Abstract
DNA methylation and histone modifications have essential roles in remodeling chromatin structure of genes necessary for multi-lineage differentiation of mammary stem/progenitor cells. The role of this well-defined epigenetic programming is to heritably maintain transcriptional plasticity of these loci over multiple cell divisions in the differentiated progeny. Epigenetic events can be deregulated in progenitor cells chronically exposed to xenoestrogen or inflammatory microenvironment. In addition, epigenetically mediated silencing of genes associated with tumor suppression can take place, resulting in clonal proliferation of undifferentiated or semidifferentiated cells. Alternatively, microRNAs that negatively regulate the expression of their protein-coding targets may become epigenetically repressed, leading to oncogenic expression of these genes. Here we further discuss interactions between DNA methylation and histone modifications that have significant contributions to the differentiation of mammary stem/progenitor cells and to tumor initiation and progression.
Mammary gland stem cells and their progeny display characteristic DNA methylation and histone modification patterns. Carcinogens and inflammation can disrupt such epigenetic programs, leading to tumorigenesis.
In the breast, a subpopulation of cells located in the basal layer of terminal end ducts have been implicated as stem cells (Dontu et al. 2003). A study using serial transplantation of this cell population in mice unequivocally showed their stem/progenitor functions (Stingl et al. 2006). These slowly dividing cells have the capacity of self-renewal and, in response to hormonal stimuli, give rise to transient populations that may undergo terminal differentiation into different epithelial lineages (Péchoux et al. 1999; Gudjonsson et al. 2002; Dontu et al. 2004, 2005; Villadsen et al. 2007). Differential transcriptional programs are used to maintain lineage specification and homeostasis of differentiated cells. Estrogen receptor α (ERα)-negative progenitors usually differentiate into myoepithelial cells, which form the basal layer of mammary ducts whereas other progenitors give rise to luminal epithelial cells, some of which appear to be ERα-positive (Dontu et al. 2004, 2005).
The maintenance of stem/progenitor cells and their differentiation fate follows a well-defined epigenetic program. Without altering nucleotide sequences or DNA copy-number, the event heritably transmits transcriptional information from progenitors to their differentiated progeny. This epigenetic program is coordinately regulated by DNA methylation, histone modifications, and noncoding RNAs (Jaenisch and Bird 2003; Jones and Baylin 2007; Feinberg 2008; Suzuki and Bird 2008; Cedar and Bergman 2009; Davalos and Esteller 2009; Guil and Esteller 2009). Increasing evidence has indicated that spatiotemporal interactions among different epigenetic components play essential roles in modulating transcriptional activation or deactivation. In a permissive environment, the chromatin structure of a target locus exists in an open conformation that allows transcription factors and coregulators to bind to their cognate sequences and initiate transcription (Cairns 2009). In a nonpermissive environment, these loci are inaccessible to these transcription factors, resulting in transcriptional silencing. DNA methylation and histone modifications are known to regulate this transcriptional plasticity, which is necessary for normal development and maintenance of mammary epithelial lineages (Dontu et al. 2005; Bloushtain-Qimron et al. 2009). Deregulation of these epigenetic programs may result in clonal proliferation of transformed progenitor cells, also called cancer-initiating cells. Subtypes of these aberrant cells may develop into diverse groups of epithelial tumors, including luminal A and B (ERα-positive), basal (ERα-negative), ERBB2-overexpressing, and normal-like phenotypes (Sorlie et al. 2001).
In this article, we discuss the importance of DNA methylation and histone modifications in modulating chromatin structure for gene transcription. The emerging role of noncoding RNA in cell differentiation and neoplasm is discussed in a preceding article. Potential uses of DNA methyltransferases and histone deacetylase inhibitors in epigenetic therapies are extensively reviewed elsewhere (Balch et al. 2005; Lo and Sukumar 2008; Batty et al. 2009). Here we focus on describing the contributions made by DNA methylation and histone modifications to mammary epithelial differentiation. We also highlight the significance of epigenetic deregulation in breast tumorigenesis.
DNA METHYLATION
Methylation Variations in Normal Tissues and Mammary Epithelial Subtypes
The human genome contains 28 million CpG dinucleotides (Table 1). Approximately 10% (2 million) of these dinucleotides are primarily distributed in GC-rich repetitive sequences (e.g., ribosomal DNA and α satellites) and single-copy CpG islands (Antequera and Bird 1993, 1999). These CpG islands (1 to 2-kb) are usually associated with protein-coding genes or noncoding RNAs, with the majority located in the promoter regions (70%) and less within intergenic regions (10%) (Jaenisch and Bird 2003; Jones and Baylin 2007). These GC-rich sequences are targets for enzymatic modifications by DNA methyltransferases (DNMTs), which add a methyl group to the fifth position of cytosine of a CpG dinucleotide and convert it into methylcytosine. De novo DNA methylation is usually carried out by DNMT3A and DNMT3B whereas maintenance of methylation is mediated by DNMT1 (Bestor 2000). Recent evidence suggests that DNMT3A and DNMT3B also cooperate with DNMT1 for the maintenance function, which serves to transmit methylated DNA patterns from parental to daughter cells (Jones and Liang 2009).
Table 1.
Information of CpG dinucleotides in the human genomea
| Total number of CpG dinucleotides | 28,340,937 |
| CpG dinucleotides within CpG islands | 2,107,414 |
| CpG dinucleotides outside of CpG islands | 26,233,523 |
| Total number of CpG islands | 27,934 |
| CpG islands within ±1 kb TSS | 14,909 |
| Gene-associated CpG island outside of ±1 kb TSS | 7,093 |
| CpG islands not associated with known genes | 5,932 |
| Total bp covered by promoter CpG islands | 13,992,078 |
| Total bp covered by gene-body CpG islands | 3,700,617 |
| Total bp covered by intergenic CpG islands | 3,663,451 |
aBased on the UCSC Human Genome Assembly hg18.
Repetitive sequences, such as Alu, LINE, and α satellites, are usually methylated in the normal genome (Jaenisch and Bird 2003; Jones and Baylin 2007). This acquisition of DNA methylation may act to repress the transcription or retrotransposition of repetitive sequences. However, CpG islands near the promoters of genes, with the exception of imprinted and inactive X-chromosome loci, are thought to remain unmethylated in the genome (Jaenisch and Bird 2003; Jones and Baylin 2007). This prevailing notion that CpG islands are normally “protected” from DNA methylation has been challenged by detailed studies on candidate loci or global screening of the genome (see examples in Table 2 for different technologies used for methylation analysis). Together, the findings indicate that normal methylation could occur in 5%–15% of human CpG islands not associated with imprinting or X-inactivation (Weber et al. 2005; Shen et al. 2007; Weber et al. 2007). In this regard, tissue-specific DNA methylated regions are present in the genome and have been proposed to repress transcripts during cell differentiation and organogenesis (Futscher et al. 2002; Ohgane et al. 2005; Brena et al. 2006b). This differential methylation may contribute to phenotypic variations of individuals, most dramatically shown in studies of monozygotic twins (Fraga et al. 2005). DNA methylation also changes subtly in normal aging cells, with a progressive loss of methylation content in repetitive sequences, but a progressive increase of density in selected CpG islands (Issa et al. 1994; Issa 2003).
