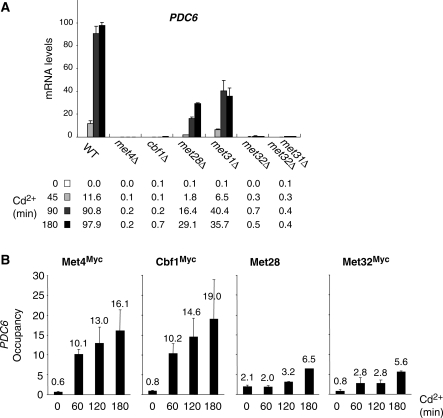
To determine whether Met4 cofactors Cbf1, Met28, Met31 and Met32 are involved in PDC6 transcriptional activation, we carried out reverse transcription quantitative PCR (RT–QPCR) analysis on mutant strains exposed to Cd2+ (Figure 1A). The results showed that PDC6 activation in response to Cd2+ was completely abolished in strains containing null alleles of Met4, Cbf1 or Met32, and partially diminished in strains containing null alleles of Met28 or Met31. Interestingly, inactivation of Met32 alone is sufficient to abolish PDC6 activation, whereas both Met31 and Met32 must be inactivated to abolish MET gene activation (data not shown) (11). Thus, similar to MET genes, PDC6 activation involves Cbf1, Met28, Met31 and Met32 alongside with Met4; however, the relative importance of these factors seems to differ between PDC6 and MET genes.
Figure 1.
PDC6 activation in response to Cd2+ is controlled by Met4 and its cofactors. (A) mRNA levels for PDC6 in mutant strains. A WT, and the indicated mutants were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. Total RNA was prepared from samples taken before and at different time points after addition of Cd2+. RNA was quantified by RT followed by real-time PCR. Values were normalized to 25S ribosomal RNA and represent the average of two independent experiments. To facilitate comparison, the maximum value was set up to 100 and other values were expressed in proportion. Error bars indicate average deviations. (B) Association of Met4 and its cofactors with PDC6 promoter. Isogenic strains expressing Myc-tagged Met4, Cbf1 or Met32 were grown and exposed to Cd2+ as in (A). Aliquots were crosslinked with formaldehyde at the time points indicated following Cd2+ addition and subjected to ChIP using antibodies to Myc or to Met28. DNA fragments were quantified by real time PCR using primer pairs specific for PDC6 upstream region (from −455 to −306 relative to ATG), and primer pairs specific for IME2 ORF as a negative control. Occupancy levels correspond to the fraction of PDC6 promoter immunoprecipitates relative to the fraction IME2 ORF immunoprecipitates. Values represent the average of two independent experiments, and error bars indicate average deviations.
ChIP was performed to determine the presence of Met4 and its cofactors at PDC6 promoter (Figure 1B). All factors were immunoprecipitated except Met31 because no antibody was available and our attempts to construct epitope-tagged versions of Met31 led to derivatives unable to support growth using sulfate as unique sulfur source in a met32Δ background, indicating that protein activity was affected (data not shown). The ChIP results for Met4, Cbf1, Met28 and Met32 showed no significant association with PDC6 promoter in YPD medium and a several fold increase in association upon Cd2+ exposure. This increase was particularly strong in the case of Met4 and Cbf1 (more than 20-fold compared to the control region), and more modest in the case of Met32 and Met28 (7- and 3-fold, respectively).These results demonstrate that Met4 and its cofactors are directly involved in PDC6 activation in response to Cd2+.
PDC6 transcription is induced in all conditions that trigger Met4 activity
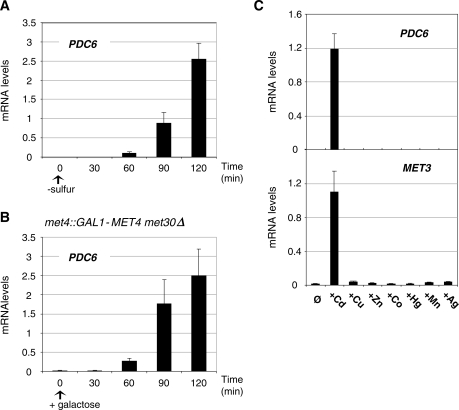
To further establish the role of Met4 in PDC6 regulation, we analyzed PDC6 transcription in sulfur limitation (Figure 2). The results showed that PDC6 transcription was strongly activated when cells were transferred to minimal medium depleted in sulfur (Figure 2A). We also monitored PDC6 transcription in a strain expressing Met4 from the inducible GAL1 promoter and lacking Met30, the negative regulator of Met4. As already reported, expression of Met4 in a met30Δ background allows induction of MET genes in rich medium (25). We found that Met4 expression in the absence of Met30 was also sufficient to induce PDC6 transcription in rich media (Figure 2B). Altogether, these results demonstrate that PDC6 activation is not limited to cadmium exposure but occurs in other conditions that induce MET genes. By contrast, no activation was observed in the presence of metals that do not induce MET genes, such as copper, cobalt, manganese, mercury, silver or zinc (Figure 2C).
Figure 2.
Analysis of PDC6 transcription under various conditions. (A) PDC6 transcription in response to sulfur limitation. WT cells were grown to early log phase into B-medium supplemented with 0.5 mM methionine, collected by filtration, and transferred into B-medium with no sulfur source at time zero. Samples were taken at the time points indicated and total RNA was extracted and quantified by RT-real time PCR. Values were normalized to 25S ribosomal RNA and represent the average of two independent experiments. Error bars indicate average deviations. (B) PDC6 transcription in response to Met4 hyperactivation. met4::GAL1-MET4 Δmet30 (CC932-8B) cells were grown in YP-raffinose medium to early log phase and supplemented with galactose at the zero time point. (C) PDC6 and MET3 transcription upon exposure to various heavy metals. Wild-type cells were grown to early-log phase in YPD medium and exposed to 0.5 mM CdCl2, 1 mM CuCl2, 1 mM ZnCl2, 2 mM CoCl2, 50 µM HgCl2, 0.5 mM MnCl2 or 30 µM AgNO3 for 90 min.
