Abstract
RAD51 is a key factor in homologous recombination (HR) and plays an essential role in cellular proliferation by repairing DNA damage during replication. The assembly of RAD51 at DNA damage is strictly controlled by RAD51 mediators, including BRCA1 and BRCA2. We found that human RAD51 directly binds GEMIN2/SIP1, a protein involved in spliceosome biogenesis. Biochemical analyses indicated that GEMIN2 enhances the RAD51–DNA complex formation by inhibiting RAD51 dissociation from DNA, and thereby stimulates RAD51-mediated homologous pairing. GEMIN2 also enhanced the RAD51-mediated strand exchange, when RPA was pre-bound to ssDNA before the addition of RAD51. To analyze the function of GEMIN2, we depleted GEMIN2 in the chicken DT40 line and in human cells. The loss of GEMIN2 reduced HR efficiency and resulted in a significant decrease in the number of RAD51 subnuclear foci, as observed in cells deficient in BRCA1 and BRCA2. These observations and our biochemical analyses reveal that GEMIN2 regulates HR as a novel RAD51 mediator.
INTRODUCTION
Double-strand breaks (DSBs) arise spontaneously and are also induced by environmental factors such as ionizing radiation (IR) and topoisomerase I and II inhibitors (reviewed in 1,2). There are two main DSB repair pathways: homologous recombination (HR) and non-homologous end-joining (NHEJ) (reviewed in 3,4). HR is preferred when DSBs occur as a consequence of replication blocks. DSBs in sister chromatids are repaired using the intact sister chromatid as a template. Null HR-deficient vertebrate cells, such as RAD51−/− cells, display extensive chromosomal breakage in mitotic cells and fail to go through even a single cell cycle (5). Thus, HR plays an essential role in the maintenance of chromosomal DNA during replication.
The current model for HR-mediated DSB repair is that DSBs are first recognized by the ATM kinase, which phosphorylates histone H2AX in their vicinity (6). The Mre11 complex, CtIP and the UBC13-mediated ubiquitylation of chromatin proteins are involved in the processing of DSBs to generate 3′ single-stranded DNA (ssDNA) tails (7, 8). These ssDNA tails are associated with the ssDNA binding protein, RPA, which is subsequently replaced by RAD51. This replacement is promoted by a number of RAD51 mediators, including the tumor-suppressor gene, i.e. Breast Cancer Susceptibility Gene 1 (BRCA1), BRCA2, and five RAD51 paralogs (RAD51B/C/D, and XRCC2/3) (reviewed in 9, 10). The resulting RAD51–DNA filament undergoes homologous pairing, strand invasion into intact homologous double-stranded DNA (dsDNA) to form a D-loop structure, and strand exchange. The RAD51-mediated D-loop formation is stimulated by RAD54 and RAD51AP1 (11–15). DNA synthesis from the invading strand, followed by dissociation from the homologous duplex DNA and re-annealing of the newly synthesized strand with the other end of the DSB, completes the DSB repair (reviewed in 16).
The assembly of RAD51 at 3′ ssDNA tails is strictly regulated by the RAD51 mediators (reviewed in 17). Thus, cells deficient in the mediators, such as RAD51 paralogs and BRCA2, consistently show a significant decrease in the number of RAD51 subnuclear foci, but not in the number of foci containing the RPA ssDNA binding protein at DNA damage. Despite the essential role of vertebrate RAD51 in cellular proliferation, cells deficient in the RAD51 mediators exhibit a less prominent phenotype, indicating that the mediators do participate in HR, presumably by controlling the activity of RAD51 (5,18–21). Recent studies have reported the successful reconstitution of in vitro HR using fission yeast RAD51 and RAD51 mediators (22,23). However, the molecular mechanisms underlying the action of mammalian RAD51 mediators are poorly understood. A mammalian RAD51 mediator, the RAD51B–RAD51C complex, has been purified (24). This RAD51 paralog complex may not function alone as the RAD51 mediator, because the genetic data suggest that the five RAD51 paralogs form a functional unit wherein each one is essential in promoting the assembly of RAD51 at DNA damage (18). Although the function of BRCA2 has been extensively studied on a cellular level, its function has not been characterized in in vitro HR reactions, since it has not been possible to purify the complete BRCA2 protein. Instead, biochemical studies of truncated BRCA2 proteins suggest that BRCA2 facilitates the assembly of RAD51, presumably by directly interacting with both the damaged DNA and RAD51 (25–31). It is still unclear how BRCA1 facilitates the accumulation of RAD51 at 3′ ssDNA tails.
We then studied RAD51-interacting proteins using yeast two-hybrid analysis, and found human GEMIN2 as a candidate protein. GEMIN2 is a 32-kDa protein with no homology to any other protein, and is known to form a stable RNA-protein complex with several other components, including the GEMIN and SMN proteins (32). The SMN protein is the product of the survival motor neuron (SMN) gene. Mutation of the SMN gene is responsible for spinal muscular atrophy (SMA), which is a neurodegenerative disease accompanied by the loss of motor neurons (33,34). The homozygous gene-disruption of either GEMIN2 or SMN is lethal to early mouse embryogenesis, while GEMIN2 knockdown using the small interference RNA (siRNA) treatment leads to a drastic decrease in the assembly activity of spliceosomal small nuclear ribonucleoprotein (snRNP) (35). The genetic and biochemical studies cited here indicate that GEMIN2 and the SMN proteins constitute a core complex that is essential for the formation of a functional SMN complex. Additionally, this complex plays an essential cellular role in the assembly of spliceosomal snRNPs, a process that is partially disrupted in SMA.
In the present study, we found that human GEMIN2 physically interacts with RAD51. Immunoprecipitation experiments indicated that GEMIN2 and RAD51 form a complex. To evaluate the DNA repair function of GEMIN2, we performed in vitro HR assays and also analyzed the phenotype of GEMIN2-depleted cells. Purified GEMIN2 protein enhances the complex formation between RAD51 and DNA, and stimulates RAD51-mediated homologous pairing and strand exchange. In agreement with these biochemical results, the depletion of GEMIN2 in the chicken B cell line DT40 (36) as well as in human cells significantly reduced the recruitment of RAD51 to DNA lesions. In summary, our observations reveal that GEMIN2 has another unexpected function: regulating the RAD51 recombinase and thereby promoting HR-dependent repair.
MATERIALS AND METHODS
Yeast two-hybrid screening of GEMIN2 cDNA
The two-hybrid analysis of the N-terminal RAD51-fragment (1–114 amino acid residues) interaction was carried out using a Matchmaker two-hybrid cDNA library from human testis (Clontech, Mountain View, CA, USA). The human GEMIN2 gene was cloned from a human testis cDNA pool (purchased from Clontech) by polymerase chain reaction (PCR). The DNA fragments containing sequences derived from RAD51 and GEMIN2 were ligated into the pACT2 and pAS2-1 vectors (Clontech), respectively. The pAS2-1 and pACT2 plasmids contain the GAL4 DNA-binding domain and the GAL4 activation domain, respectively, just upstream of their cloning sites. The plasmids for the positive control experiment (pGBKT7-53 and pGADT7-T) and for the negative control experiment (pGBKT7-Lam and pGADT7-T) were supplied by the manufacturer (Clontech). The plasmids containing the RAD51 and GEMIN2 sequences were introduced into the yeast strain AH109. The two-hybrid interaction between RAD51 and GEMIN2 was then tested according to the manufacturer’s protocol (Clontech Matchmaker GAL4 protocol). The interaction between RAD51 and GEMIN2 induced the expression of the HIS3, ADE2 and LacZ reporter genes, which allow the yeast strain to grow on a synthetic dextrose minimal medium plate without histidine and adenine, and to produce a blue color in the presence of X-Gal.
Protein purification
The human GEMIN2 gene was ligated into the NdeI site of the pET15b plasmid (Novagen, Darmstadt, Germany). The His6-tagged GEMIN2 protein was overexpressed in the Escherichia coli strain BL21(DE3), which also carried an expression vector for the minor tRNAs [Codon( + )RIL, Novagen]. The E. coli cells containing the GEMIN2-expressing plasmid were cultured at 30°C. At the logarithmic phase of growth (A600 = 0.6), the GEMIN2 production was induced with 1 mM isopropyl-1-thio-β-D-galactopyranoside (final concentration), and the cells were further cultured for 12 h at 18°C. The cells were disrupted by sonication in buffer A [50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 2 mM 2-mercaptoethanol and 10% glycerol]. The cell debris was clarified by centrifugation, and the supernatant was mixed with 4 ml of ProBond resin (Invitrogen, Carlsbad, CA, USA). The sample was gently mixed for 1 h at 4°C, and the resin was washed with buffer A. The resin was packed in a column and washed with 200 ml of buffer A, containing 30 mM imidazole. The resin was further washed with 40 ml of 50 mM Tris-HCl buffer (pH 7.5), containing 2 mM 2-mercaptoethanol, 30 mM imidazole and 10% glycerol, and the protein was eluted by a linear gradient of imidazole from 30 mM to 500 mM. The ProBond fractions containing the His6-tagged GEMIN2 protein were collected and fractionated on a Superdex 75pg column (GE Healthcare Biosciences, Uppsala, Sweden), which was previously equilibrated with buffer B [20 mM Tris-HCl (pH 8.0), 1 M NaCl, 0.25 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol]. The His6-tagged GEMIN2 protein eluted from the Superdex 75 pg column was treated with 2.5 U/mg of thrombin protease (GE Healthcare Biosciences) during dialysis against buffer C [20 mM Tris-HCl (pH 8.0), 0.25 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol]. After removal of the His6 tag, GEMIN2 was further purified by MonoQ column chromatography (GE Healthcare Biosciences). Purified GEMIN2 was stored in 20 mM HEPES-NaOH buffer (pH 7.5), containing 200 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol at −80°C. To prepare Cy5-labeled GEMIN2, purified GEMIN2 (1.3 mg) and Cy5 maleimide mono-reactive dye (0.2 mg, GE Healthcare Biosciences) were incubated at 4°C for 1 h. Free Cy5 dye was removed by NAP10 gel filtration column chromatography (GE Healthcare Biosciences). Human RAD51 was purified as described earlier (37).