Table 2.
Summary of candidate gene and genome-wide techniques for DNA methylation analysisa
| Categories | Techniques |
|---|---|
| Candidate gene, cloning, and gel-based approaches | Bisulfite sequencing; MSP (methylation-specific PCR); COBRA (combined bisulfite restriction analysis); MethyLight; Pyrosequencing; MassARRAY; MS-AP PCR (methylation-sensitive arbitrarily primed PCR) MSRF (methylation-sensitive restriction fingerprinting); MS-RDA (methylation-sensitive representational analysis); and RLGS (restriction landscape genome scanning) |
| Microarray-based approaches | DMH (differential methylation hybridization): MTA (methylation tissue array); MSO (methylation-specific oligonucleotide); HELP (HpaII tiny fragment enrichment by ligation-mediated PCR); AIMS (amplification of intermethylated sites); MSNP (methylation single nucleotide polymorphism chip-based method); MMASS (microarray-based methylation assessment of single sample); PMAD (promoter-associated methylated DNA amplification); MSDK (methylation-specific digital karyotyping) MIAMI (microarray-based integrated analysis of methylation); MCAM (methylated CpG island amplification and microarray); MeDIP-chip (methylated DNA immunoprecipitation on microarray); MIRA-chip (methylated-CpG island recovery assay on microarray); McrBC; MethylScope; Pharmacologic unmasking analysis; and Infinium BeadArray |
| Next generation sequencing | RRBS (Reduced representation bisulfite sequencing); BS-seq; MethylC-seq; Padlock probes coupled with whole-genome bisulfite sequencing; MRE-seq; HELP- seq; MeDIP-seq; and MIRA-seq |
aDescriptions of these techniques are provided in review articles (Balch et al. 2008; Brena et al. 2006b; Chang et al. 2008; Zuo et al. 2009).
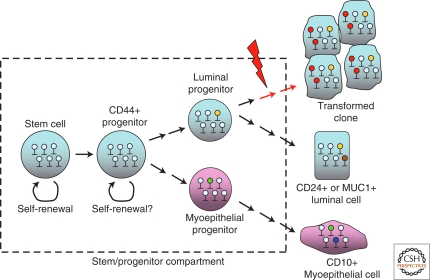
Cell type-specific methylation patterns are similarly found in the mammary gland. Polyak and colleagues (Bloushtain-Qimron et al. 2008, 2009) used the methylation-specific digital karyotyping (MSDK) method to determine DNA methylation profiles in subtypes of mammary epithelial cells. Based on the characteristics of cell surface markers, four different types—myoepithelial (CD10+), luminal epithelial (CD24+ and MUC1+), and progenitor (CD44+) were isolated for methylation profiling (Bloushtain-Qimron et al. 2008). CD44+ cells display stem/progenitor characteristics and, on signaling activation, undergo lineage-specific differentiation. Genes important for pluripotency and self-renewal were hypomethylated and highly expressed in CD44+ cells compared with the other three differentiated cells (Bloushtain-Qimron et al. 2008). Examples are SUZ12 gene targets in which promoter methylation was found in CD24+ cells, but not in CD44+ cells. SUZ12, a member of polycomb-repressor complex 2, is known to regulate the transcription of genes essential for maintaining the pluripotency and self-renewal of stem/progenitor cells (Lee et al. 2006; Pasini et al. 2007). Acquired DNA methylation may result in permanent silencing of SUZ12 targets during mammary epithelial differentiation (see Fig. 1). Epigenetic control of gene silencing, marked by promoter methylation, therefore is an essential step of mammary epithelial differentiation.
Figure 1.
Model of mammary gland differentiation and differential accumulation of DNA methylation in normal and transformed epithelial cells. A hierarchy of stem/progenitor cells, analogous to that in the hematopoietic system, may exist in mammary gland. Stem cells undergo self-renewal and differentiation within their compartment. The primitive progenitor cells give rise to myoepithelial and luminal progenitors, which then differentiate into different epithelial subtypes (Note: other cell types, such as alveolar epithelial cells, are omitted in this model). Acquired DNA methylation in promoter CpG islands occurs in genes required for differentiation of progenitor cells. The resulting epigenetic repression (marked in “green” and “yellow” lollipops for myoepithelial and luminal epithelial cells, respectively) is also essential for maintaining the homeostasis of mature epithelial cells. Deregulation of differentiation signaling may disrupt this epigenetic programming (e.g., aberrant acquisition of promoter methylation marked in “red” lollipops), leading to neoplastic transformation and clonal proliferation.
DNA Methylation Alterations in Transformed Breast Epithelial Cells
It has been hypothesized that mammary stem/progenitor cells are primary targets for cellular transformation. Compared to terminally differentiated cells that have a high turnover rate, these cells have a long life span and are slowly dividing within the stem/progenitor compartment (Dontu et al. 2003; Cheng et al. 2008). These primitive cells are therefore susceptible to molecular injuries because of persistent exposures to different environmental or inflammatory stimulants (Fenton 2006; Yager and Davidson 2006; Cheng et al. 2008). Environmental chemicals, such as xenoestrogens, can mimic estrogenic actions that aberrantly influence epithelial differentiation in mammary glands (Cheng et al. 2008; Hsu et al. 2009). The action is mediated in part through nuclear hormone receptors (e.g., ERα) that activate or deactivate the transcription of target genes (Cheng et al. 2008; Hsu et al. 2009). Alternatively, xenoestrogens may stimulate phosphorylation of membrane-bound proteins, which subsequently activate different kinase signal transduction pathways that are required for transcriptional regulation (Jensen and Jordan 2003).
Persistent exposure to these environmental chemicals therefore increases breast cancer risk. Animal and epidemiologic studies suggest an imprinting phenomenon in which early exposure to xenoestrogens may promote a carcinogenic process observed later in adult life (Fenton 2006; Maffini et al. 2006). Specifically, long-term exposure to low-dose xenoestrogens may alter epigenetic reprogramming of stem/progenitor cells during epithelial differentiation (Ho et al. 2006; Cheng et al. 2008). This molecular alteration may deregulate transcriptional programs important for epithelial differentiation. Repressive chromatin is subsequently established and sets the stage for heritable silencing of target genes (Cheng et al. 2008; Hsu et al. 2009). De novo DNA methylation occurs and gradually spreads from pre-existing methylated CpG sites to neighboring areas in the genome (Cheng et al. 2008; Hsu et al. 2009). These methylated sites may serve as seeds to propagate the repressive information to the core regions of targeted CpG islands. A ChIP-chip analysis in breast cancer cells using antibodies against methyl-CpG binding domain proteins, which bind like “magnets” to methylated cytosines, identified a large number of hypermethylated CpG islands (Ballestar et al. 2003). More recently, using the methylation-dependent immunoprecipitation on microarray (MeDIP-chip, see Table 2) approach, our study found that ∼0.5% of human CpG islands become hypermethylated in epithelial cells derived from estrogen-exposed mammary progenitors compared with the nonestrogen control cells in an in vitro model (Cheng et al. 2008). Ontological analysis revealed significant methylation enrichment in CpG islands of polycomb protein-regulated targets, which are known to regulate pluripotency and self-renewal of stem/progenitor cells (Bracken et al. 2006).