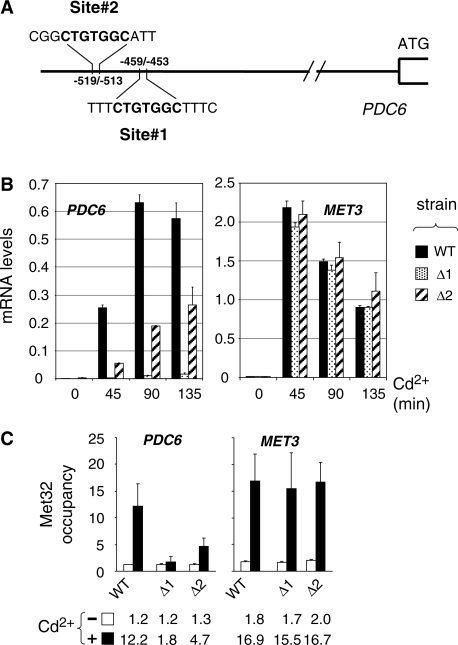
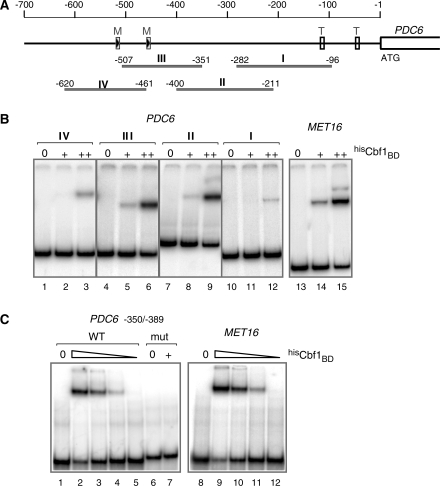
PDC6 activation involves Met31 and Met32 recruitment to truncated, suboptimal DNA-binding sites
PDC6 upstream region does not contain the consensus binding sites for Cbf1 or Met31/32 usually found upstream the sulfur metabolic genes controlled by Met4 (TCACGTG and AAACTGTGGC, respectively; see Supplemental Figure S2). However, PDC6 contains two repetitions of the sequence CTGTGGC corresponding to a truncated version of the Met31/32 binding site (site #1 between positions −453 and −459, and site #2 between positions −513 and −519; Figure 3A). To determine the requirement of these two sites for PDC6 activation, mRNA analysis was performed on yeast strains lacking one or the other (Figure 3B). We found that PDC6 transcription was almost completely abolished by deletion of site #1 and diminished by 2–4-fold by deletion of site #2 (Figure 3B). ChIP analysis was carried out in parallel (Figure 3C). In good agreement with the transcriptional analysis, deletion of site #1 decreased Met32 association with PDC6 promoter to background level whereas deletion of site #2 caused a 60% decrease (Figure 3C). We concluded that the proximal CTGTGGC motif (site #1) served as binding site for Met32, whereas the distal CTGTGGC motif (site #2) was not able to bind Met32 by its own but had a stimulatory effect on Met32 association with the first site.
Figure 3.
Characterization of PDC6 upstream activating sequences (UAS). (A) Schematic of PDC6 promoter. (B) Effect of deletion of the CTGTGGC sequences on PDC6 transcription. Two mutants containing at the chromosome a PDC6-Kan derivative lacking either the first (Δ1) or the second (Δ2) CTGTGGC sequence, and the isogenic WT strain, were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. Total RNA was extracted from samples taken at the time points indicated and RNA levels were quantified by RT-real time PCR. Values were normalized to 25S ribosomal RNA levels and represent the average of two independent experiments. Error bars indicate average deviations. (C) ChIP assay on Met32. The same strains as in (B) were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. ChIPs were performed on aliquots crosslinked with formaldehyde before (−) or 180 min after Cd2+ addition (+) using antibodies to Met32. DNA fragments were quantified by real time PCR using primer pairs for the promoter region of PDC6 (−620/−461 relative to ATG) and MET3. Occupancy levels were calculated as in Figure 1B. Values represent the average of two independent experiments, and error bars indicate average deviations.
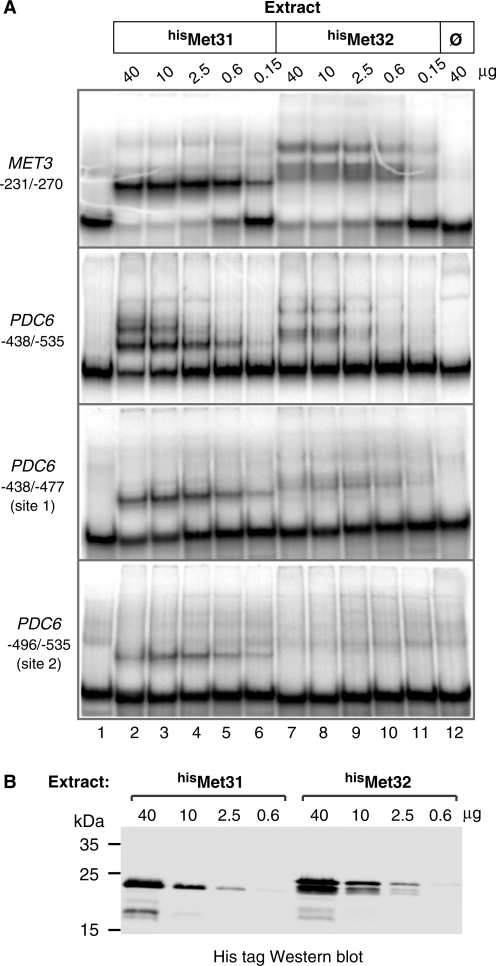
To gain information on Met31 binding to PDC6 and better characterize Met32 binding, gel shift assays were carried out using Escherichia coli cell extracts containing or not polyhistidine-tagged Met31 and Met32 proteins (hisMet31 and hisMet32; Figure 4). The result clearly showed hisMet31 and hisMet32 binding to the PDC6 fragment encompassing the two TGTGGC sites (Figure 4, second panel). However binding to PDC6 was much less efficient compared to MET3, even though MET3 contains only one CTGTGGC motif (Figure 4, first panel). Gel shift assays with shorter PDC6 fragments containing either of the two CTGTGGC sites revealed that Met32 was not able to bind to site #2 (Figure 4, fourth panel), which was consistent with the ChIP results in Figure 3C. Moreover, the fraction of fragment with site #1 shifted in the presence of hisMet31 was significantly higher than the fraction shifted in the presence of hisMet32 (Figure 4, third panel). Since gels shift were carried out with similar amounts of hisMet31 and hisMet32 (see western blot in Figure 4B), we concluded that Met31 and Met32 had differential affinities for PDC6.
Figure 4.
In vitro associations of Met31 and Met32 with PDC6 promoter. (A) Gel shift assays. The indicated amounts of E. coli cell extracts expressing polyhistidine-tagged Met31 and Met32 (lanes 2–6 and 7–11, respectively) or not (lane 12) were incubated with equimolar amounts of the indicated 32P-labeled DNA fragments. The MET3 fragment contains AAACTGTGGC. The PDC6 fragments contain both CTGTGG sites (second panel) and either of them (third and fourth panel). Fragments were resolved by 5% polyacrylamide gel electrophoresis and visualized by PhosphorImager analysis. (B) Western blot. Extracts used in (A) containing hisMet31 or hisMet32 were separated by 12% SDS–polyacrylamide gel electrophoresis and analyzed by immunoblotting with a monoclonal antibody to the his-tag (Novagen). The calculated molecular weights of hisMet31 and hisMet32 are 23 226.5 and 25 441, respectively.