Assay for RAD51–GEMIN2 binding
RAD51 (40 µg) was covalently conjugated to Affi-Gel 10 beads (75 µl, Bio-Rad Laboratories, Hercules, CA, USA), according to the manufacturer’s instructions, and the Affi-Gel 10-protein matrices were adjusted to 50% slurries. GEMIN2 (8 µg) was incubated with RAD51-conjugated Affi-Gel 10 beads (2.5 µg of RAD51) in 100 µl of 20 mM potassium phosphate buffer (pH 7.5), containing 0.05% Triton X-100. After incubation at room temperature for 2.5 h, the RAD51 beads were extensively washed with 20 mM potassium phosphate buffer (pH 7.5), containing 150 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol. Proteins bound to the RAD51 beads were fractionated by a 15% SDS–PAGE. Bands were visualized by Coomassie Brilliant Blue staining.
Immunoprecipitation
We used 3 × 107 HeLa cells for the RAD51 binding Flag-tagged chicken GEMIN2 immunoprecipitation experiments. The Flag-chicken GEMIN2 expression vector was transfected into HeLa cells by Lipofectamine 2000 (Invitrogen). At 48 h after the transfection, the cells were washed three times with cold PBS buffer and harvested. After centrifugation, collected cells were mixed with 400 µl of lysis buffer [300 mM NaCl, 50 mM Tris-HCl (pH. 7.5), 1 mM NaF, 0.5% NP-40 and 10% Glycerol], supplemented with a protease inhibitor cocktail (Complete Mini, Roche, Mannheim, Germany) and kept on ice for 5 min. After addition of BSA (1% final concentration), the resulting cell lysates were each sonicated five times for 30 sec and centrifuged at 4°C. The supernatant was collected as a whole-cell extract. One microgram of antibody RAD51 (Ab-1, Calbiochem, Darmstadt, Germany) and 20 µl of DYNA-protein G beads (DYNAL, Carlsbad, CA, USA) were mixed and incubated overnight in PBS containing 1% BSA at 4°C to conjugate the antibody and beads. The mixture was then washed with the lysis buffer. A 360 µl aliquot of the whole-cell extract was mixed for 120 min with the pre-treated beads for immunoprecipitation at 4°C. The resulting precipitates were washed five times with the lysis buffer (containing 0.1% BSA). The washed precipitates were mixed with 40 µl of the standard SDS final sample buffer, boiled for 5 min and subjected to SDS gel-electrophoresis.
DNA substrates for in vitro reactions
Single-stranded ϕX174 viral ( + ) strand DNA and double-stranded ϕX174 replicative form I DNA were purchased from New England Biolabs (Ipswich, MA, USA). The linear dsDNA was prepared from the ϕX174 replicative form I DNA by PstI digestion. The dsDNA fragments were prepared from pGsat4 DNA (38) by Alw44I digestion. High-pressure liquid chromatography-purified oligodeoxyribonucleotides were purchased from Nihon Gene Research Laboratory. The DNA sequences used in this study are: 49-mer ssDNA: 5′-CCA ATG CGC TTG GTA AAC ATG CTC AGG ATT GCG ACG CTA GCA AGT GAT C -3′, and 49-mer dsDNA: 5′-GTC CCA GGC CAT TAC AGA TCA ATC CTG AGC ATG TTT ACC AAG CGC ATT G-3′, 5′-CAA TGC GCT TGG TAA ACA TGC TCA GGA TTG ATC TGT AAT GGC CTG GGA C-3′ and 5′-CCA ATG CGC TTG GTA AAC ATG CTC AGG ATT GCG ACG CTA GCA AGT GAT C-3′.
Measurement for binding of RAD51 and GEMIN2 to ssDNA and dsDNA
The reaction mixtures (10 µl) contained 28 mM HEPES buffer (pH 7.5), 300 mM NaCl, 0.04 mM EDTA, 0.8 mM 2-mercaptoethanol, 1 mM ATP, 1 mM DTT, 100 µg/ml BSA, 1 mM MgCl2 and 4% glycerol. RAD51 was incubated in the presence or absence of GEMIN2 or Cy5-labeled GEMIN2 at 37°C for 5 min, and then either the ϕX174 ssDNA (20 µM) or the linearized ϕX174 dsDNA was added. After incubation for 15 min, the protein–DNA complexes were analyzed by 0.8% agarose gel electrophoresis in 1 × TAE buffer (40 mM Tris-acetate and 1 mM EDTA) at 3.3 V/cm for 2.5 h. The bands were visualized by ethidium bromide staining. Cy5-labeled GEMIN2 was visualized using a Typhoon9410 imaging analyzer (GE Healthcare Biosciences).
Measurement of RAD51 assembly onto DNA
The RAD51 protein was incubated in the presence or absence of the GEMIN2 protein at 37°C for 10 min, and then the ssDNA 49-mer (10 µM) or the dsDNA 49-mer (6 µM) was added. Reactions were performed in 10 µl of 20 mM potassium phosphate buffer (pH 7.5), containing 70 mM NaCl, 1 mM ATP, 1 mM DTT, 100 μg/ml BSA, 1 mM MgCl2 and 2% glycerol. After a 10-min incubation, the RAD51–ssDNA and Rad51–dsDNA filaments were analyzed by 10% and 8% polyacrylamide gel electrophoresis in 1 × TBE buffer (90 mM Tris, 90 mM boric acid and 2 mM EDTA) at 7 V/cm for 1.5 h, respectively. The bands were visualized by SYBR Gold (Invitrogen) staining, and quantitated using an LAS-1000 imaging analyzer with the Image Gauge software (Fujifilm, Tokyo, Japan).
Assay for RAD51–DNA filament disassembly
RAD51 (2 µM) was incubated in the presence or absence of GEMIN2 (4 µM) at 37°C for 10 min in 28 mM HEPES buffer (pH 7.5), containing 535 mM NaCl, 0.04 mM EDTA, 0.8 mM 2-mercaptoethanol, 1 mM ATP, 1 mM DTT, 100 µg/ml BSA, 1 mM MgCl2 and 4% glycerol. The linearized ϕX174 dsDNA was then added, and the volume of the reaction mixture was adjusted to 10 µl. After an incubation at 37°C for 10 min, these mixtures were incubated with various concentrations of the Alw44I-digested pGsat4 dsDNA fragments (0–50 µM) at 37°C for 2 h. The products were fractionated by 0.8% agarose gel electrophoresis in TAE buffer at 3.3 V/cm for 2.5 h, and visualized by staining with ethidium bromide and quantitated using an LAS-1000 imaging analyzer with the Image Gauge software (Fujifilm).
Measurement of RAD51 disassembly by the BRC4 polypeptide
The BRC4 polypeptide used in this assay was purchased from Toray Research Center, Inc. (Tokyo, Japan). The amino acid sequence of the BRC4 polypeptide is as follows: LLGFHTASGKKVKIAKESLDKVKNLFDE. RAD51 (2 µM) was incubated in the presence or absence of GEMIN2 (4 µM) at 37°C for 10 min in 20 mM potassium phosphate buffer (pH 7.5), containing 30 mM NaCl, 0.04 mM EDTA, 0.8 mM 2-mercaptoethanol, 1 mM ATP, 1 mM DTT, 100 µg/ml BSA, 1 mM MgCl2 and 4% glycerol. The linearized ϕX174 dsDNA was then added, followed by an incubation with the BRC4 polypeptide (0, 0.5, 1 and 2 µM) at 37°C for 1 h. The volume of the reaction mixture was 10 µl. The protein–DNA complexes were analyzed by 0.8% agarose gel electrophoresis at 3.3 V/cm for 2.5 h in 1 × TAE buffer, and the bands were visualized by ethidium bromide staining and then quantitated using an LAS-1000 imaging analyzer with the Image Gauge software (Fujifilm).