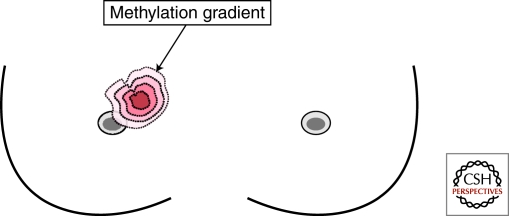
This methylation alteration may precede morphologic transformation of normal breast epithelia and is an early event of tumorigenesis. As a result, a large field defect of premalignant cells is generated in an affected breast area (Fig. 2). Additional accumulation of genetic and epigenetic damages is needed in the affected area to generate invasive carcinomas. Methylation alterations have frequently been found in histologically normal breast tissues adjacent to primary tumor sites (Yan et al. 2003, 2006). Overall methylation levels of two candidate loci, RASSF1A and RUNX3, are significantly higher in these adjacent tissues compared to control samples obtained from individuals undergoing reduction mammoplasties (Yan et al. 2006; Cheng et al. 2008). These methylation levels, however, are lower than those of primary tumor sites, and there is a gradient pattern in which higher degrees of methylation are seen in normal tissues closest to the tumor site compared to those which are 2–4 cm away (Yan et al. 2006; Cheng et al. 2008).
Figure 2.
Model of epigenetic field cancerization occurring in the human breast. Depicted here is a large affected area of methylation gradient in the upper-left side of the breast. Chronic exposures to xenoestrogens may alter epigenetic programs of normal differentiation. Methylation accumulation of promoter CpG islands takes place in silenced genes associated with neoplastic development. A tumor site may sever as the epicenter (depicted in red) from which the highest number of methylated CpG islands and the highest density of methylation within an island are observed in the breast.
DNA Hypermethylation in Epithelial Breast Tumors
Increased methylation of promoter CpG islands and increased numbers of methylated islands are associated with tumor progression (Nephew and Huang 2003; Novak et al. 2009). From a geographic perspective, a tumor site can serve as the “epicenter center” from which a maximum accumulation of promoter methylation can take place in the human breast (see Fig. 2) (Yan et al. 2006; Cheng et al. 2008). These methylation accumulations are known to contribute to different stages of neoplastic development, including initiation, invasion, metastasis, and endocrine resistance. As of today, candidate gene and global screening approaches have now identified >150 loci frequently hypermethylated in breast cancer, including genes encoding for cell cycle regulation (e.g., p16INK4a, p15, and p14ARF), DNA repair (e.g., MLH1 and GST3), tumor suppression (e.g., BRCA1 and VHL), tissue remodeling (e.g., TIMP3 and E-cadherin), and hormone receptor (e.g., ESR1 and ESR2) (Tables 3 and 4). In addition to protein-coding genes, microRNAs with tumor and metastasis suppressor functions also undergo CpG island methylation-associated silencing in transformed cells (Saito et al. 2006; Lujambio et al. 2007, 2008; Hsu et al. 2009).
Table 3.
Summary of genes commonly hypermethylated or hypomethylated in breast cancera
| Functions | Genes |
|---|---|
| Angiogenesis | CXCR4, HIF1A, IL2, IL10, NOS3, and VEGF |
| Apoptosis | APAF1, BAK1, BAX, BIM, BMF, BCL2, BCL2L1, BCL2L2, CASP8, DR4, DR5, FADD, MCL1, SFRP1, PYCARD, TRAIL, and XIAP |
| Binding protein | CEBPD, FABP3 (MDGI), GATA3, GNAL, HRAS, IGFBP3, IGFBP7, LDLRAP1, LRP2, PRKCDBP, and SOCS1 |
| Cell cycle regulation | CCNA1, CCNA2, CCND1, CCND2, CDKN1A, CDKN1B, CDKN1C, CDKN2A, CTPS, DBC1, DNAJC15, GADD45, EPB41L3, GREM1, IGF2, RAD9A, SFN, RNR1, SCGB3A1, and TYMS |
| Proteases, transcripatses, and enzymes | KLK6, KLK10, hTERT, ABL1, AK5, AKT1, DAPK, FLT3, RAF1, TEK, PTGS2, CYP1B1, HS3ST2, HSD17B4, GSTP1, NAT1, SAT2, SULT1A1, SYK, TDH, and WRN |
| DNA repair/detoxify carcinogen | MGMT, MLH1, PRKDC, RPA2, TYMS, XRCC5, and XRCC6 |
| Enhancers and transcription factors | F0XA2, H0XA5, H0XD11, IRF7, ID4, MY0D1, PAX5, PAX6, P0U3F1, RUNX3, SIM1, SIM2, TWIST1, and WT1 |
| Hormone receptors and kinases | AR, CALCA, EPHA3, EPO, EGFR, ERBB2, ESR1, ESR2, GALR2, LTB4R, PGR, RARB, STK11, TGFB1, TGFBR2, TNFRSF12, and UPA |
| Immune responses | CsADMI (TSLC1), CD40, CD44, CD80, CD86, and ICAM1 |
| Invasion/metastasis suppressors | COL9A1, MMP2, MMP9, MT1A, PLS3, RECK, R0B01, S100A4, SLIT2, THBS1, TIMP1, TIMP2, TIMP3, TJP2, TPM1, and VCAN |
| Transporters, glycoproteins, and membrane-associated proteins | ABCB1, AMN, CDCP1, CDH1, CDH3, CDH13, CFTR, DSC3, EDNRB, GJB2, GPC3, LAMA3, LAMB3, LAMC2, MUC2, NEFL, PCDH10, RARRES1, RBP1 TMEFF2, TSPAN2, SLC5A5, and SLC6A20 |
| Tumor suppressors | APC, BRCA1, CAV1, CST6, DAB2, DCC, DLC1, FHIT, GSN, H19, HIC1, LATS1/LATS2, PLAGL1, PTEN, RASSF1A, RB1, RBL1, PRDM2, SNCG, SERPINB5, TP53, TSC1, TSC2, TUSC3, VHL, and WIIF1 |
| Tumor antigens | ABO, MAGEA1, and MME |
aSee also www.pubmeth.org
Table 4.