Cbf1 association with PDC6 promoter occurs through a non-canonical binding site
The absence of TCACGTG sequence upstream of PDC6 raised the question whether Cbf1 could bind PDC6 by itself or was recruited through interactions with Met4 and/or other factors involved in PDC6 activation. To address this point, we first performed gel shift assays using a recombinant derivative of Cbf1 containing the bHLH domain (residues 210–351) fused to a polyhistidine tag (hisCbf1BD; Figure 5). This derivative was incubated with four overlapping DNA fragments covering PDC6 from positions −96 to −620 (Figure 5A), and with a fragment of MET16 containing the TCACGTG sequence as a control. The results in Figure 5B showed that hisCbf1BD did associate with PDC6 by its own. The fact that fragments II and III of PDC6 produced band shifts of similar intensities encouraged us to examine more carefully the overlapping region, which led us to notice the sequence TCACGTT between positions −368 and −374. To determine whether Cbf1 was able to bind this sequence, we performed additional gel shift assays with two short 40 base-pair DNA fragments containing either the TCACGTT sequence of PDC6 or a mutated version. For comparison, a MET16 fragment containing the bona fide TCACGTG site was included. The results did show hisCbf1BD association with the WT PDC6, but not with the mutant (Figure 5C). Moreover, hisCbf1BD affinity for PDC6 was comparable to its affinity for MET16 (Figure 5C).
Figure 5.
In vitro association of Cbf1 with PDC6 promoter. (A) Schematic of PDC6 showing the DNA fragments used in the gel shift assay. ‘M’ boxes represent the CTGTGGC motifs. ‘T’ boxes represent the TATA elements. (B) Gel shift assays with large DNA fragments. The 32P-labeled DNA fragments indicated were incubated with 0, 50 and 200 ng of a recombinant derivative of Cbf1 containing the bHLH domain fused to a polyhistidine tag (hisCbf1BD). Fragments were resolved by 5% polyacrylamide gel electrophoresis and visualized by PhosphorImager analysis. The MET16 fragment covers positions from –126 to –270 and contains TCACGTG. (C) Gel shift assays with 40 bp DNA fragments. The 32P-labeled DNA fragments indicated were incubated with 0, 200, 100, 50 and 25 ng of hisCbf1BD (only 0 and 200 ng in the case of mut PDC6). The PDC6 fragment covers positions from –350 to –389 and contains the TCACGTT sequence. The MET16 fragment covers positions from –155 to –194 and contains the TCACGTG sequence. mut PDC6 contains TCCAGTT instead of TCACGTT.
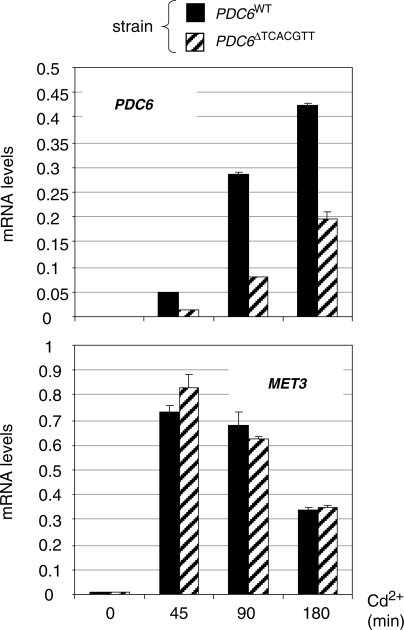
To determine whether this TCACGTT sequence was required for PDC6 activation in vivo, mRNA analysis was performed on a yeast strain containing a mutated derivative of PDC6 lacking the TCACGTT sequence (Figure 6). We found that PDC6 transcription was still strongly induced in the absence of TCACGTT, but at a level 2–3-fold lower compared to the WT. Therefore, since Cbf1 inactivation almost completely abolishes PDC6 induction (Figure 1), we concluded that Cbf1 can be recruited to PDC6 in vivo without this TCACGTT sequence.
Figure 6.
PDC6 transcription in the absence of the TCACGTT motif. pdc6Δ strains containing at the URA3 locus a WT PDC6 allele or a derivative lacking the TCACGTT sequence were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. Total RNA was extracted from samples taken at the time points indicated and RNA levels for PDC6 and MET3 were quantified by RT-real time PCR and normalized to 25S ribosomal RNA. Values represent the average of two independent experiments and error bars indicate average deviations.
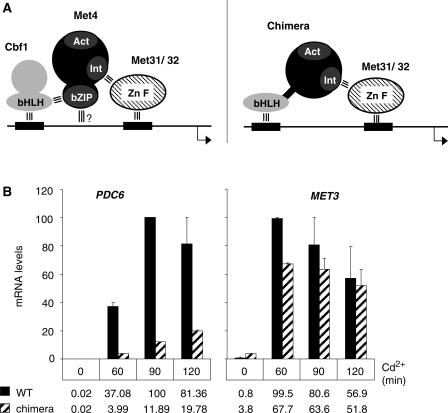
To gain more insight into the mechanism of association of Cbf1 with PDC6 in vivo, we used a strain expressing a chimera in which the carboxy-terminal bZIP of Met4 was replaced by the bHLH of Cbf1 (Figure 7A). A previous study showed that expression of this chimera in place of the full length Met4 in a cbf1Δ strain allowed activation of the sulfur amino acid biosynthetic genes and, as a result, supported cell growth on a medium containing sulfate as unique sulfur source (7). We asked whether this chimera would also support activation of PDC6 transcription in response to cadmium. The results showed that PDC6 transcription was diminished by several fold in the cbf1Δ strain expressing the chimera compared to the WT strain expressing the full length Met4 (Figure 7B). For comparison, MET3 was induced at similar levels in both strains, confirming that the chimera was able to activate MET genes in response to cadmium as efficiently as the WT Met4. Two conclusions could be drawn from these results: first, in vivo the TCACGTT sequence present in PDC6 does not bind Cbf1 bHLH as efficiently as the TCACGTG sequence present in MET genes; secondly, assembly of the Met4 activation complex at PDC6 involves a mechanism that is different from the mechanism involved at MET genes.
Figure 7.
Replacement of Met4 bZIP by Cbf1 bHLH. (A) Model scheme for association to MET promoters of Met4 and a Met4-Cbf1 chimera containing residues 1–590 of Met4 fused to residues 210–351 of Cbf1. ‘Act’ represents Met4 activation domain and ‘Int’ represents Met4 interaction domain with Met31/32. The three parallel bars represent known protein–protein and protein–DNA interactions. (B) Transcription analysis in WT and cbf1Δ cells expressing the Met4-Cbf1 derivative described in (A) from MET4 endogenous promoter. Cells were cultivated, exposed to cadmium, and RNA levels were quantified by RT real-time PCR as in Figure 1A. For comparison, the maximum level for each gene was set to 100. Error bars indicate average deviations from two independent experiments.