The D-loop formation assay
RAD51 and GEMIN2 were incubated at 37°C for 5 min in 8 µl of reaction buffer, containing 24 mM HEPES-NaOH (pH 7.5), 35 mM NaCl, 0.02 mM EDTA, 0.4 mM 2-mercaptoethanol, 2% glycerol, 1 mM MgCl2, 1 mM DTT, 2 mM ATP, 2mM CaCl2 and 0.1 mg/ml BSA. After this incubation, 1 µl of a 32P-labeled 50-mer oligonucleotide (1 µM, 5′-GGA ATT CGG TAT TCC CAG GCG GTC TCC CAT CCA AGT ACT AAC CGA GCC CT-3′) was added, and the samples were further incubated at 37°C for 5 min. The reactions were then initiated by the addition of 1 µl of the pB5Sarray superhelical dsDNA (20 µM), and were continued at 37°C for 30 min. The pB5Sarray DNA contained 11 repeats of a sea urchin 5S rRNA gene (207-bp fragment) within the pBlueScript II SK(+) vector, and was prepared by a method avoiding alkaline treatment of the cells harboring the plasmid DNA, to prevent the dsDNA substrates from undergoing irreversible denaturation. The reactions were stopped by the addition of 0.1% SDS and 1.6 mg/ml proteinase K (Roche Applied Science, Basel, Switzerland) and were further incubated at 37°C for 15 min. After adding 6-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1 × TAE buffer at 4 V/cm for 2 h. The gels were dried, exposed to an imaging plate and visualized using an FLA-7000 imaging analyzer (Fujifilm).
Assay for strand exchange
The ϕX174 circular ssDNA (20 µM) was incubated with or without 2 µM RPA at 37°C for 10 min in 10 µl of 20 mM potassium phosphate buffer (pH 7.5), containing 75 mM NaCl, 0.04 mM EDTA, 0.8 mM 2-mercaptoethanol, 4% glycerol, 1 mM MgCl2, 1 mM DTT, 1 mM ATP, 100 µg/ml BSA, 20 mM creatine phosphate and 75 µg/ml creatine kinase. After this incubation, RAD51 (4 µM) and the indicated amounts of GEMIN2, which were pre-incubated at 37°C for 5 min, were added to the reaction mixture. The reactions were then initiated by the addition of 100 mM (NH4)2SO4 and 20 µM ϕX174 linear dsDNA and were continued for 30 min. The reactions were stopped by the addition of 0.1% SDS and 1.97 mg/ml proteinase K (Roche Applied Science), and were further incubated at 37°C for 15 min. After adding 6-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1 × TAE buffer at 3.3 V/cm for 4 h. The products were visualized by SYBR Gold (Invitrogen) staining and quantitated using an LAS-1000 imaging analyzer with the Image Gauge software (Fujifilm).
Surface plasmon resonance analysis
The surface plasmon resonance (SPR) signals were measured with Biacore X100 instrument (GE Healthcare). Flow cells were maintained at 25°C during the measurement, and the instrument was operated at the mid-flow rate (∼30 µl/min). The RAD51 or GEMIN2 protein was conjugated to the activated surface of the CM5 sensor chips (GE Healthcare), using the standard amine coupling conditions recommended by the manufacturer. The level of the conjugated RAD51 and GEMIN2 protein was 18 000 and 6500 resonance units (RU). SPR signals of the flow cell containing a sensor chip without the proteins were subtracted from those of the SPR signals of the flow cell containing the RAD51 or GEMIN2-conjugated sensor chip. The running buffer was 10 mM HEPES-NaOH (pH 7.4), 150 mM NaCl, 3 mM EDTA and 0.05% Tween 20. For each binding assay, 10 µM BRC4 polypeptide was injected for 2 min.
Supercoiling assay
The indicated amounts of GEMIN2 or RAD51 (8 µM) were pre-incubated at 37°C for 10 min in 9 µl of 24 mM HEPES-NaOH buffer (pH 7.5), containing 1 mM ATP, 1 mM MgCl2, 1 mM DTT, 2 mM CaCl2, 4% glycerol, 0.04 mM EDTA, 0.8 mM 2-mercaptoethanol, 70 mM NaCl, 0.1 µg/ml BSA, 20 mM creatine phosphate and 75 µg/ml creatine kinase. The ϕX174 superhelical dsDNA (20 µM) was incubated with topoisomerase I at 37°C for 1 h in 15 µl of 20 mM HEPES-NaOH buffer (pH 7.5), containing 1 mM MgCl2, 1 mM DTT, 100 µg/ml BSA, 20 mM creatine phosphate and 75 µg/ml creatine kinase. The reactions were then initiated by the addition of 20 µM ϕX174 relaxed dsDNA, which was treated with wheat germ topoisomerase I. After an incubation at 37°C for 30 min, the reactions were stopped by the addition of 0.1% SDS and 1.97 mg/ml proteinase K (Roche Applied Science), and were further incubated at 37°C for 15 min. After adding 6-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1× TAE buffer at 1.3 V/cm at 4°C for 16 h. The products were visualized by SYBR Gold (Invitrogen) staining.
Cloning of chicken GEMIN2 cDNA
The full-length cDNA encoding for the GEMIN2 proteins was obtained from the NCBI data bank. The Gene Bank accession numbers are AF027150 and NM001039302 for the human and chicken GEMIN2 genes, respectively. The amino acid sequences of the human and chicken GEMIN2 cDNAs share 79% identity. The human GEMIN2 gene was cloned from the human testis cDNA library (Clontech) by PCR.
Cell culture
DT40 cells were cultured in RPMI-1640 medium supplemented with 10−5 M β-mercaptoethanol, penicillin streptomycin, 10% fetal calf serum (FCS) and 1% chicken serum (Sigma-Aldrich, St Louis, MO, USA) at 39.5°C. HeLa and U2OS cells were cultured in DMEM supplemented with 10% FCS at 37°C.
Construction of targeting vectors
To generate targeting vectors for the chicken GEMIN2 gene, 1.5 and 3.0 kb GEMIN2 genomic fragments were obtained by PCR using DT40 genomic DNA as a template, and were used as 5′ and 3′ arms, respectively. The primers used were 5′-ggg gac aac ttt gta tag aaa agt tgG TTT TGG GCT CCG TGT GGC AGT AA-3′ and 5′-ggg gac tgc ttt ttt gta caa act tgC ACT TGA GCG TTG GAG AGT ATC CT-3′ for the 5′ arm and 5′-ggg gac agc ttt ctt gta caa agt ggA CAA GCA CAG AAA TCA CTG GCG GT-3′ and 5′-ggg gac aac ttt gta taa taa agt tgA GAA AAC CCG TGT GTT TCC TCT GT-3′ for the 3′ arm. Underlined sequences denote the recognition sequences in the Gateway system (Invitrogen). By using the MultiSite Gateway® system with pENTR-lox-HisD, pENTR-lox-puro and pDEST-DTA-MLS (39), a floxed HisDR or PuroR gene was inserted between the 5′ and 3′ arms on a plasmid carrying a DT-A gene, thus yielding the two targeting vectors: GEMIN2-HisDR/loxP and GEMIN2-PuroR/loxP.
Generation of the GEMIN2 conditional mutant cells
To generate GEMIN2−/− cells, linearized GEMIN2 targeting vectors (PuroR and HisDR) were transfected sequentially by electroporation (Bio-Rad Laboratories). The genomic DNA of the transfectants was digested with EcoRV and the targeted clones were confirmed by Southern-blot analysis. The probe was a PCR-amplified fragment derived from chicken DT40 genomic DNA, using the primer set 5′-CGC ATT TGT GGT GGC TTT CAG CTT GAG ACA-3′ and 5′-CTT TAA GTG GCC TCT TCT TGT CAG TGG CAG-3′. The probe was labeled using the Alkphos Direct Labelling Module (GE Healthcare Biosciences). To generate GEMIN2+/− cells, wild-type (WT) DT40 cells were transfected with the GEMIN2 targeting vector (PuroR). Next, one of the GEMIN2+/− clones was transfected with the tetracycline-controlled trans-activator (tTA) gene (Invitrogen) and the conditional human GEMIN2 expression construct to obtain GEMIN2+/−tetGEMIN2 cells carrying the two transgenes at random sites on the chromosome. Expression of human GEMIN2 and tTA was confirmed by western blotting. To generate GEMIN2−/−tetGEMIN2 cells, GEMIN2+/−tetGEMIN2 cells were transfected with the GEMIN2 targeting vector (HisDR). Expression of the chicken GEMIN2 in GEMIN2−/−tetGEMIN2 cells was measured by employing Reverse-transcription PCR (RT–PCR) using the SuperScriptTM III First-Strand system (Invitrogen) and the following primers: 5′-GAG TAT ACT CAG AAA TAG ATG CAG-3′ and 5′-CTA GGA GGG TTC ATC AGC TAA-3′.
siRNA transfection and colony formation assay
We depleted GEMIN2 in HeLa and U2OS cells using the following method. The GEMIN2 siRNA used in this study was provided by Dharmacon. The control siRNA sequences used in this study were 5′-CCC GGA CCA CAA CGC UCU G-3′. Twenty-four h prior to transfection, 2 × 105 or 3 × 106 cells were seeded in 35 mm or 100 mm culture dishes, and transfection was carried out using HiperFect Transfection Reagent (Qiagen, Hilden, Germany) and antibiotic-free media. Assays were performed 48 h after siRNA transfection.