Analysis of histone modifications and nucleosome positioning using candidate gene and global approachesa
| Category | Examples |
|---|---|
| Histone modifications | ChIP-qPCR or -cloning (chromatin immunoprecipitation followed by quantitative PCR of candidate genes or cloning); ChIP-PET (ChIP-paired-end-tagging); ChAP (ChIP coupled to arbitrarily primed PCR); SACO (serial analysis of chromatin occupancy); ChIP-seq (ChIP coupled to massively parallel sequencing); and Mass spectrometry |
| Nucleosome positioning | Nuclease protection assay; DNse-chip or -seq (mapping of DNase I hypersensitive sites on microarray or with massively parallel sequencing); and Micrococcal nuclease array |
aDescriptions of these techniques are provided in review articles (Lo and Sukumar 2008; Balch et al. 2008; Wu et al. 2006).
Methylation-mediated silencing of these loci may contribute, in part, to the development of malignant phenotypes. For example, hypermethylation of CpG islands in the p16INK4a, or CDKN2A, gene is known to be associated with cell cycle deregulation in breast cancer cells (Herman et al. 1995). This gene normally encodes a cyclin-dependent kinase which is responsible for the maintenance of the retinoblastoma protein in an active state for transcriptional regulation of downstream targets (Kamb et al. 1994). Lack of the p16INK4a expression promotes aberrant proliferation of primary mammary epithelial cells that normally undergo senescence in vitro (Huschtscha et al. 1998; Hinshelwood and Clark 2008; Hinshelwood et al. 2009). This epigenetic silencing may acquire additional genetic and epigenetic changes in cancer cells (Romanov et al. 2001; Tlsty et al. 2001). Specifically, the p16INK4a silencing results in the up-regulation of polycomb repressors, EZH2 and SUZ12, which are known to recruit DNA methyltransferases to target genes leading to de novo methylation of their promoters (Reynolds et al. 2006).
Another gene that undergoes methylation-mediated gene silencing is BRCA1 and this event disrupts cellular functions which are required for DNA repair, protein ubiquitination, and chromatin remodeling (Narod and Foulkes 2004). Inactivation of one BRCA1 allele, i.e., the first hit of Knudsen's hypothesis, is attributed to genetic mutations or loss of heterozygosity in breast neoplasm (Meric-Bernstam 2007). Hypermethylation of BRCA1 may be a second hit during breast tumorigenesis (Birgisdottir et al. 2006) and in invasive carcinomas it is frequently associated with shorter patient survival (Xu et al. 2009).
In addition to single-copy genes, hypermethylation occurs in 18S and 28S ribosomal genes in breast cancer (Yan et al. 2000). Approximately 400 copies of ribosomal loci, arranged in “head-to-tail” arrays, are located on the short arms of human acrocentric chromosomes. The transcription domain of these loci displays sequence characteristics of a CpG island. Increased methylation in these repetitive GC-rich sequences may result in down-regulation of ribosomal gene expression in breast cancer cells (Yan et al. 2000; Chan et al. 2005). Because this aberrant event is preferentially found in undifferentiated and ERα-negative tumors, hypermethylation of ribosomal DNA can be an additional prognostic biomarker for breast cancer (Yan et al. 2000).
DNA Hypomethylation in Epithelial Tumors
Global hypomethylation is frequently observed in cancer cells because of a deficiency in the production of S-adenosylmethionine (SAM) (Ehrlich 2002, 2006). SAM is the principal methyl donor for DNA methylation reactions in normal cells. As a result of this deficiency in cancer cells, hypomethylation of repeat elements (e.g., Alu, LINE, and α satellites) occurs, contributing to reactivation of transposable elements, and the promotion of chromosomal translocation, deletion, and duplication, as well as genomic instability (Ehrlich 2002, 2006). This aberrant epigenetic event has been known to correlate with advanced clinical stage, tumor size, and histological grade in breast cancer (Soares et al. 1999). Several lines of investigation also indicate hypomethylation of CpG islands may contribute to loss of imprinting and reactivation of testis-antigen related genes (Cho et al. 2003; De Smet et al. 2004; Kaneda and Feinberg 2005). Promoter hypomethylation may be linked to reactivation of proto-oncogenes (e.g., synuclein γ, ID4, and annexin A4) that are associated with tumor metastasis and endocrine resistance (Gupta et al. 2003; Pakneshan et al. 2004; Fan et al. 2006). In some cases, hypo- and hyper-methylation of the same CpG island co-exist in advanced tumors. This heterogeneity has been found in the E-cadherin promoter, suggesting that, in breast cancer cells, DNA methylation is a dynamic and plastic event which accompanies progression to a metastatic state (Graff et al. 2000).
Hypomethylation of promoter CpG islands may require a functional DNA demethylase yet to be characterized in cancer cells. In this case, a demethylation event is supposed to occur in an inactive, methylated promoter, leading to transcriptional reactivation of an oncogene. However, experimental proof for genuine promoter hypomethylation is frequently inconclusive and difficult to obtain because the outgrowth of a subpopulation of cancer cells may confound this epigenetic observation. For example, the oncogene of interest may have never been silent in a minor population of cancer-initiating cells while the majority of other cells display promoter hypermethylation of the gene. The increased expression of this oncogene may simply result from rapid expansion of these few cells that eventually take over the whole population during tumor progression. If this scenario indeed occurs, it cannot be a bona fide demethylating event for oncogene activation. Alternatively, active demethylation may be mediated by the 5-methyl-CpG binding domain protein 4 (MBD4) (Zhu 2009). Though MBD4 functions as a T-G mismatch glycosylase, it has recently been found to induce active demethylation of methylated CpG sites (Kim et al. 2009; Zhu 2009). MBD4 mutant with a deletion in the glycosylase catalytic domain abrogates this demethylating ability (Kim et al. 2009). Clearly, elucidating the functions of MBD4 in promoter hypomethylation during mammary gland differentiation and breast tumorigenesis is an important area in need of further investigation.
HISTONE MODIFICATIONS
Combinatorial Variations Associated with Gene Transcription
Histone modification is another epigenetic mechanism important for chromatin remodeling during stem/progenitor differentiation. The basic subunit of chromatin is the nucleosome, a complex consisting of 146 bp of genomic DNA wrapped around a core histone octomer composed of one histone H3-H4 tetramer and two histone H2A-H2B dimers (Kouzarides 2007). The histone amino-terminal “tails” extending from a nucleosome octamer are frequently subject to posttranslational modifications, including acetylation, phosphorylation, ubiquitination, and methylation. Among these various modifications, histone methylation and acetylation are shown to be relatively stable and therefore considered potential marks for carrying epigenetic information through cell divisions (Barski et al. 2007; Wang et al. 2008). The sum total of these covalent alterations is referred to as the histone code that can be “written” by histone modifying enzymes (e.g., acetyltransferases, deacetylases, methyltransferases, or demethylases) and read by various binding proteins that act to further modify chromatin structure (Wang et al. 2009). Among these proteins, EZH2, a known component of polycomb repressor complex, can cooperatively work with histone deacetylases and methyltransferases to modify histone tails for initiating transcriptional repression. EZH2 also works with DNA methyltransferases for subsequent maintenance of gene silencing (Sparmann and van Lohuizen 2006). Table 4 lists techniques for analysis of histone modifications and chromatin structure of active and inactive genes.