PDC6 activation is delayed compared with MET genes
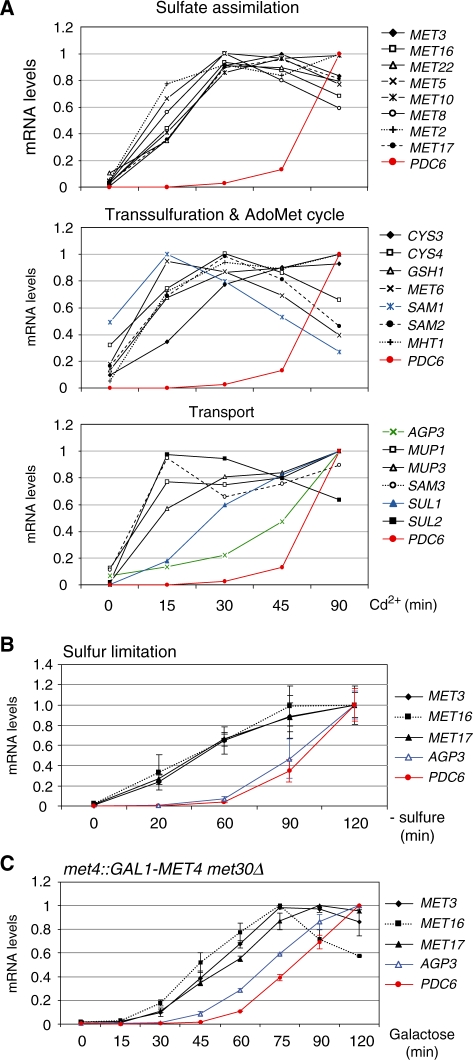
The unusual structure of PDC6 promoter raises the question of whether PDC6 and MET gene regulations are identical. To address this point, we compared the kinetics of induction of PDC6 in response to cadmium with that of known MET genes involved in sulfate assimilation, transsulfuration, glutathione biosynthesis, S-adenosylmethionine cycle, or uptake of sulfur compounds (Figure 8A and Supplementary Table S1). This study revealed that the majority of MET genes showed a similar profile of mRNA accumulation upon cadmium induction and, unquestionably, this profile was very different from the profile of PDC6. For most MET genes, the maximum level of transcription was reached within 30 min upon exposure to cadmium, whereas PDC6 was transcribed at only a few percents of its maximum level at this time point. Among MET genes, only AGP3, which encodes a methionine transporter, showed a notable delay in transcript accumulation, but this delay was still not as strong as in the case of PDC6. So, the whole sulfur metabolism gene network is induced in a quite synchronous manner and PDC6 clearly stands apart. We also analyzed the transcriptional kinetics of PDC6 and several representative MET genes in response to sulphur limitation as well as in the GAL1-MET4 met30Δ strain after addition of galactose (Figure 8B and C). We also observed a clear delay in accumulation of PDC6 transcripts compared to MET gene transcripts. Therefore the differential transcriptional kinetics of PDC6 and MET genes is not limited to the case where induction is triggered by cadmium.
Figure 8.
Transcriptional kinetics of PDC6 versus MET genes. (A) In response to cadmium. wild-type cells were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. Total RNA was extracted from samples taken at the time points indicated. RNA levels were quantified by RT-real time PCR and normalized to 25S ribosomal RNA. For direct comparison among genes, the maximum value for each gene was set up to 1. Data represent the average of two independent experiments and average deviations were 25% at maximum (error bars were omitted for clarity). Numbers are given in Supplementary Table S1. (B) In response to sulfur limitation. Cells were cultivated and RNA was analyzed as in Figure 2A. (C) In the GAL1-MET4 met30Δ strain. Cells were cultivated and RNA was analyzed as in Figure 2B.
PDC6 and MET genes have distinctive thresholds of induction
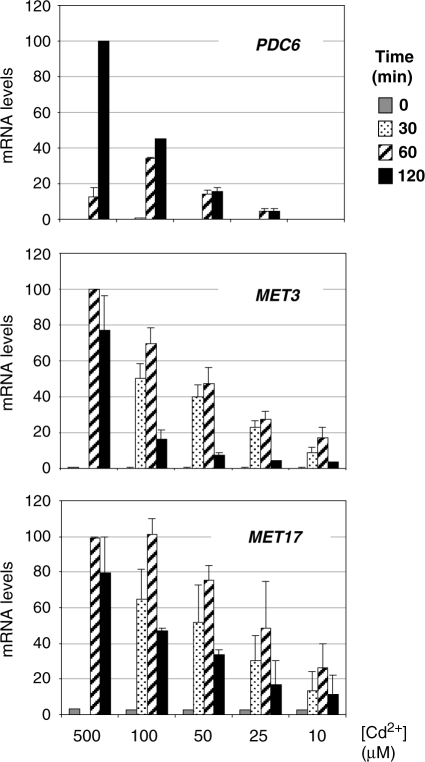
To better apprehend the logic of PDC6 regulation, we compared the dose response relations between the concentration of cadmium in the medium and the level of transcription of PDC6 and MET genes. WT cells were exposed to concentrations ranging from 10 to 500 µM and RNA levels of PDC6 and two representative MET genes were monitored for up to 3 h (Figure 9). For all three genes, we observed an overall positive correlation between the concentration of cadmium and the intensity of the transcriptional response. However, PDC6 transcription fell more abruptly than MET3 and MET17 transcription when cadmium concentration was diminished. For instance, the maximum of PDC6 transcripts at 50 µM was almost 10-fold lower than its maximum at 500 µM whereas, comparatively, the maxima of MET3 and MET17 transcripts were reduced by maximum 2-fold. Moreover, PDC6 transcription was undetectable at 10 µM cadmium whereas MET3 and MET17 were still transcribed at, respectively, 17 and 26% of their maximum levels. These results suggested that the threshold concentration of cadmium necessary to induce PDC6 was higher than that necessary to induce MET genes.
Figure 9.
Dose–response relations between cadmium concentrations and transcription levels. WT cells grown to early log phase in YPD medium were exposed to various concentrations of Cd2+. Total RNA was extracted from samples taken at the time points indicated (no sample was taken at 30 min in the culture exposed to 500 µM Cd2+). RNA levels for PDC6, MET3 and MET17 were quantified by RT-real time PCR and normalized to 25S ribosomal RNA. Maximum values for each gene were set to 100 to facilitate comparisons. Error bars indicate average deviations from two independent experiments.