Western blotting and antibodies
Cells were lysed in an SDS lysis buffer (25 mM Tris-HCl pH 6.5, 1% SDS, 0.24 M 2-mercaptoethanol, 0.1% Bromophenol Blue and 5% glycerol) and boiled for 5 min. The aliquots were subjected to SDS–PAGE and transferred to an Immobilon-P transfer membrane (Millipore, Billerica, MA, USA). Antibodies specific to human GEMIN2 [anti-gemin2 (clone 2E17), Immuquest, Seamer, North Yorkshire, UK], RAD51 (Ab-1), mouse Ig [horseradish peroxidase (HRP)-conjugated, GE Healthcare Biosciences] and rabbit Ig (HRP-conjugated; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used for the detection.
Microscopy imaging and generation of DNA damage
Immunocytochemical and laser irradiative analysis were carried out as described by Takata et al. (18) and Murakawa et al. (40). Before laser treatment, HeLa cells were cultured in BrdU (10 µM) containing medium for 48 h and exposed to a 405 nm laser from the confocal microscopy, FV-1000 (Olympus, Tokyo, Japan), to induce DSBs. During laser treatment, cells were incubated in phenol red-free Opti-medium (GIBCO, Carlsbad, CA, USA) to prevent the absorption of the laser wavelength. γ-irradiation was performed using 137Cs (Gammacell 40, Nordion, Ottawa, ON, Canada). Antibodies against RAD51 (Ab-1), FK2 (Nippon Biotest Laboratories, Tokyo, Japan), RPA32/RPA2 (abcam, Cambridge, MA, USA), rabbit Ig (Alexa 488 conjugated, Molecular Probes) and mouse Ig (Alexa 594 conjugated, Molecular probes) were used for visualization. Fluorescence images were obtained and processed using fluorescence microscopy, IX81 (Olympus).
Chromosome aberration analysis
Karyotype analysis was performed as described by Sonoda et al. (5).
RESULTS
GEMIN2 directly interacts with RAD51
To identify unknown RAD51 mediators, we searched for RAD51-interacting proteins, using a human testis cDNA library for yeast two-hybrid screening. We used the N-terminal RAD51 fragment (1–114 amino acid region) as bait. The N-terminal RAD51 region forms a distinct domain, and is accessible for interactions with other HR factors in RAD51–DNA filaments (41,42). From ∼4.8 × 105 transformants, we identified eight clones that activated both ADE2 and HIS3 expression. The nucleotide sequence analysis of these positive clones revealed that one clone encoded GEMIN2. The interaction between RAD51 and GEMIN2 in the yeast two-hybrid system was re-assessed by analyzing the expression of LacZ (blue coloring on X-Gal) and the prototrophy for adenine and histidine (Supplementary Figure S1A). To test whether or not RAD51 directly interacts with GEMIN2, GEMIN2 was expressed in bacteria and purified as a recombinant protein (Supplementary Figure S1B). We performed a pull-down assay, using beads associated with RAD51. GEMIN2 indeed co-pelleted with the RAD51 beads, whereas bovine serum albumin (BSA) did not bind to RAD51 (Supplementary Figure S1C). We therefore conclude that GEMIN2 binds specifically to RAD51.
To analyze the interaction between GEMIN2 and RAD51 in vivo, we transiently expressed Flag-tagged GEMIN2 in HeLa cells, exposed the cells to Camptothecin (1), and isolated cellular extracts. Immunoprecipitation of the extracts with anti-GEMIN2 (Flag) and RAD51 antibodies co-precipitated RAD51 and GEMIN2, respectively (Supplemental Figure S1D), indicating a physical interaction between GEMIN2 and RAD51. Moreover, this interaction was significantly increased by having exposed the cells to Camptothecin, which stimulates HR (1). These data support the idea that GEMIN2 physically associates with RAD51 in vivo as well as in vitro.
GEMIN2 enhances the formation of RAD51–DNA complexes
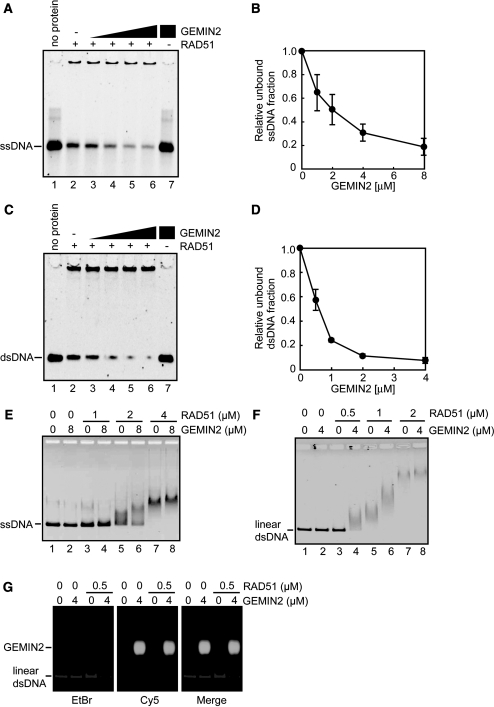
To explore the function of GEMIN2 in HR, we tested whether or not GEMIN2 can stimulate the assembly of RAD51 onto DNA in vitro. To this end, RAD51 was incubated with 49-mer ssDNA in the presence or absence of GEMIN2, and the incubated products were subjected to electrophoresis on polyacrylamide gels (Figure 1A and B). Remarkably, pre-incubation of RAD51 with GEMIN2, prior to the addition of ssDNA, significantly reduced the amount of free ssDNA in the GEMIN2 concentration dependent manner (Figure 1A, lanes 3–6, and Figure 1B). Similar results were obtained using 49-mer dsDNA (Figure 1C and D) instead of ssDNA. Hence, GEMIN2 may indeed enhance the formation of the RAD51–DNA complexes in vitro. We next performed the RAD51–DNA binding assay with the ϕX174 ssDNA (5386 bases) or dsDNA (5386 bp). Consistently, the addition of GEMIN2 to RAD51 prior to the addition of the ϕX174 ssDNA or dsDNA caused prominent retardation of their migration (Figure 1E and F). This retardation may not be due to the association of GEMIN2 with the RAD51–DNA complex, because it did not carry a detectable amount of fluorescently labeled GEMIN2 (Figure 1G). We therefore conclude that GEMIN2 enhances the formation of the RAD51–DNA complex, without tightly associating with this complex.
Figure 1.
GEMIN2 stimulates RAD51–DNA filament formation. (A) Polyacrylamide gel electrophoresis to examine the formation of the RAD51–DNA filament. RAD51 (4 µM) and GEMIN2 were incubated with 10 µM 49-mer ssDNA. DNA was visualized by SYBR Gold (Invitrogen) staining. The GEMIN2 concentrations were 0 µM (lane 2), 1 µM (lane 3), 2 µM (lane 4), 4 µM (lane 5) and 8 µM (lanes 6 and 7). Under these experimental conditions, 90% of the input ssDNA was estimated as being in the RAD51-bound fraction in the absence of the GEMIN2 protein. (B) Quantification of experiments shown in panel A. The amounts of complexes formed were estimated from the residual free DNA substrates, and unbound ssDNA fractions relative to lane 2 of panel A were plotted. Average values of three independent experiments are shown with standard deviation values. (C) Polyacrylamide gel electrophoresis, as in panel A. RAD51 (2 µM) and GEMIN2 were incubated with 6 µM 49-mer dsDNA. DNA was visualized by SYBR Gold (Invitrogen) staining. The GEMIN2 concentrations were 0 µM (lane 2), 0.5 µM (lane 3), 1 µM (lane 4), 2 µM (lane 5) and 4 µM (lanes 6 and 7). (D) Quantification of experiments shown in panel C. The amounts of complexes formed were estimated from the residual free DNA substrates, and unbound dsDNA fractions relative to lane 2 of panel C were plotted. Average values of three independent experiments are shown with standard deviation values. (E) Agarose gel electrophoresis to examine the formation of the RAD51-ssDNA filament. RAD51 was incubated in the presence or absence of the GEMIN2 protein, followed by addition of ϕX174 ssDNA (20 µM). DNA was visualized by ethidium bromide staining. (F) Agarose gel electrophoresis to examine the formation of the RAD51–dsDNA filament. RAD51 was incubated in the presence or absence of the GEMIN2 protein, followed by addition of linear ϕX174 dsDNA (10 µM). Results presented as in panel E. (G) Agarose gel electrophoresis to assess the complex formation between the RAD51-dsDNA filament and GEMIN2. GEMIN2 was labeled with Cy5 and dsDNA was stained with EtBr. Note that GEMIN2 facilitated the formation of the RAD51-dsDNA filament, but did not bind to the filament.
It has been shown that all yeast and mammalian RAD51 mediators, including BRCA2 and the RAD51B–RAD51C complex, can bind to DNA in vitro (24,25). We thus tested whether GEMIN2 binds to DNA. In contrast to the RAD51 mediators reported previously, GEMIN2 did not affect the mobility of ssDNA or dsDNA (Figure 1E and F, lane 2). GEMIN2 did not induce topological changes in supercoiled DNA in the absence of RAD51 (Supplementary Figure S2). Therefore, GEMIN2 did not exhibit DNA binding under the conditions used in this assay. This observation indicates that GEMIN2 is a unique RAD51 mediator, in the sense that it can stimulate the assembly of RAD51 on DNA without closely associating with the DNA.