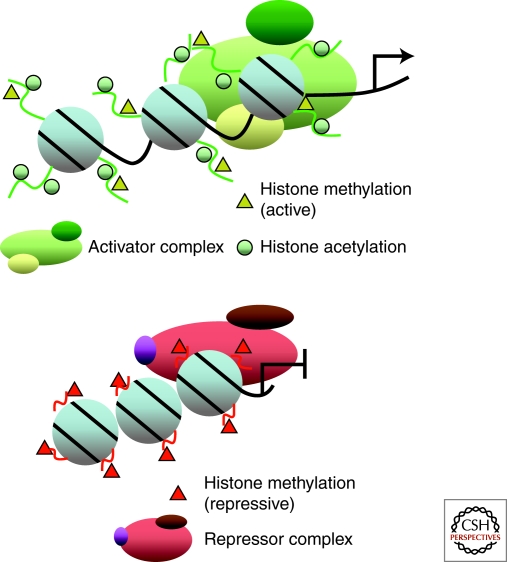
Both packaging of DNA into chromatin and nucleosome positioning influence gene transcription by either enhancing or inhibiting the accessibility of general and site-specific transcription factors to target loci. In general, acetylation of histone lysine residues is associated with open chromatin structure and active transcription whereas the methylation of these residues can be associated with either an active or repressive chromatin structural conformation (Fig. 3) (see examples in Table 5). For example, active transcription of a promoter can be marked by acetylation at specific lysine residues of histone H3 (e.g., lysine 5, 8, and 12), histone H4 lysine 16, and dimethylation of histone H3 lysine 4. In contrast, when genes are silenced, these active marks are replaced by repressive marks, including mono-, di-, and tri-methylation of H3K9, H3K27, and H4K20. Combinatorial epigenetic alterations likely mark differential degrees of gene silencing, starting from a transient to a more rigid state of repression.
Figure 3.
Models of open or closed chromatin configurations associated with gene transcription status. Binding of regulatory factors and their co-factors to a target locus results in the formation of either an activator or repressor complex. This complex may act as docking molecules for recruitment of acetyltransferases, deacetylases, methyltransferases, or demethylases that post-translationally modify the amino-terminal tail of histone. Modified histone marks are associated with specific transcription status of a target gene. Active transcription is usually associated with histone acetylation of H3K9 or methylation of H3K4 residues, whereas gene repression is linked to methylation of H3K9, H3K27 and H4K20 residues.
Table 5.
Representative examples of histone tail methylation and its associated genomic functionsa
| Histone methylation mark | Function | Genomic location |
|---|---|---|
| H3K4me1 | Active | TSS, enhancer |
| H3K4me2 | Active | TSS |
| H3K4me3 | Bivalent | TSS, enhancer |
| H3K9me1 | Active | TSS |
| H3K9me2 | Repressive | 10-kb of TSS |
| H3K9me3 | Repressive | 10-kb of TSS |
| H3K27me1 | Active | Downstream of TSS |
| H3K27me2 | Repressive | — |
| H3K27me3 | Bivalent | Promoter, gene-body |
| H3K36me1 | Active | TSS |
| H3K36me3 | Active (?) | Downstream of TSS |
| H3K79me1 | No preference | — |
| H3K79me2 | No preference | — |
| H3K79me3 | Repressive | Promoter, gene-body |
| H3R2me1 | Modestly active | — |
| H3R2me2 | Modestly active | — |
| H4K20me1 | Active | Downstream of TSS |
| H4K20me3 | No preference | — |
| H2A+H4R3me2 | No preference | — |
| H2BK5me1 | Active | Downstream of TSS |
aSee a publication by Zha and colleagues (Barski et al. 2007). K: lysine; R: arginine; TSS: transcription start site.
When binding target sequences in the chromatin, transcription factors may act as docking molecules for recruitment of other DNA- and histone-modifying proteins to target genes for transcriptional regulation (Jenuwein and Allis 2001). For example, Wang and coworkers (Ko et al. 2008) showed that YY1 physically interacts with SUZ12 and acts as a mediator in the recruitment of polycomb group proteins and DNA methyltransferases that promote transcriptional repression. Several studies have found that open chromatin regions are usually correlated with gene-dense regions and that closed chromatin regions exist in both euchromatic G-bands and heterochromatic C-bands (Gilbert et al. 2004; Weil et al. 2004; Crawford et al. 2006). These studies also show active genes can exist within closed chromatin domains.
Histone Modifications in Normal Epithelial Differentiation and Breast Tumorigenesis
The fact that somatic cells possess an identical genome, yet display diverse phenotypes, can partly be attributed to combinatorial variations of histone modifications that are functionally associated with gene transcription (Balch et al. 2007). Recent advances in technologies have allowed for the examination of global histone modifications within a cell, and this accomplishment has provided insight into the organization of the genome (Balch et al. 2008). In human tumors, modifications of histone H4 generally result in a loss of monoacetylated K16 and trimethylated K20 forms (Fraga et al. 2005). These histone changes are associated with DNA hypomethylation in various tumor types, including breast cancer (Fraga et al. 2005; Tryndyak et al. 2006). More recently, Zhao and colleagues (Barski et al. 2007) have comprehensively examined 20 histone marks, RNA polymerase II, and other binding proteins across the human T-cell genome. This study found that combinatorial histone variations contribute to the maintenance of transcriptional plasticity and to the fate of specific cell lineages during differentiation. In a separate study, Bernstein and co-workers (Mikkelsen et al. 2007) constructed global chromatin maps for mouse embryonic stem cells (ESCs) and their differentiated cells. A large number of genes important for differentiation were found to possess a “bivalent” feature, consisting of post-translational histone modifications commonly found in transcriptionally active (H3K4me2) and a repressive (H3K27me3) chromatin. Coexistence of these histone marks is likely to maintain progenitor cells in a “ready state” for lineage-specific gene activation or repression. For example, the Sox2 gene possesses these bivalent modifications in ESCs, but only has the H3K27me3 modification in differentiated cells, indicating that this locus is poised for repression during terminal differentiation (Mikkelsen et al. 2007).
At present, chromatin-state maps have not been systematically generated for breast progenitor cells and their differentiated progeny. It is likely that bivalent genes are present to maintain pluripotency of the progenitor genome. During epithelial differentiation, these bivalent loci may acquire activating or repressive chromatin marks that specify specific cell lineages (Balch et al. 2007). Terminally differentiated cells may therefore display unique chromatin signatures distinctly associated with their developmental fates. In an initial study, Polyak and coworkers (Bloushtain-Qimron et al. 2008) have further shown that bivalent loci identified in ESCs might also be present in CD44+ breast progenitor cells, as genes highly expressed in CD44+ compared to CD24+ cells were enriched in Suz12 targets in ESCs. As discussed earlier, CpG islands of these genes were found to be hypomethylated in CD44+ cells compared to those of differentiated CD24+ cells. Although the aforementioned bivalent marks play an important role in maintaining transcriptional plasticity in CD44+ cells, it remains to be determined whether other histone modifications also contribute to this ready state for activation and deactivation of genes during mammary epithelial differentiation.