We analyzed PDC6 transcription in response to additional metals that induce MET gene transcription: arsenic and chromium (26,27). As expected, exposition to arsenic and chromium led to MET3 activation, even though activation levels were weaker compared to cadmium exposition, especially in the case of chromium (Figure 10, top panel). PDC6 was also activated in response to arsenic, but transcripts levels were low compared to cadmium, and it was not in response to chromium (Figure 10, bottom panel). These results show similarities with the results obtained in the presence of low concentrations of cadmium (Figure 9), suggesting that PDC6 has a threshold of induction distinct from MET genes.
Figure 10.
Response to cadmium versus arsenic and chromium. WT cells grown to early log phase in YPD medium were exposed to 0.5 mM Cd(CH3COO)2, 2.5 mM NaAsO2 or 5 mM CrO3. Total RNA was extracted from samples taken before or 45 and 90 min after exposure. RNA levels for MET3 and PDC6 were quantified by RT-real time PCR and normalized to 25S ribosomal RNA. Maximum values for each gene were set to 100 to facilitate comparisons. Error bars indicate average deviations from two independent experiments.
The delay in PDC6 induction is due to a delay in the recruitment of the activator
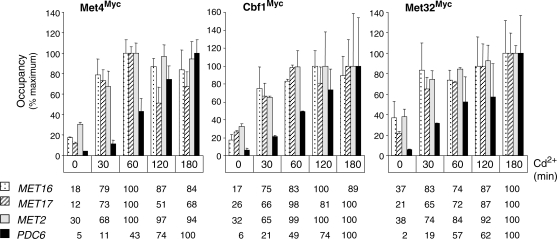
We speculated that the particular regulation of PDC6 was in direct relation with the presence of non-canonical sites for Cbf1 and Met31/32 within the promoter. To support this hypothesis, we first carried out ChIP experiments to examine association of Met4, Cbf1 and Met32 with PDC6 and MET promoters during the course of time following cadmium addition (Figure 11). The ChIP results showed a marked delay in association of all three factors with PDC6 promoter compared to MET promoters. In agreement with the transcription results, MET promoters became maximally occupied by Met4 within 30 to 60 minutes following the exposure to cadmium, whereas at the same time point, PDC6 remained less than half occupied. Therefore, the delay in accumulation of PDC6 mRNA was most likely due to a delay in the recruitment of the activator Met4.
Figure 11.
Association of Met4, Cbf1 and Met32 with promoters in response to Cd2+. Isogenic strains expressing Myc-tagged versions of Met4, Cbf1 or Met32 were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. ChIPs were performed on samples crosslinked with formaldehyde at the time points indicated using Myc antibodies. DNA fragments were quantified by real-time PCR using primers for the indicated promoters and for IME2 ORF as a control. Occupancy levels were calculated as in Figure 1B but are given for each gene as a percentage of the maximum value obtained in the time course. Error bars indicate average deviations from two independent experiments.
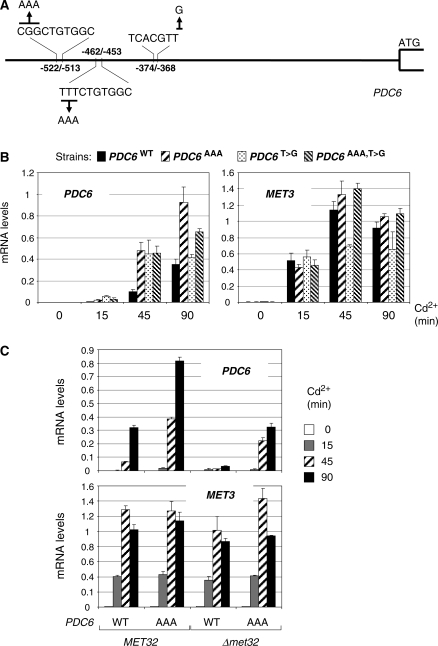
We next mutated the promoter of PDC6 to transform the binding sites for Met31/32 and Cbf1 into canonical sites, either separately or altogether (Figure 12A). As shown in Figure 12B, these mutations resulted in acceleration of PDC6 transcription in response to cadmium, especially in the case of Met31/32 binding sites, so that mRNA levels at 45 min in the mutated strains were similar to those at 90 min in the WT (Figure 12B). However, mRNA levels at 15 min remained low, indicating that the mutations did accelerate PDC6 transcription but were not sufficient to make the transcriptional kinetics of PDC6 similar to that of MET genes. One likely explanation for this observation is that binding of Cbf1 and Met31/32 to their target promoters and recruitment of Met4 involve sequences adjacent to the AAACTGTGGC and TCACGTG sites. The existence of additional promoter elements recognized by some repressive factors cannot be completely ruled out either.
Figure 12.
Mutagenesis of PDC6 promoter. (A) Schematic diagram showing base changes introduced in PDC6 promoter. (B) pdc6Δ strains containing at the URA3 locus a WT allele of PDC6 or mutant derivatives containing either a A-triplet upstream the two CTGTGGC motifs, or TCACGTG instead of TCACGTT, or both changes, were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. Total RNA was extracted from samples taken at the time points indicated. RNA levels for PDC6 and MET3 were quantified by RT-real time PCR and normalized to 25S ribosomal RNA. Values represent the average of two independent experiments and error bars indicate average deviations. (C) MET32 and met32Δ strains containing the WT allele of PDC6 or the derivative with the AAACTGTGGC motifs were manipulated as in (B).
Since inactivation of Met32 is sufficient to stop PDC6 transcription and not MET gene transcription, we wondered whether inserting AAA just before the two CTGTGGC motifs would have an effect on PDC6 transcription in a met32Δ strain (Figure 12C). The results clearly showed that the AAA insertions in the met32Δ strain restore PDC6 transcripts to WT levels in addition to accelerate the transcription kinetics. These results demonstrate that the presence of the minimal CTGTGGC sequence generates a strict requirement for Met32 in addition to slow down transcriptional induction.