GEMIN2 inhibits the RAD51–DNA dissociation
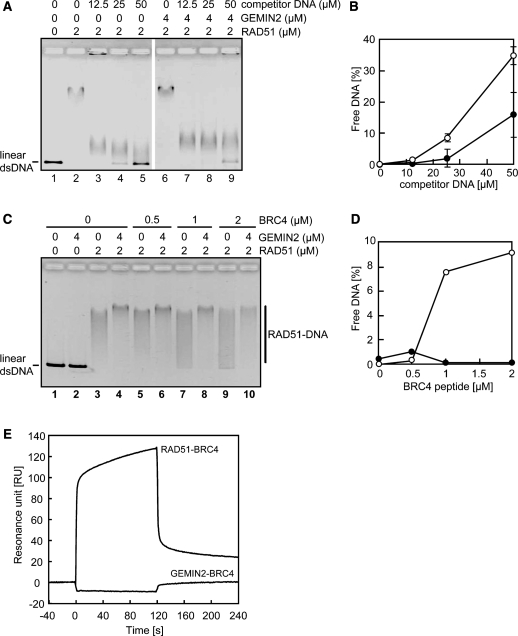
To test whether GEMIN2 inhibited the dissociation of RAD51 from DNA, we generated the RAD51–DNA complex and subsequently measured the stability of this complex in the presence of excess amounts of either competitor DNA (975–1246 bp) (Figure 2A and B) or the BRCA2-derived BRC4 polypeptide (Figure 2C and D), which is known to disassemble the RAD51–DNA filament (43–46). In these assays, we used ϕX174 linear dsDNA (5386 bp) associated with RAD51 as the RAD51–DNA complex. We evaluated the stability by measuring the amount of free dsDNA using agarose gel electrophoresis. Excess amounts of competitor DNA disrupted the RAD51–DNA complex, as evidenced by the fact that substantial amounts of free ϕX174 linear dsDNA were detected, as the amount of competitor DNA was increased (Figure 2A, lanes 2–5). The disruption of the RAD51–DNA complex was significantly suppressed by the addition of GEMIN2 (Figure 2A, lanes 6–9 and Figure 2B), indicating that GEMIN2 may inhibit the RAD51–DNA dissociation. We also analyzed the impact of the BRC4 polypeptide on the RAD51–DNA complex with and without GEMIN2. The BRC4 polypeptide dissociated free RAD51 from ϕX174 linear dsDNA. This inhibitory effect of BRC4 on the RAD51–DNA complex was significantly reversed by the addition of GEMIN2 (Figure 2C and D). SPR analysis revealed that the BRC4 polypeptide did not bind to GEMIN2 (Figure 2E), eliminating the possibility that GEMIN2 masked the BRC4 polypeptide from the RAD51–BRC4 interaction. We therefore conclude that GEMIN2 facilitates the formation of the RAD51–DNA complex, probably by inhibiting the dissociation of RAD51 from damaged DNA sites.
Figure 2.
GEMIN2 stabilizes the RAD51–DNA filament. (A) Complex formation of RAD51 and dsDNA was evaluated by electrophoresis of unbound free DNA in agarose gel. Increased concentrations of competitor DNA were incubated with 2 µM of RAD51 in the presence or absence of 4 µM of GEMIN2, prior to the addition of ϕX174 dsDNA. (B) Quantification of results from panel A. The relative amounts of RAD51-unbound DNA are shown. Closed and open circles indicate experiments with and without GEMIN2. Average values and standard deviation were calculated from three independent experiments. (C) Complex formation of RAD51 and dsDNA in the presence of the BRC4 polypeptide. The experiments were done as described for panel A. (D) Quantification of the data from panel C. (E) Surface plasmon resonance analysis. The RAD51- or GEMIN2-conjugated sensor chips were used. Sensorgrams of RAD51-BRC4 and GEMIN2-BRC4 interactions are presented. The BRC4 polypeptide concentration was 10 µM. Time 0 of the horizontal axis indicates the initiation time of the peptide injection.
GEMIN2 may enhance RAD51-mediated homologous pairing and strand exchange in vitro
To verify that GEMIN2 plays a role in HR, we tested the effect of GEMIN2 on an in vitro HR reaction: RAD51-mediated homologous pairing (Figure 3A). To this end, we measured the formation of the D-loop structure between a 32P-labeled 50-mer oligonucleotide and superhelical dsDNA. Consistent with a previous observation (47), RAD51 alone promoted homologous pairing (Figure 3A and B, lane 2). The addition of GEMIN2 significantly increased the amount of D-loop in a dose-dependent manner (Figure 3A and B, lanes 3–5). Therefore, we conclude that GEMIN2 promotes RAD51-mediated homologous pairing.
Figure 3.
GEMIN2 enhances the homologous-pairing and strand-exchange activities of RAD51. (A) GEMIN2 stimulates the RAD51-mediated homologous pairing. RAD51 and GEMIN2 were incubated at 37°C for 5 min. After this incubation, a 32P-labeled 50-mer oligonucleotide (1 µM) was added, and the samples were further incubated at 37°C for 5 min. The reactions were then initiated by the addition of the pB5Sarray superhelical dsDNA (20 µM), and were continued at 37°C for 30 min. The reactions were stopped by the addition of SDS and proteinase K, and the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1× TAE buffer. The gels were dried, exposed to an imaging plate and visualized using an FLA-7000 imaging analyzer (Fujifilm, Tokyo, Japan). The reactions were conducted with 100 nM RAD51 in the presence of increasing amounts of GEMIN2. A schematic representation of the homologous pairing is presented on the top of the panel. (B) Graphic representation of the experiments shown in panel A. Amounts of D-loops relative to that of the RAD51 alone are plotted. The average values of three independent experiments are shown with the SD values. (C) Schematic representations of strand-exchange reactions. (i) The RAD51-ssDNA complexes are formed before the RPA addition. (ii) The RPA-ssDNA complexes are formed before the RAD51 addition. (D) Strand-exchange reactions where RPA was added to ϕX174 circular ssDNA (20 µM), after [lanes 1–4, panel C(i)] or before [lanes 5–8, panel C(ii)] incubation of the ssDNA with RAD51. Strand-exchange reactions were initiated by the addition of ϕX174 linear dsDNA (20 µM) and (NH4)2SO4 (100 mM), and incubated for 30 min. The deproteinized products of the reaction mixtures were separated using 1% agarose gel electrophoresis and were visualized by SYBR Gold (Invitrogen) staining. (E) GEMIN2 enhances strand exchange. ssDNA was incubated with RPA and then with RAD51 [panel C(ii)]. The indicated amounts of GEMIN2 were pre-incubated with RAD51, and subsequently added to the reaction mixture containing the ssDNA and RPA. (F) Quantification of panel E. The band intensities of the joint molecule (jm) products were quantified as the percentage of the entire input of the ssDNA and dsDNA molecules. Average values of three independent experiments are shown with standard deviation values.
We next focused on RAD51-mediated strand exchange. It is known that the effect of RPA on in vitro strand exchange reaction differs depending on the experimental method used (Figure 3C) (48,49). As expected, when RPA was added after the formation of RAD51–ssDNA filaments, it supported to promote the RAD51-mediated strand exchange in vitro [Figure 3C(i) and D, lanes 1–4]. On the other hand, when RPA was pre-incubated with ssDNA before the addition of RAD51 to ssDNA [Figure 3C(ii)], RPA did not stimulate the RAD51-mediated strand exchange, because RPA competed with RAD51 in ssDNA binding (Figure 3D, lanes 5–8). We found that GEMIN2 enhanced the RAD51-mediated strand exchange in the latter case (Figure 3E and F). In nucleus, RAD51 is considered to compete with RPA in the ssDNA binding at the DSB site, and this suppressive effect by RPA may be overcome by the RAD51 mediators. Therefore, GEMIN2 may function as a RAD51 mediator to support the RAD51–ssDNA binding at the DSB sites in the presence of RPA. Similar enhancing effect was previously reported for yeast RAD52 (50–52), a key enzyme of HR as well as for some mammalian RAD51 paralog proteins (24).