As indicated earlier, propagation of neoplastic lesions can be initiated by CD44+ progenitor cells, which have been exposed to aberrant microenvironments. In this regard, deregulation of TGF-β (Hinshelwood et al. 2007) and AKT1 (Lin et al. 2008) signaling has been implicated in abnormal differentiation of mammary epithelial cells. In vitro studies have found that cultured mammary epithelial cells routinely enter a phase of growth arrest, but a subpopulation of progenitor cells may continue to proliferate into preneoplastic clones (Hinshelwood and Clark 2008). It has been hypothesized also that aberrant progenitor cells retain bivalent features of differentiation-control genes, but many of these genes encoding tumor suppressor functions may undergo epigenetic silencing during neoplastic development. In addition to H3K27me3, two additional repressive marks, H3K9me2 and H3K9me3, may be present in these genes, leading to permanent silencing of tumor-suppressor genes and subsequent propagation of transformed phenotypes (Bloushtain-Qimron et al. 2008; Cheng et al. 2008; Hsu et al. 2009). Transition from bivalent to fully repressive histone modifications therefore contributes to the development of breast cancer. A systematic cataloging of different histone marks for normal progenitors and cancer-initiating cells is an important task for future research of mammary gland development and tumorigenesis.
Connecting Histone Modifications and DNA Methylation for Gene Silencing
The interplay between histone modifications and DNA methylation has a critical role in nucleosome remodeling and subsequent regulation of gene transcription (Ballestar and Esteller 2005; Esteller 2006). The aforementioned polycomb target genes, known to be associated with stem/progenitor differentiation, provide the best example of how these two epigenetic components cooperate to initiate and maintain gene silencing (Cheng et al. 2008). These genes are generally associated with promoter CpG islands and are usually protected from de novo DNA methylation in stem/progenitor cells (Meissner et al. 2008). During epithelial differentiation, EZH2, as part of the polycomb repressor complex 2, catalyzes trimethylation of H3K27 on target nucleosomes (Sparmann and van Lohuizen 2006). This modification may serve as a dock site for the recruitment of a second repressor complex, PRC1, which additionally modifies target loci into compact chromatin configuration. These polycomb complexes may be constitutively present and can readily reattach to target sequences following DNA replication of daughter cells. Further recruitment of DNA methyltransferases and methyl-CpG binding protein may occur in promoter CpG islands for de novo DNA methylation (Leu et al. 2004; Hinshelwood et al. 2009). This epigenetic repression occurs in a subset of PRC2-mediated target genes essential for tumor suppression (Cheng et al. 2008) and can be heritably maintained over multiple divisions in the differentiated progeny.
This epigenetic reprogramming can be disrupted in mammary progenitor cells as a result of chronic exposure to xenoestrogens or inflammatory microenvironment. Deregulated signaling related to epithelial differentiation may aberrantly up-regulate the expression of EZH2 and other polycomb repressor proteins, such as SUZ12, BMI1, G9a, and SUV39H (Dimri et al. 2002; Schultz et al. 2002; Kleer et al. 2003; Lehnertz et al. 2003). As a result, polycomb-mediated silencing of genes associated with tumor suppressor functions takes place, resulting in clonal proliferation of undifferentiated or semi-differentiated cells. Further accumulation of DNA methylation likely causes the permanent silencing of these genes in proliferating cells. In addition to tumor-suppressor genes, epigenetic silencing may lead to up-regulation of oncogenes. In this regard, microRNAs that negatively regulate the expression of their target loci may become transcriptionally silent, leading to aberrant de-repression of these oncogenes in cancer cells (Huang et al. 2009).
The cooperation between histone methylation and DNA methylation is highly dynamic in different cancer cell types. In some cancer types, epigenetically silent loci are highly enriched for trimethylation of H3K27 (H3K27me3), but show low levels of DNA methylation (Kondo et al. 2008). In other cases, this trimethylation mark can be lost while DNA methylation is accumulated in silent loci (Gal-Yam et al. 2008). It is possible that these two repressive marks become more independent of each other in terms of maintenance silencing during cancer development, because genetic knockdown of EZH2 does not change profiles of de novo DNA methylation in a transfection study (McGarvey et al. 2007). Therefore, H3K27me3 can be replaced by DNA methylation for long-term repression. This reprogramming may be more widespread in cancer cells that show high degrees of genomic instability.
Concluding Remarks
Epigenetic disruption is a characteristic of human cancer. The reduction of the total amounts of DNA methylation compared with their normal counterparts has been the first epigenetic alteration described in human tumors. Global DNA hypomethylation contributes to the origin of cancer cells by generation of chromosomal instability, reactivation of transposable elements, and loss of imprinting. Most importantly, CpG methylation can be acquired in promoter regions of tumor-suppressor genes, such as BRCA1 and p16INK4a, leading to the inactivation of these cancer-protecting proteins. Furthermore, methylation mediated-silencing was observed in microRNA loci with tumor-suppressive functions. Human tumors also show a distorted histone code. Breast tumors undergo massive and adaptive changes in their natural history, i.e., these cancer cells can metastasize to distant sites where they create new blood and lymph vessels to feed on and eliminate metabolites. They can also phenotypically alter their response to treatment with drugs, hormones, or radiation. In this regard, these cancer cells may have a limited ability to undergo fast genetic changes to adapt to hostile microenvironments. However, the expansion of a subset of breast cancer cells can occur through rapidly occurring epigenetic changes. For example, the cell adherence E-cadherin gene becomes methylated and silenced in breast cancer cells once they have metastasized to other organs or tissues. One of the essential differences between genetic and epigenetic alterations in cancer cells is that, unlike the former, DNA methylation and histone modifications are reversible under the right circumstances. Thus, epigenetic changes can be one of weak points in self-defense mechanisms of cancer cells, because those hypermethylated tumor-suppressor genes in their long “sleep” can be awakened and reactivated with the right drug regimen and exert their normal growth-inhibitory functions. Two families of epigenetic drugs-DNA demethylating agents and histone deacetylase inhibitors hold great promise for future cancer treatments.
ACKNOWLEDGMENT
The authors wish to thank Drs. Yi-Wen Huang and Gregory Singer for technical assistance. This work was supported by the National Institutes of Health grants (U01ES015986, U54CA113001, R01CA069065, and R01ES017594), the Ohio State University Comprehensive Cancer Center, the Health Department of the Catalan Government, the Spanish Association Against Cancer, and the grants by Fondo de Investigaciones Sanitarias PI08-1345 and Consolider MEC09-05.