PDC6 transcriptional regulation is not conserved in S. cerevisiae close relatives
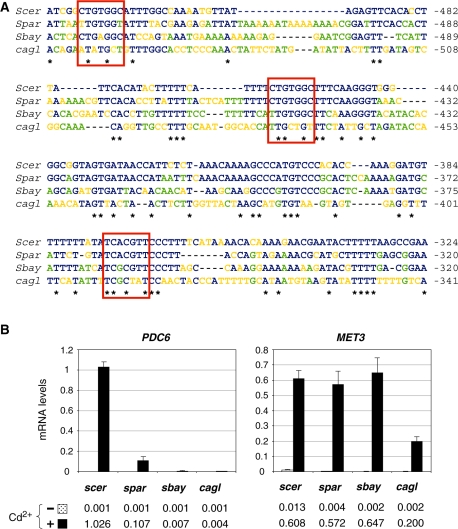
Syntenic orthologs of PDC6 are present in S. paradoxus, S. bayanus and C. glabrata (28,29), three species that diverged from S. cerevisiae after the whole-genome duplication (WGD). Alignment of the promoter regions showed that Met31/32 and Cbf1 binding sites were conserved in S. paradoxus, the closest relative of S. cerevisiae, but not in S. bayanus and C. glabrata, which are more distantly related (Figure 13A). Indeed, S. paradoxus PDC6 possesses the TCACGTT sequence and the first CTGTGGC site, whereas S. bayanus and C. glabrata possess only divergent counterparts. Pattern search using the fuzznuc program (available at http://mobyle.pasteur.fr) confirmed that neither the TCACGTT nor the CTGTGGC sequences were present upstream of S. bayanus and C. glabrata PDC6 orthologs (data not shown). Transcriptional analysis was carried out in S. paradoxus, S. bayanus and C. glabrata strains exposed to cadmium (Figure 13B). The results showed that PDC6 was significantly induced in response to cadmium exposure in S. paradoxus, but almost not in S. bayanus and C. glabrata. Interestingly, the level of PDC6 induction was 10-fold lower in S. paradoxus compared to S. cerevisiae, which is in good agreement with the fact that S. paradoxus PDC6 does not contain the second CTGTGGC site.
Figure 13.
Transcriptional regulation of PDC6 in S. cerevisiae relatives. (A) Sequence alignment. PDC6 orthologs in S. cerevisiae (YGR087C), S. paradoxus (spar6-g11.1), S. bayanus (sbayc642-g17.1) and C. glabrata (CAGL02937) were aligned with Clustal W. Fully conserved positions are marked by an asterisk. Positions conserved among S. cerevisiae and at least one other species are marked in blue. Positions conserved among several species except S. cerevisiae are marked in green. Positions occupied by different nucleotides are marked in orange. Binding sites for Met31/32 and Cbf1 are marked by red boxes. (B) Saccharomyces cerevisiae (W303-1A), S. paradoxus (CLIB228), S. bayanus (CLIB181) and C. glabrata (CLIB298) strains were grown to early log phase in YPD medium and exposed to 0.5 mM Cd2+. Total RNA was extracted from samples taken just before (−) or 120 min (+) after the addition of cadmium. RNA levels for PDC6 and MET3 orthologs were quantified by RT real-time PCR and normalized to 25S ribosomal RNA. Values represent the average of three independent experiments and error bars indicate average deviations.
DISCUSSION
The Met4 activator is known as a master regulator of the sulfur metabolism, coordinating transcription of the genes encoding the multiple enzymatic activities (more than 15) required for assimilation of sulfate and biosynthesis of essential organosulphur compounds such as methionine, cysteine, AdoMet and GSH (see Supplementary Figure S1) (1). Met4 also controls genes encoding transporters for sulfur-containing compounds (30,31), as well as genes encoding regulatory factors of the sulfur metabolism (4,6). As a whole, the Met4 regulon comprises more than 25 genes whose expression is regulated in a coordinated manner. We demonstrate here that PDC6, which encodes an isoform of pyruvate decarboxylase, is another direct target of Met4. Pyruvate decarboxylase is the key enzyme of alcoholic fermentation, catalyzing the conversion of the end product of glycolysis, pyruvate, to acetaldehyde and carbon dioxide. Pyruvate decarboxylase is also involved in amino acid catabolism and production of higher alcohol (32) but, to date, is not known for having a role in the sulfur metabolism. One striking characteristic of Pdc6 is its low content in sulfur atoms compared to the two other pyruvate decarboxylase isoforms, Pdc1 and Pdc5. It has been proposed that Pdc6 represents an isoform designed to spare sulfur when high amounts of glutathione need to be synthesized, such as in the presence of cadmium (17). However, our finding that PDC6 is also activated in response to sulfur amino acid limitation supports the possibility that the low sulfur content of Pdc6 may represent a form of molecular adaptation to environments where sulfur containing amino acids are scarce. Such hypothesis has already been proposed to explain why enzymes involved in sulfur amino acid biosynthesis in S. cerevisiae and E. coli contain fewer sulfur atoms compared to total proteins (33).
Recruitment of Met4 to its target promoter: same set of factors, but different mechanisms
Previous studies led to the model that Met4 was recruited to its target promoters through two distinct anchoring platforms consisting of Cbf1 bound to TCACGTG and Met31/32 bound to AAACTGTGGC (9,11,16). The present study introduces a novel mechanism of recruitment of Met4 to promoters and shed new lights on the properties of bZIP, bHLH and zinc finger factors.
One remarkable characteristic of the mechanism of Met4 association with PDC6 is that it involves non-canonical sites for both Cbf1 and the Met31/32 pair. Using the motif research software MEME (34,35), we were able to identify sequences matching the Met31/32 motif characterized by Blaiseau et al. (11) upstream of all Met4-regulated genes involved in the sulfur metabolism (see Supplemental Figure S2A). The logo derived from these sequences shows a bipartite motif consisting of a highly conserved CTGTGGC box preceded by a short A-rich stretch (see Supplemental Figure S2C). Therefore, PDC6 stands clearly apart from MET genes in that it contains two CTGTGGC boxes with no adjacent A-residues (sites #1 and #2; Figure 3A).
Our in vitro DNA-binding studies show that both Met31 and Met32 are able to bind PDC6 by their own (see Figure 4), which means that the A-rich stretch flanking the CTGTGGC box is not an essential determinant for Met31/32 binding. In line with this conclusion, the Met31/32 binding site identified by Kellis et al. (28) in their comparative analysis of Saccharomyces species is the sequence SKGTGGS (where S = C/G and K = G/T). Several lines of evidence indicate that the nucleotides surrounding the CTGTGGC sites have nevertheless decisive importance. First, sites #1 and #2 are not functionally equivalent in vivo since deletion of site #1 is sufficient to completely stop PDC6 transcription, whereas deletion of site #2 leads to a decrease in transcription (Figure 3). Secondly, Met31 can bind both sites in vitro whereas Met32 can bind only site #1 (Figure 4). Thirdly, Met31 and Met32 have less affinity for PDC6 than for MET3 which contains the canonical AAACTGTGGC sequence (Figure 4), strongly suggesting that the AAA stretch in front of CTGTGGC stabilizes Met31 and Met32 DNA binding. Our finding that insertion of AAA in front of the CTGTGGC sites of PDC6 is sufficient to restore PDC6 transcription in the met32Δ strain highlights the functional importance of the flanking sequences (Figure 12). Therefore, the absence of A-stretch may serve to delimit among Met4-regulated genes a subset of genes strictly dependent on Met32, thereby offering the possibility of differential regulation. Interestingly, these results remind the earlier observation that deletion of MET32 can suppress the Met4-dependent cell cycle arrest caused by MET30 inactivation (25), whereas deletion of MET31 cannot, leading to the suggestion that Met32 had a distinctive role in the regulation of genes involved in the cell cycle control [see ref. (36)]. In light of our results with PDC6, it is tempting to speculate that the unidentified genes that are deregulated in the met30Δ strain contain CTGTGGC boxes in their promoter.