GEMIN2 is essential for cellular proliferation
To determine the function of GEMIN2 in vivo, we had to disrupt the GEMIN2 gene. We began by designing gene-targeting constructs to delete the two exons that encode residues 86 to 101 of the protein (Supplementary Figure S3A and B). However, we failed to generate GEMIN2−/− cells from the chicken DT40 B cell line (36), suggesting that the inactivation of GEMIN2 is lethal to cells. We therefore conditionally disrupted the GEMIN2 gene using a human GEMIN2 transgene under the control of a tetracycline-(doxycycline) repressible promoter (tetGEMIN2 transgene). We independently isolated two GEMIN2−/−tetGEMIN2 clones and confirmed that they showed the same phenotype. We then examined the GEMIN2−/−tetGEMIN2 cells for their levels of endogenous GEMIN2 transcripts and human GEMIN2 protein (Supplementary Figure S3C and D) as well as their growth properties (Supplementary Figure S4A) in the absence and presence of doxycycline. Without doxycycline, the GEMIN2−/−tetGEMIN2 cells tended to grow more slowly than the WT cells, presumably because human GEMIN2 is not fully functional in chicken cells (compare Supplementary Figure S4A). Two days after the addition of doxycycline, the level of human GEMIN2 protein had been dramatically reduced—down below the endogenous level of the HeLa cells (Supplementary Figure S3D)—and the GEMIN2−/−tetGEMIN2 cells had begun to stop proliferating (Supplementary Figure S4A). Over time, fractions of the GEMIN2−/−tetGEMIN2 cells accumulated in the G1 and G2 phases (Supplementary Figure S4B). These cells showed a moderate increase in the number of un-repaired DSBs, as measured by the number of spontaneous chromosomal breaks (Supplementary Table S1) (53). By Day 5, most of the GEMIN2−/−tetGEMIN2 cells had stopped growing and exhibited a large population of dead cells. These observations indicate that GEMIN2 is essential for the cell viability.
GEMIN2 facilitates HR-mediated DSB repair in DT40 cells
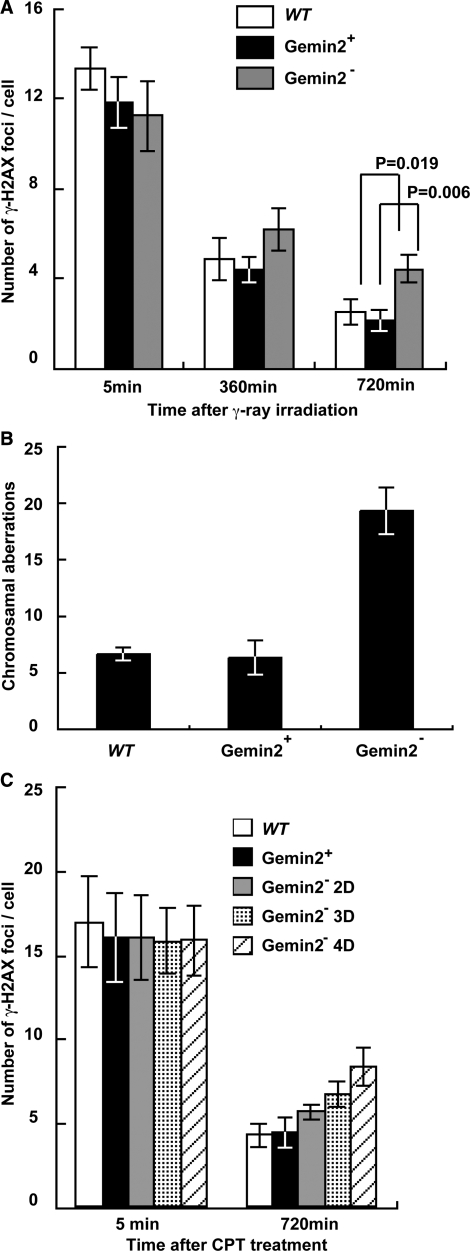
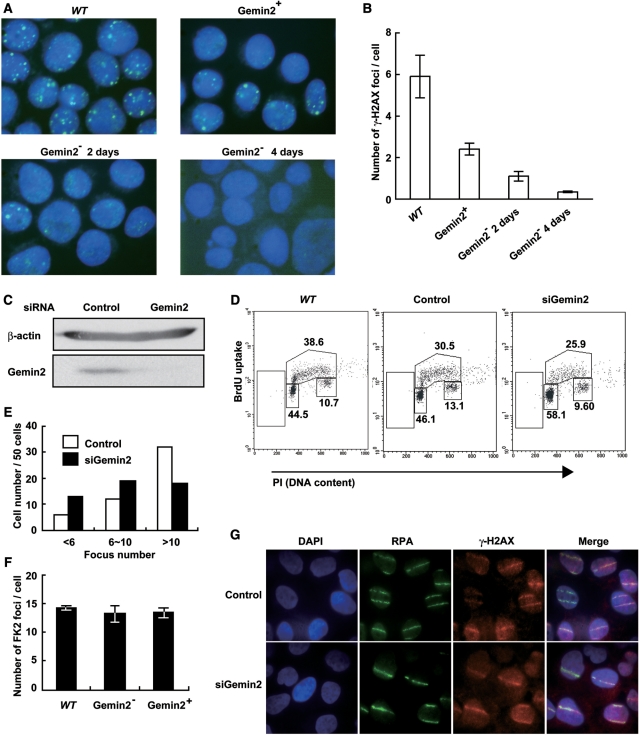
To assess the role of GEMIN2 in DSB repair, we exposed GEMIN2−/−tetGEMIN2 cells to γ-rays, and measured the kinetics of DSB repair by counting the number of phosphorylated histone H2AX (γ-H2AX) foci (Figure 4A) as well as by counting chromosomal breaks in the subsequent M phase (Figure 4B). To this end, 4 days after adding the doxycycline, we exposed an asynchronous population of cells to 4Gy γ-rays. The numbers of induced γ-H2AX foci were comparable irrespective of GEMIN2 expression (Figure 4A), indicating that GEMIN2 does not affect the kinetics of γ-induced DSB formation. However, at 6 and 12 h after irradiation, larger numbers of DSBs were left unrepaired in the GEMIN2-deficient cells than in the GEMIN2+ cells, indicating that GEMIN2 promotes DSB repair. We also harvested mitotic cells after 6 h and measured the number of chromosomal breaks (Figure 4B). Note that cells irradiated in the S to G2 phases can enter the M phase within 6 h after γ-irradiation (54). GEMIN2-deficient cells exhibited a ∼3-fold increase in the number of induced chromosome breaks in comparison with WT and GEMIN2+ GEMIN2−/−tetGEMIN2 cells. Since DSB repair is primarily carried out by HR in DT40 cells (55), the compromised DSB repair in the S to G2 phases raises the possibility that GEMIN2 is required for HR-mediated DSB repair.
Figure 4.
Defective repair of DSBs induced by γ-rays and camptothecin in GEMIN2-deficient cells. (A) Time line of γ-H2AX foci per cell at the indicated time after treatment with 4Gy γ-rays. Results for WT cells and for GEMIN2−/−tetGEMIN2 untreated (GEMIN2+) and treated (GEMIN2−) with doxycycline for 4 days are shown. Error bars represent standard deviation. Statistical analysis was performed using the t test. (B) Ionizing-radiation-induced chromosome aberrations. Mitotic cells were harvested at 6 h after exposure of an asynchronous population of cells to 4Gy γ-rays. Irradiated cells were treated with colcemid for the last 3 h before harvest to enrich the mitotic cells. Results from WT and GEMIN2−/−tetGEMIN2 cells untreated with doxycycline (GEMIN2+) and treated (GEMIN2−) with doxycycline for 4 days are shown. The number of chromosomal aberrations per 50 mitotic cells is shown on the Y-axis. Error bars represent standard deviation. (C) Time course of the formation of γ-H2AX foci per cell at the indicated times after treatment with camptothecin (CPT, 100 ng). Results for WT cells and for GEMIN2−/−tetGEMIN2 untreated (GEMIN2+) and treated (2 days, GEMIN2− 2D; 3 days, GEMIN2− 3D; 4 days, GEMIN2− 4D) with doxycycline are shown. CPT is supposed to induce DSBs only in cells at the S phase, and the number of γ-H2AX foci was counted only in γ-H2AX foci positive cells. Error bars represent standard deviation.
To further examine HR-dependent DSB repair, we monitored the repair kinetics by measuring the disappearance of phosphorylated histone H2AX (γ-H2AX) foci following the exposure of cells to camptothecin, a topoisomerase I poison (Figure 4C). Camptothecin stabilizes the single strand cleavage-topoisomerase I complex, which interferes with replication and thereby induces DSBs in one of the sister chromatids [reviewed in (1)] The resulting DSBs are repaired by HR with the intact sister chromatid (56–59). To monitor HR-dependent repair of camptothecin-induced DSBs, we treated GEMIN2−/−tetGEMIN2 cells with doxycycline for 2–4 days. Similar numbers of γ-H2AX foci appeared at 5 min after camptothecin treatment in GEMIN2-deficient cells (Figure 4C), indicating that the loss of GEMIN2 does not affect camptothecin-induced DSBs. Remarkably, the camptothecin-induced γ-H2AX foci disappeared with retarded kinetics in GEMIN2-depleted cells, in comparison with GEMIN2+ cells. Hence, GEMIN2 may facilitate HR-mediated DSB repair.