Footnotes
Editors: Mina J. Bissell, Kornelia Polyak, and Jeffrey Rosen
Additional Perspectives on Mammary Gland Biology available at www.cshperspectives.org
REFERENCES
- Antequera F, Bird A 1993. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci 90: 11995–11999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Bird A 1999. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol 9: R661–667 [DOI] [PubMed] [Google Scholar]
- Balch C, Huang TH-M, Nephew KP 2008. High-throughput analysis of epigenomics in human disease. In Genome Sequencing Technology and Algorithms (eds. Kim S., Tang H., Mardis E.R.). Artech House Publishers [Google Scholar]
- Balch C, Montgomery JS, Paik HI, Kim S, Huang TH-M, Nephew KP 2005. New anti-cancer strategies: epigenetic therapies and biomarkers. Front Biosci 10: 1897–1931 [DOI] [PubMed] [Google Scholar]
- Balch C, Nephew KP, Huang TH-M, Bapat SA 2007. Epigenetic “bivalently marked” process of cancer stem cell-driven tumorigenesis. Bioessays 29: 842–845 [DOI] [PubMed] [Google Scholar]
- Ballestar E, Esteller M 2005. The epigenetic breakdown of cancer cells: From DNA methylation to histone modifications. Prog Mol Subcell Biol 38: 169–181 [DOI] [PubMed] [Google Scholar]
- Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, Cigudosa JC, Huang TH-M, Esteller M 2003. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J 22: 6335–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Batty N, Malouf GG, Issa JP 2009. Histone deacetylase inhibitors as anti-neoplastic agents. Cancer Lett 280: 192–200 [DOI] [PubMed] [Google Scholar]
- Bestor TH 2000. The DNA methyltransferases of mammals. Hum Mol Genet 9: 2395–2402 [DOI] [PubMed] [Google Scholar]
- Birgisdottir V, Stefansson OA, Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG, Eyfjord JE 2006. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res 8: R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloushtain-Qimron N, Yao J, Shipitsin M, Maruyama R, Polyak K 2009. Epigenetic patterns of embryonic and adult stem cells. Cell Cycle 8: 809–817 [DOI] [PubMed] [Google Scholar]
- Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, Hu M, Chen H, Ustyansky V, Antosiewicz JE, et al. 2008. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci 105: 14076–14081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brena RM, Huang TH-M, Plass C 2006a. Quantitative assessment of DNA methylation: Potential applications for disease diagnosis, classification, and prognosis in clinical settings. J Mol Med 84: 365–377 [DOI] [PubMed] [Google Scholar]
- Brena RM, Huang TH-M, Plass C 2006b. Toward a human epigenome. Nat Genet 38: 1359–1360 [DOI] [PubMed] [Google Scholar]
- Cairns BR 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y 2009. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet 10: 295–304 [DOI] [PubMed] [Google Scholar]
- Chan MW, Wei SH, Wen P, Wang Z, Matei DE, Liu JC, Liyanarachchi S, Brown R, Nephew KP, Yan PS, et al. 2005. Hypermethylation of 18S and 28S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin Cancer Res 11: 7376–7383 [DOI] [PubMed] [Google Scholar]
- Chang JW, Huang TH-M, Wang YC 2008. Emerging methods for analysis of the cancer methylome. Pharmacogenomics 9: 1869–1878 [DOI] [PubMed] [Google Scholar]
- Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J, et al. 2008. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res 68: 1786–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B, Lee H, Jeong S, Bang YJ, Lee HJ, Hwang KS, Kim HY, Lee YS, Kang GH, Jeoung DI 2003. Promoter hypomethylation of a novel cancer/testis antigen gene CAGE is correlated with its aberrant expression and is seen in premalignant stage of gastric carcinoma. Biochem Biophys Res Commun 307: 52–63 [DOI] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. 2006. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing. Genome Res 16: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Esteller M 2009. MicroRNAs and cancer epigenetics: A macrorevolution. Curr Opin Oncol 22: 35–45 [DOI] [PubMed] [Google Scholar]
- De Smet C, Loriot A, Boon T 2004. Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol Cell Biol 24: 4781–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Martinez JL, Jacobs JJ, Keblusek P, Itahana K, Van Lohuizen M, Campisi J, Wazer DE, Band V 2002. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res 62: 4736–4745 [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS 2003. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17: 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, El-Ashry D, Wicha MS 2004. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab 15: 193–197 [DOI] [PubMed] [Google Scholar]
- Dontu G, Liu S, Wicha MS 2005. Stem cells in mammary development and carcinogenesis: Implications for prevention and treatment. Stem Cell Rev 1: 207–213 [DOI] [PubMed] [Google Scholar]
- Ehrlich M 2002. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J Nutr 132: 2424S–2429S [DOI] [PubMed] [Google Scholar]
- Ehrlich M 2006. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol 310: 251–274 [DOI] [PubMed] [Google Scholar]
- Esteller M 2006. CpG island methylation and histone modifications: Biology and clinical significance. Ernst Schering Res Found Workshop 57: 115–126 [DOI] [PubMed] [Google Scholar]
- Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, Salisbury JD, Cheng ASL, Li L, Abbosh PH, et al. 2006. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res 66: 11954–11966 [DOI] [PubMed] [Google Scholar]
- Feinberg AP 2008. Epigenetics at the epicenter of modern medicine. JAMA 299: 1345–1350 [DOI] [PubMed] [Google Scholar]
- Fenton SE 2006. Endocrine-disrupting compounds and mammary gland development: Early exposure and later life consequences. Endocrinology 147: S18–S24 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400 [DOI] [PubMed] [Google Scholar]
- Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE 2002. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet 31: 175–179 [DOI] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A 2008. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci 105: 12979–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA 2004. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell 118: 555–566 [DOI] [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG 2000. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem 275: 2727–2732 [DOI] [PubMed] [Google Scholar]
- Guil S, Esteller M 2009. DNA methylomes, histone codes and miRNAs: Tying it all together. Int J Biochem Cell Biol 41: 87–95 [DOI] [PubMed] [Google Scholar]
- Gudjonsson T, Villadsen R, Nielsen HL, Rønnov–Jessen L, Bissell MJ, Petersen OW 2002. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev 16: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J 2003. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res 63: 664–673 [PubMed] [Google Scholar]
- Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB 1995. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 55: 4525–4530 [PubMed] [Google Scholar]
- Hinshelwood RA, Clark SJ 2008. Breast cancer epigenetics: Normal human mammary epithelial cells as a model system. J Mol Med 86: 1315–1328 [DOI] [PubMed] [Google Scholar]
- Hinshelwood RA, Huschtscha LI, Melki J, Stirzaker C, Abdipranoto A, Vissel B, Ravasi T, Wells CA, Hume DA, Reddel RR, et al. 2007. Concordant epigenetic silencing of transforming growth factor-beta signaling pathway genes occurs early in breast carcinogenesis. Cancer Res 67: 11517–11527 [DOI] [PubMed] [Google Scholar]
- Hinshelwood RA, Melki JR, Huschtscha LI, Paul C, Song JZ, Stirzaker C, Reddel RR, Clark SJ 2009. Aberrant de novo methylation of the p16INK4A CpG island is initiated post gene silencing in association with chromatin remodelling and mimics nucleosome positioning. Hum Mol Genet 18: 3098–3109 [DOI] [PubMed] [Google Scholar]