It is striking that Met31 cannot support PDC6 transcription since it binds both CTGTGGC sites in vitro (Figure 4). The possibility that Met31 would not bind PDC6 in vivo seems unlikely if one considers that Met31 has a stimulating role on PDC6 transcription (Figure 1A). Moreover, the fact that inactivation of Met31 has a similar effect as deletion of the distal CTGTGGC box (compare Figures 1A and 3A) strongly suggests a functional relationship. A more likely explanation is that Met31 bound to CTGTGGC does not form an adequate platform to recruit Met4, whereas Met31 bound to AAACTGTGGC does, possibly because the presence of the A-stretch stabilizes the DNA–Met31–Met4 complex (see model Figure 14B). Therefore, the only role of Met31 at PDC6 would be to stimulate or facilitate Met32 binding to DNA. These results offer a novel example of interplay between two homologous zinc finger proteins. They also show for the first time that Met31 and Met32 have distinct binding specificities despite the high homology of their DNA-binding domains (40 identical residues plus three conservative changes in the 51-amino acid long domain containing the two C2H2 motifs).
Figure 14.
Models for Met4 recruitment to MET (A) versus PDC6 (B) promoters. Met4 recruitment involves direct interactions with Cbf1 (through the bZIP domain) and Met31 or Met32 (through the ‘Int’ domain). Met28 associates with Met4 bZIP and participates in Met4 recruitment through stabilization of the Met4–Cbf1–DNA complex (see text for details and references). The question marks at the extremity of the arrows originating from Met4 bZIP indicate that, even though direct interactions with promoter DNA are likely [see ref. (16)], the precise points of contact remain to be established. At PDC6 promoter, Met31 and Met32 bind to two distinct sites. Met4 recruitment to this promoter requires interaction with Met32 specifically, and Met31 participates by assisting Met32 association with its binding site. The width of the arrows pointing toward Cbf1 and Met31/32 DNA binding sites translate the strength of the interaction, which differs between MET and PDC6 promoters.
Another interesting characteristic of PDC6 regulation concerns the bHLH factor Cbf1. We have found that Cbf1 associates with PDC6 promoter and plays an essential role in its transcription (Figures 1, 5 and 6). However, its association does not occur through the sequence TCACGTG classically found upstream of MET genes (see Supplementary Figure S2) but, at least in part, through the related sequence TCACGTT. Noteworthily, this sequence does not include the palindromic motif CANNTG characteristic of bHLH transcription factors. A parallel can be made with the other yeast bHLH transcription factor Pho4, which recognizes both the sequences CACGTG and CACGTT found in PHO promoters (37). It is important to note that Cbf1 and Pho4 have slightly different specificities of DNA binding, as illustrated by the fact that, in PHO promoters, CACGT(G/T) motifs are preferentially flanked by a 5′G residue (37) and, the presence of a 5′T residue dramatically affects Pho4 binding (38). The gel shift assays indicates that Cbf1 has comparable affinities for CACGTT and CACGTG in vitro (Figure 5). This is quite unexpected considering that, in the existing crystal structures, the outer C:G base pair makes critical contacts with amino acid residues conserved among bHLH factors, including Cbf1 (39–42). On the other hand, our experiment with the Met4-Cbf1 chimera suggests that the bHLH of Cbf1 does not associate strongly with the CACGTT motif found in PDC6 promoter in vivo, as evidenced by the important decrease in PDC6 transcription when Met4 bZIP is replaced by Cbf1 bHLH, whereas transcription of MET genes is not much affected (Figure 7). Therefore, the CACGTT sequence does not seem to bind Cbf1 bHLH as efficiently as the CACGTG sequence in vivo. It is also interesting to note that deletion of the CACGTT sequence has only a modest effect on PDC6 activation compared to inactivation of Cbf1, which suggests an additional mechanism of recruitment that does not involve a classical DNA-binding site.
A last point addressed by our study concerns the role of Met4 bZIP in assembly of the Cbf1/Met4/Met31/32 complex. The experiment with the chimera shows that a direct connection between Met4 and the bHLH of Cbf1 can bypass the need for Met4 bZIP at MET genes but not at PDC6 (Figure 7). This result, in combination with our previous results (10,16), strongly suggests that Met4 makes direct contacts with DNA, most likely as a heterodimer with Met28, and these contacts are critical for the stability of the Cbf1/Met4/Met28/Met31/Met32 complex when binding sites for Met31/32 and Cbf1 are not optimal. Yet it cannot be fully excluded at this point that the N-terminal part of Cbf1, which is absent in the Met4-Cbf1 chimera, may have a role in the binding of Cbf1 to PDC6. All these show that Met4 recruitment to its target genes involves a sophisticated network of protein–protein and protein–DNA interactions (see model Figure 14).
An original mechanism allowing transcriptional plasticity within a regulon
One of the major questions regarding the regulation of the sulfur metabolism in yeast concerns the number of cofactors required to recruit its transcriptional activator and the reason for such complexity has remained speculative. This study presents data providing possible answers to this question. Indeed, we show that Met4 can be differentially recruited to its target promoters (Figure 11), with as a result differences in both the timing and the threshold of transcription induction among the genes it regulates (Figures 8 and 9). Moreover, we also provide evidence that this differential recruitment most likely originates from the structure of the binding sites for Met4 cofactors (Figure 12B). Although PDC6 offers the most striking example of differential regulation among Met4-dependent genes, it is not the only one. The genes SUL1 and AGP3, encoding transporters for, respectively, sulfate and methionine, are also activated with a delay compared to other MET genes (Figure 8); and as for PDC6, SUL1 and AGP3 contain non-canonical sites for Cbf1 and Met31/32 (see Supplementary Figures S2 and S3). Interestingly, Met4 controls one other sulfate transporter (SUL2) and two other methionine transporters (MUP1 and MUP3), which are all activated following kinetics comparable to other MET genes (Figure 8), leading to the suggestion that the delay in SUL1 and AGP3 transcription reflects a cellular strategy of gradual expression of sulfate and methionine transporters. Altogether these results provide an original example of how transcriptional plasticity can be achieved within a network of coordinately expressed genes using a single activator recruited to promoters through multiple DNA-binding cofactors.