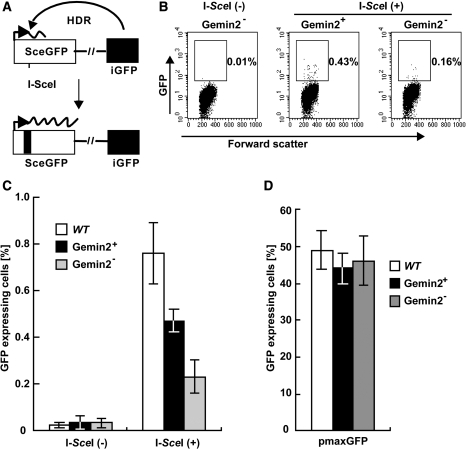
To directly assess the role of GEMIN2 in HR, we measured the HR-dependent repair of site-specific DSBs generated by the I-SceI nuclease acting on an artificial substrate inserted into the OVALBUMIN locus (60,61). The efficiency of HR-dependent repair can be evaluated by measuring the percentage of green fluorescent protein (GFP)-positive cells (Figure 5A). The percentage of GFP+ cells was three times lower in GEMIN2-deficient than in WT and GEMIN2+ GEMIN2−/−tetGEMIN2 cells (Figure 5B and C), though the efficiency of transient transfection in these cells was comparable (Figure 5D). In summary, this observation, as well as the defective repair of DSBs induced by γ-rays and camptothecin, indicates that GEMIN2 indeed facilitates HR-dependent DSB repair.
Figure 5.
HR-mediated DSB repair is impaired in GEMIN2-deficient cells. (A) Schematic of the HR-dependent DSB repair assay. Transient expression of the I-SceI restriction enzyme generates a specific DSB in an artificial HR substrate, DR-GFP, inserted into the endogenous OVALBUMIN locus. The SceGFP gene does not produce functional green fluorescent protein (GFP) because of a frame-shift mutation at the I-SceI recognition sequences. Functional GFP is produced only when the recognition sequences are eliminated by HR with the downstream internal GFP fragment (iGFP). (B) Distribution of GEMIN2−/−tetGEMIN2 cells with or without GFP expression. The population of the cells for GFP expression was measured at 12 h after transfection of I-SceI-expression plasmid [I-Sce(+)] or control plasmid [I-Sce(−)] in GEMIN2−/−tetGEMIN2 cells untreated (GEMIN2+) and treated (GEMIN2−) with doxycycline for 3 days. (C) Percentage of cells expressing GFP in the indicated cell line was calculated. Data shown are the mean of three experiments. Error bars indicate standard deviation. (D) Transient transfection efficiency of an intact GFP expression plasmid in the indicated cell lines. Error bars indicate standard deviation.
GEMIN2 promotes assembly of RAD51 at resected single-strand DSB tails
To determine at what stage GEMIN2 acts in HR, we analyzed the recruitment of RAD51 to γ-ray-induced DSBs 2 and 4 days after the addition of doxycycline. Clear RAD51 foci appeared in WT cells 3 h after γ-ray irradiation (Figure 6A). In contrast, the number of RAD51 foci decreased in GEMIN2-deficient cells (Figure 6A and B). This impaired recruitment is not due to a decrease in the amount of RAD51, as evidenced by the stable RAD51 protein level in the GEMIN2-depleted cells (Supplementary Figure S3D). This suggests that GEMIN2 increases the efficiency of HR by promoting the assembly of RAD51 at DNA damage sites.
Figure 6.
IR-induced Rad51 subnuclear foci are decreased in the absence of GEMIN2. (A) Immunostaining of irradiated WT and GEMIN2−/−tetGEMIN2 cells with the Rad51 antibody. Cells were fixed 3 h after irradiation of 4Gy γ-ray. GEMIN2−/−tetGEMIN2 cells untreated (GEMIN2+) and treated (GEMIN2−, 2 or 4 days) with doxycycline are shown. (B) Quantification of Rad51 foci in individual cells of the indicated genotype. Data shown are the means of three experiments. Error bars indicate standard deviation. (C) U2OS cells were transfected with siRNA oligos specific to GEMIN2 or with control siRNA. Cell extract was prepared at 48 h and immunoblotted as indicated. (D) Cell-cycle distribution 48 h after addition of control (GFP) and GEMIN2-specific siRNAs. Cells were pulse-labeled with BrdU for 10 min and subsequently stained with FITC-conjugated anti-BrdU antibody (Y-axis, log scale) and propidium iodide (PI) (X-axis, linear scale). The upper gate indicates cells incorporating BrdU (S-phase); the lower middle gate indicates G1 cells; and the lower right gate displays G2/M cells. The sub G1 fraction (lower left gates) indicates dead cells. The numbers given in each gate indicate the percentages of gated events. (E) Rad51 focus formation in human cells depleted of GEMIN2 by siRNA. Cells were immunostained 3 h after 4Gy IR exposure. The histogram shows the percentage of cells displaying less than 6 RAD51 foci, 6 to 10 foci, and more than 10 foci per cell. >100 cells were analyzed for each data point. (F) FK2-conjugated ubiquitin focus formation following IR exposure is normal in GEMIN2-depleted DT40 cells. GEMIN2 depletion was performed by treating GEMIN2−/−tetGEMIN2 cells with doxycycline for 4 days. Error bars indicate standard deviation. (G) The RPA ssDNA binding protein normally accumulates at DSBs 60 min after exposure to a 405 nm pulse laser in HeLa cells treated with siRNA against GEMIN2 or control siRNA for 48 h.
To test the reproducibility of this result in human cells, we measured γ-ray-induced RAD51 foci in U2OS cells treated with siRNA against GEMIN2. The siRNA treatment diminished the level of GEMIN2 at least 3-fold (Figure 6C) without significantly affecting the cell cycle (Figure 6D). The depletion of GEMIN2 by siRNA in U2OS cells also compromised the formation of RAD51 foci following γ-ray irradiation (Figure 6E). These results confirm that GEMIN2 is required for the efficient recruitment of RAD51 to sites of DNA damage in vertebrate cells.
To explore the cause of the reduced RAD51 focus formation, we analyzed conjugated ubiquitin foci at γ-ray-induced DSBs, and RPA accumulation at laser-induced DSB sites in DT40 cells (7,8,62). We previously showed that UBC13 E2 ubiquitin ligase is required for the processing of DSBs, as evidenced by the loss of conjugated ubiquitin foci and the severely attenuated accumulation of RPA at laser-induced DSBs in UBC13−/− cells (8). In contrast, GEMIN2-depleted and WT cells exhibited the same numbers of conjugated ubiquitin foci (Figure 6F). Similarly, RPA was efficiently recruited to laser-induced DSB sites 60 min after treatment in GEMIN2-depleted and WT cells (Figure 6G). Collectively, these data suggest that the HR defect in GEMIN2-deficient DT40 cells is due to a failure to efficiently load RAD51 at resected DSBs.
DISCUSSION
GEMIN2 forms a complex with SMN and plays a critical role in the motor neuron survival, by controlling the assembly of spliceosomal snRNP (32). Here we have identified an unexpected function of GEMIN2 in HR. We initially identified GEMIN2 by using a two-hybrid screening with RAD51 as bait (Supplementary Figure S1A). We confirmed that GEMIN2 binds RAD51 in vitro (Supplementary Figure S1C) and forms a complex with RAD51 in vivo (Supplementary Figure S1D). More importantly, we found that purified GEMIN2 promotes the assembly of RAD51 on ssDNA and dsDNA (Figure 1). Consistent with these biochemical observations, GEMIN2 depletion reduced the amount of RAD51 focus formation in human and chicken DT40 cells, but not the accumulation of the ssDNA binding protein RPA at DNA damage sites (Figure 6). Purified GEMIN2 enhanced RAD51-mediated homologous pairing and strand exchange in vitro (Figure 3). This in vitro observation is also in agreement with the phenotypic analysis of GEMIN2 depleted cells, which showed the diminished efficiency of HR-mediated DSB repair (Figures 4 and 5). These results demonstrate that GEMIN2 is a novel RAD51 mediator, as are BRCA1, BRCA2 and the five RAD51 paralogs. Thus, GEMIN2 has dual functions: the regulation of spliceosomal snRNP formation and the control of RAD51-dynamics at DNA damage.
Among proteins involved in splicing, we previously found that human PSF, which was originally identified as a component of spliceosomes (63), directly interacts with RAD51, and modulates the RAD51-mediated homologous pairing and strand exchange in vitro (64). PSF was also identified as a DSB repair protein (65), and found in a screen for proteins that interact with the RAD51 paralogs (66). In the present study, we found that another splicing factor, GEMIN2, also regulates the RAD51-mediated homologous pairing and strand exchange in vitro. These findings may imply developmental link between the splicing and homologous-recombination reactions in cells. Both GEMIN2 and PSF directly interacted with RAD51, but their consequences are different. GEMIN2 enhances the RAD51–DNA complex formation by suppressing the RAD51 disassembly from the complex. On the other hand, PSF does not inhibit the RAD51 disassembly, but modulates the RAD51-mediated homologous pairing and strand exchange, probably through its DNA-binding activity (64,67). These differences between GEMIN2 and PSF in the RAD51-mediated recombination reactions may reflect their roles in HR and recombinational repair of DSB in cells.