- Ho SM, Tang WY, Belmonte de Frausto J, Prins GS 2006. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res 66: 5624–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Deatherage DE, Rodriguez BA, Liyanarachchi S, Weng YI, Zuo T, Liu J, Cheng ASL, Huang TH-M 2009. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res 69: 5936–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, Miller DS, Huang TH-M 2009. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res 69: 9038–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huschtscha LI, Noble JR, Neumann AA, Moy EL, Barry P, Melki JR, Clark SJ, Reddel RR 1998. Loss of p16INK4 expression by methylation is associated with lifespan extension of human mammary epithelial cells. Cancer Res 58: 3508–3512 [PubMed] [Google Scholar]
- Issa JP 2003. Age-related epigenetic changes and the immune system. Clin Immunol 109: 103–108 [DOI] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB 1994. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7: 536–540 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33: S245–S254 [DOI] [PubMed] [Google Scholar]
- Jensen EV, Jordan VC 2003. The estrogen receptor: A model for molecular medicine. Clin Cancer Res 9: 1980–1989 [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Liang G 2009. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10: 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH 1994. A cell cycle regulator potentially involved in genesis of many tumor types. Science 264: 436–440 [DOI] [PubMed] [Google Scholar]
- Kaneda A, Feinberg AP 2005. Loss of imprinting of IGF2: A common epigenetic modifier of intestinal tumor risk. Cancer Res 65: 11236–11240 [DOI] [PubMed] [Google Scholar]
- Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, et al. 2009. DNA demethylation in hormone-induced transcriptional derepression. Nature 461: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci 100: 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM 2008. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem 283: 30919–30932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng ASL, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 40: 741–750 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Leu YW, Yan PS, Fan M, Jin VX, Liu JC, Curran EM, Welshons WV, Wei SH, Davuluri RV, Plass C, et al. 2004. Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res 64: 8184–8192 [DOI] [PubMed] [Google Scholar]
- Lin HJ, Zuo T, Lin CH, Kuo CT, Liyanarachchi S, Sun S, Shen R, Deatherage DE, Potter D, Asamoto L, et al. 2008. Breast cancer-associated fibroblasts confer AKT1-mediated epigenetic silencing of Cystatin M in epithelial cells. Cancer Res 68: 10257–10266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo PK, Sukumar S 2008. Epigenomics and breast cancer. Pharmacogenomics 9: 1879–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Montserrat Sanchez-Cespedes M, Gitt A, et al. 2007. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67: 1424–1429 [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero R, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. 2008. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci 105: 13556–13561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini MV, Rubin BS, Sonnenschein C, Soto AM 2006. Endocrine disruptors and reproductive health: The case of bisphenol-A. Mol Cell Endocrinol 254–255: 179–186 [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB 2007. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res 67: 5097–5102 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F 2007. Heterogenic loss of BRCA in breast cancer: The “two-hit” hypothesis takes a hit. Ann Surg Oncol 14: 2428–2429 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod SA, Foulkes WD 2004. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer 4: 665–676 [DOI] [PubMed] [Google Scholar]
- Nephew KP, Huang TH-M 2003. Epigenetic gene silencing in cancer initiation and progression. Cancer Lett 190: 125–133 [DOI] [PubMed] [Google Scholar]
- Novak P, Jensen TJ, Garbe JC, Stampfer MR, Futscher BW 2009. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res 69: 5251–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgane J, Hattori N, Shiota K 2005. Analysis of tissue-specific DNA methylation during development. Methods Mol Biol 289: 371–382 [DOI] [PubMed] [Google Scholar]
- Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA 2004. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J Biol Chem 279: 31735–31744 [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péchoux C, Gudjonsson T, Rønnov–Jessen L, Bissell MJ, Petersen OW 1999. Human mammary luminal epithelial cells contain progenitors to myoepithelial cells. Dev Biol 206: 88–99 [DOI] [PubMed] [Google Scholar]
- Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, Hong C, Fridlyand J, Costello JF, Tlsty TD 2006. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem 281: 24790–24802 [DOI] [PubMed] [Google Scholar]
- Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD 2001. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature 409: 633–637 [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA 2006. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9: 435–443 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ III 2002. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3: 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares J, Pinto AE, Cunha CV, Andre S, Barao I, Sousa JM, Cravo M 1999. Global DNA hypomethylation in breast carcinoma: Correlation with prognostic factors and tumor progression. Cancer 85: 112–118 [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M 2006. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ 2006. Purification and unique properties of mammary epithelial stem cells. Nature 439: 993–997 [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A 2008. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Tlsty TD, Romanov SR, Kozakiewicz BK, Holst CR, Haupt LM, Crawford YG 2001. Loss of chromosomal integrity in human mammary epithelial cells subsequent to escape from senescence. J Mammary Gland Biol Neoplasia 6: 235–243 [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Kovalchuk O, Pogribny IP 2006. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther 5: 65–70 [DOI] [PubMed] [Google Scholar]
- Villadsen R, Fridriksdottir AJ, Rønnov–Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW 2007. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol 177: 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Weil MR, Widlak P, Minna JD, Garner HR 2004. Global survey of chromatin accessibility using DNA microarrays. Genome Res 14: 1374–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Smith LT, Plass C, Huang TH-M 2006. ChIP-chip comes of age for genome-wide functional analysis. Cancer Res 66: 6899–6902 [DOI] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Zhang Y, Cho YH, Wetmur JG, Bradshaw PT, Garbowski G, Hibshoosh H, Teitelbaum SL, Neugut AI, et al. 2009. Gene promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat 115: 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager JD, Davidson NE 2006. Estrogen carcinogenesis in breast cancer. N Engl J Med 354: 270–282 [DOI] [PubMed] [Google Scholar]
- Yan PS, Rodriguez FJ, Laux DE, Perry MR, Standiford SB, Huang TH-M 2000. Hypermethylation of ribosomal DNA in human breast carcinoma. Br J Cancer 82: 514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan PS, Shi H, Rahmatpanah F, Hsiau TH, Hsiau AH, Leu YW, Liu JC, Huang TH-M 2003. Differential distribution of DNA methylation within the RASSF1A CpG island in breast cancer. Cancer Res 63: 6178–6186 [PubMed] [Google Scholar]
- Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, et al. 2006. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res 12: 6626–6636 [DOI] [PubMed] [Google Scholar]
- Zhu JK 2009. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 43: 143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T, Tycko B, Liu TM, Lin HJ, Huang TH-M 2009. Methods in DNA methylation profiling. Epigenomics 1: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]