Met4-dependent regulation of PDC6 could result from a recent evolution event
It is most likely that the Met4 regulon has emerged before the WGD that occurred in hemiascomycetes 100-million-years-ago (43,44). Indeed, orthologs of Met4 and its DNA-binding cofactors Cbf1, Met28 and Met31/32 are also present in several species which diverged from S. cerevisiae prior SGD, for example Kluyveromyces lactis. Search of putative DNA-binding sites for Cbf1 and Met31/32 upstream of MET genes in K. lactis reveals that most contain the exact TCACGTG motif as well as sequences matching the CTGTGGC box (see Supplementary Figure S4). Interestingly, compared to S. cerevisiae, A-residues are less frequent in front of the CTGTGGC boxes found in MET promoters of K. lactis (compare Supplementary Figures S2 and S4). Considering that K. lactis contains only one ortholog of Met31/32 (KLLA0D11902g), one can hypothesize that after duplication of the ancestral MET31/32 gene, one copy has specialized in regulation of the sulfur metabolism, whereas the other has kept a broader range of action.
PDC6 is a more recent addition to the Met4 regulon. Indeed, PDC6 was not present in the yeast ancestor that existed prior to WGD and, therefore, one can infer that it was gained on the S. cerevisiae lineage since WGD (29). Moreover, our results show that PDC6 orthologs in S. cerevisiae close relatives S. paradoxus, S. bayanus and C. glabrata are only weakly, or not at all, induced upon cadmium exposure (Figure 13), which suggests that PDC6 was added to the Met4 regulon during the recent history of the S. cerevisiae lineage. These results provide a new example of evolution of a transcriptional regulatory network.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique and grants from Association pour la Recherche sur le Cancer. program ToxNuc-E and Fondation pour la Recherche Médicale (to L.C.). Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jean Labarre for discussions and advices throughout this study; Rick Young for strains; Mike Tyers for antibodies; and Thierry Lacombe and Eileen Poh for critical reading of the manuscript.
REFERENCES
- 1.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas D, Jacquemin I, Surdin-Kerjan Y. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol. Cell Biol. 1992;12:1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dormer UH, Westwater J, McLaren NF, Kent NA, Mellor J, Jamieson DJ. Cadmium-inducible expression of the yeast GSH1 gene requires a functional sulfur-amino acid regulatory network. J. Biol. Chem. 2000;275:32611–32616. doi: 10.1074/jbc.M004167200. [DOI] [PubMed] [Google Scholar]
- 4.Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30)complex. EMBO J. 2000;19:282–294. doi: 10.1093/emboj/19.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 6.Kuras L, Rouillon A, Lee T, Barbey R, Tyers M, Thomas D. Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol. Cell. 2002;10:69–80. doi: 10.1016/s1097-2765(02)00561-0. [DOI] [PubMed] [Google Scholar]
- 7.Menant A, Baudouin-Cornu P, Peyraud C, Tyers M, Thomas D. Determinants of the ubiquitin-mediated degradation of the Met4 transcription factor. J. Biol. Chem. 2006;281:11744–11754. doi: 10.1074/jbc.M600037200. [DOI] [PubMed] [Google Scholar]
- 8.Leroy C, Cormier L, Kuras L. Independent recruitment of mediator and SAGA by the activator Met4. Mol. Cell Biol. 2006;26:3149–3163. doi: 10.1128/MCB.26.8.3149-3163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaiseau PL, Thomas D. Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J. 1998;17:6327–6336. doi: 10.1093/emboj/17.21.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. Independent recruitment of mediator and SAGA by the activator Met4. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 11.Blaiseau PL, Isnard AD, Surdin-Kerjan Y, Thomas D. Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol. Cell Biol. 1997;17:3640–3648. doi: 10.1128/mcb.17.7.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellor J, Jiang W, Funk M, Rathjen J, Barnes CA, Hinz T, Hegemann JH, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker RE, Masison DC. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai M, Davis RW. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 15.Hieter P, Pridmore D, Hegemann JH, Thomas M, Davis RW, Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 16.Kuras L, Barbey R, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1997;16:2441–2451. [PMC free article] [PubMed] [Google Scholar]
- 17.Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- 18.Momose Y, Iwahashi H. Bioassay of cadmium using a DNA microarray: genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Environ. Toxicol. Chem. 2001;20:2353–2360. doi: 10.1897/1551-5028(2001)020<2353:bocuad>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Baudouin-Cornu P, Labarre J. Molecular evolution of protein atomic composition. Biochimie. 2006;88:1673–1685. [Google Scholar]
- 20.Barbey R, Baudouin-Cornu P, Lee TA, Rouillon A, Zarzov P, Tyers M, Thomas D. Inducible dissociation of SCF(Met30) ubiquitin ligase mediates a rapid transcriptional response to cadmium. EMBO J. 2005;24:521–532. doi: 10.1038/sj.emboj.7600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen JL, Su NY, Kaiser P. The yeast ubiquitin ligase SCFMet30 regulates heavy metal response. Mol. Biol. Cell. 2005;16:1872–1882. doi: 10.1091/mbc.E04-12-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Kuras L, Thomas D. Identification of the yeast methionine biosynthetic genes that require the centromere binding factor 1 for their transcriptional activation. FEBS Lett. 1995;367:15–18. doi: 10.1016/0014-5793(95)00528-h. [DOI] [PubMed] [Google Scholar]
- 24.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patton EE, Peyraud C, Rouillon A, Surdin-Kerjan Y, Tyers M, Thomas D. SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)-S transition. EMBO J. 2000;19:1613–1624. doi: 10.1093/emboj/19.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Van Houten B. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 2004;5:R95. doi: 10.1186/gb-2004-5-12-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin YH, Dunlap PE, McBride SJ, Al-Refai H, Bushel PR, Freedman JH. Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet. 2008;4:e1000053. doi: 10.1371/journal.pgen.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2003;423:241–254. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 29.Gordon JL, Byrne KP, Wolfe KH. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menant A, Barbey R, Thomas D. Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J. 2006;25:4436–4447. doi: 10.1038/sj.emboj.7601330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur J, Bachhawat AK. Yct1p, a novel, high-affinity, cysteine-specific transporter from the yeast Saccharomyces cerevisiae. Genetics. 2007;176:877–890. doi: 10.1534/genetics.107.070342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008;74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baudouin-Cornu P, Surdin-Kerjan Y, Marliere P, Thomas D. Molecular evolution of protein atomic composition. Science. 2001;293:297–300. doi: 10.1126/science.1061052. [DOI] [PubMed] [Google Scholar]
- 34.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 35.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaiser P, Su NY, Yen JL, Ouni I, Flick K. The yeast ubiquitin ligase SCFMet30: connecting environmental and intracellular conditions to cell division. Cell Div. 2006;1:16. doi: 10.1186/1747-1028-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshima Y, Ogawa N, Harashima S. Regulation of phosphatase synthesis in Saccharomyces cerevisiae–a review. Gene. 1996;179:171–177. doi: 10.1016/s0378-1119(96)00425-8. [DOI] [PubMed] [Google Scholar]
- 38.Fisher F, Goding CR. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferre-D'Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 40.Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferre-D'Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 44.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 45.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.