The proteins involved in HR promote the action of RAD51 in two different manners. First, BRCA1, BRCA2 and the five RAD51 paralogs promote the nucleation of RAD51 on DNA (17). Defects in these mediators decrease the level of RAD51 focus formation at DNA damage sites. Second, mammalian RAD54 and RAD51AP1 stimulate RAD51-mediated homologous pairing (11–15). Our study reveals that, unlike the first or second type of HR factors, GEMIN2 enhances the RAD51–DNA complex formation probably by inhibiting the dissociation of RAD51 from the RAD51 polymer-DNA complex. It remains elusive how GEMIN2 regulates RAD51–DNA dynamics. In contrast to RAD51 mediators previously reported, GEMIN2 is a unique RAD51 mediator, in the sense that GEMIN2 can promote the loading of RAD51 onto DNA without directly associating with the damaged DNA (Figure 1H). Taking these points together, one possible scenario for the action of GEMIN2 is that it might transiently bind to RAD51 polymerized on DNA and might thereby prevent RAD51 from dissociating from DNA.
In bacteria, the RAD51 homologue, RecA, is the central enzyme, which catalyzes homologous pairing and strand exchange. Modulators that regulate the RecA recombinase activity at many levels have been found (68). Among them, DinI, which functions in the SOS response in bacteria (69–71), reportedly enhanced stability of the RecA filament (72). DinI binds to the RecA filament without significantly inhibiting DNA binding and strand exchange by RecA (72,73). These biochemical characteristics of bacterial DinI are reminiscent of those of GEMIN2.
In the present study, we showed that GEMIN2 enhances the RAD51–ssDNA complex formation by inhibiting the RAD51 dissociation from the complex. This is consistent with in vivo observations that GEMIN2 stimulates RAD51 assembly onto the ssDNA at the DSB sites. However, our biochemical studies also showed that GEMIN2 enhances formation of the RAD51–dsDNA complex, which is considered to be inactive for homologous pairing. This apparent discrepancy between in vitro and in vivo observations may be due to the chromatin structure in cells. In nuclei, ssDNA regions produced at the DSB sites may be exposed from chromatin, but, in contrast, dsDNA remains in the higher ordered chromatin structure. The enhancement of the RAD51–dsDNA binding may be suppressed by the chromatin structure in cells, therefore, the stimulation of the RAD51–ssDNA binding by GEMIN2 may dominantly function in chromatin context.
Vertebrate cells require more sophisticated control of RAD51-mediated HR than do yeast species, because in vertebrate cells, HR must (i) undergo homology search for a considerably longer distance than in yeast, (ii) occur only in the S to G2 phases to prevent hetero-allelic HR and (iii) probably not interfere with NHEJ, even in the S to G2 phases (3). To meet these requirements, a greater number of RAD51 mediators have evolved for vertebrate cells. In the future, we must address the distinctive and collaborative functions of the growing number of RAD51 mediators, i.e. BRCA1, BRCA2, RAD51B, RAD51C, RAD51D, XRCC2, XRCC3 and GEMIN2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to H.K. and S.T., partial), and grants from the Special Coordination Funds for Promoting Science and Technology, Collaborative Research among Japan, China, Korea and Vietnam on Technical Development to Assess Environmental Pollution Prevention (to Y.Q. and S.T.). H.K. is supported as a research fellow in the Waseda Research Institute for Science and Engineering. Funding for open access charge: Waseda University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Professor Paul Crocker and Dr Diana Otto (University of Dundee) for providing the Gateway plasmids along with advice on the Gateway system.
REFERENCES
- 1.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 2.Sung PA, Libura J, Richardson C. Etoposide and illegitimate DNA double-strand break repair in the generation of MLL translocations: new insights and new questions. DNA Repair. 2006;5:1109–1118. doi: 10.1016/j.dnarep.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Takeda S, Nakamura K, Taniguchi Y, Paull TT. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol. Cell. 2007;28:351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 7.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Thacker J, Zdzienicka MZ. The mammalian XRCC genes: their roles in DNA repair and genetic stability. DNA Repair. 2003;2:655–672. doi: 10.1016/s1568-7864(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 10.Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair. 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 12.Mazina OM, Mazin AV. Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+ J. Biol. Chem. 2004;279:52042–52051. doi: 10.1074/jbc.M410244200. [DOI] [PubMed] [Google Scholar]
- 13.Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, van Steeg H, van Benthem J, Wassenaar E, Baarends WM, et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 18.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatanaka A, Yamazoe M, Sale JE, Takata M, Yamamoto K, Kitao H, Sonoda E, Kikuchi K, Yonetani Y, Takeda S. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol. Cell. Biol. 2005;25:1124–1134. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonetani Y, Hochegger H, Sonoda E, Shinya S, Yoshikawa H, Takeda S, Yamazoe M. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 2005;33:4544–4552. doi: 10.1093/nar/gki766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67:9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol. 2008;6:e88. doi: 10.1371/journal.pbio.0060088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murayama Y, Kurokawa Y, Mayanagi K, Iwasaki H. Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases. Nature. 2008;451:1018–1021. doi: 10.1038/nature06609. [DOI] [PubMed] [Google Scholar]
- 24.Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 26.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 27.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 28.Shivji MK, Davies OR, Savill JM, Bates DL, Pellegrini L, Venkitaraman AR. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 30.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 31.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 33.Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J. Child Neurol. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 37.Ishida T, Takizawa Y, Sakane I, Kurumizaka H. The Lys313 residue of the human Rad51 protein negatively regulates the strand-exchange activity. Genes Cells. 2008;13:91–103. doi: 10.1111/j.1365-2443.2007.01143.x. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka Y, Tawaramoto-Sasanuma M, Kawaguchi S, Ohta T, Yoda K, Kurumizaka H, Yokoyama S. Expression and purification of recombinant human histones. Methods. 2004;33:3–11. doi: 10.1016/j.ymeth.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 39.Iiizumi S, Nomura Y, So S, Uegaki K, Aoki K, Shibahara K, Adachi N, Koyama H. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques. 2006;41:311–316. doi: 10.2144/000112233. [DOI] [PubMed] [Google Scholar]
- 40.Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H, et al. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67:8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 41.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 42.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 43.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 44.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 45.Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc. Natl Acad. Sci. USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nomme J, Takizawa Y, Martinez SF, Renodon-Cornière A, Fleury F, Weigel P, Yamamoto K, Kurumizaka H, Takahashi M. Inhibition of filament formation of human Rad51 protein by a small peptide derived from the BRC-motif of the BRCA2 protein. Genes Cells. 2008;13:471–481. doi: 10.1111/j.1365-2443.2008.01180.x. [DOI] [PubMed] [Google Scholar]
- 47.Tracy RB, Baumohl JK, Kowalczykowski SC. The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes Dev. 1997;11:3423–3431. doi: 10.1101/gad.11.24.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 49.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 50.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 51.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–140. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 52.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 53.Sedelnikova OA, Horikawa I, Redon C, Nakamura A, Zimonjic DB, Popescu NC, Bonner WM. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell. 2008;7:89–100. doi: 10.1111/j.1474-9726.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- 54.Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M, et al. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 57.Adachi N, So S, Koyama H. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 2004;279:37343–37348. doi: 10.1074/jbc.M313910200. [DOI] [PubMed] [Google Scholar]
- 58.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, Taniguchi Y, Murakawa Y, Zeng W, Yokomori K, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol. Cell. Biol. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukushima T, Takata M, Morrison C, Araki R, Fujimori A, Abe M, Tatsumi K, Jasin M, Dhar PK, Sonoda E, et al. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 2001;276:44413–44418. doi: 10.1074/jbc.M106295200. [DOI] [PubMed] [Google Scholar]
- 62.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 2006;25:2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 64.Morozumi Y, Takizawa Y, Takaku M, Kurumizaka H. Human PSF binds to RAD51 and modulates its homologous-pairing and strand-exchange activities. Nucleic Acids Res. 2009;37:4296–4307. doi: 10.1093/nar/gkp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 66.Rajesh C, Gruver AM, Basrur V, Pittman DL. The interaction profile of homologous recombination repair proteins RAD51C, RAD51D and XRCC2 as determined by proteomic analysis. Proteomics. 2009;9:4071–4086. doi: 10.1002/pmic.200800977. [DOI] [PubMed] [Google Scholar]
- 67.Akhmedov AT, Lopez BS. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000;28:3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cox MM. Regulation of Bacterial RecA Protein Function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 69.Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 1998;17:3207–3216. doi: 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yasuda T, Morimatsu K, Kato R, Usukura J, Takahashi M, Ohmori H. Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. EMBO J. 2001;20:1192–1202. doi: 10.1093/emboj/20.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voloshin ON, Ramirez BE, Bax A, Camerini-Otero RD. A model for the abrogation of the SOS response by an SOS protein: a negatively charged helix in DinI mimics DNA in its interaction with RecA. Gene Dev. 2001;15:415–427. doi: 10.1101/gad.862901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lusetti SL, Voloshin ON, Inman RB, Camerini-Otero RD, Cox MM. The DinI protein stabilizes RecA protein filaments. J. Biol. Chem. 2004;279:30037–30046. doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- 73.Yoshimasu M, Aihara H, Ito Y, Rajesh S, Ishibe S, Mikawa T, Yokoyama S, Shibata T. An NMR study on the inter- action of Escherichia coli DinI with RecA-ssDNA complexes. Nucleic Acids Res. 2003;31:1735–1743. doi: 10.1093/nar/gkg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.