Abstract
Investigations into the organization of transcription have their origins in cell biology. Early studies characterized nascent transcription in relation to discernable nuclear structures and components. Advances in light microscopy, immunofluorescence, and in situ hybridization helped to begin the difficult task of naming the countless individual players and components of transcription and placing them in context. With the completion of mammalian genome sequences, the seemingly boundless task of understanding transcription of the genome became finite and began a new period of rapid advance. Here we focus on the organization of transcription in mammals drawing upon information from lower organisms where necessary. The emerging picture is one of a highly organized nucleus with specific conformations of the genome adapted for tissue-specific programs of transcription and gene expression.
Active genes probably migrate to “transcription factories” where DNA is reeled through RNA polymerases—rather than the polymerases moving along the DNA.
Much of what is known about eukaryotic transcription is dominated by decades of advances in in vitro biochemistry with whole-cell extracts, or subfractionated and recombinant proteins on purified DNA templates. These reductionist approaches have lead to seminal findings describing the basic DNA sequence regulatory elements and enzymatic machinery of transcription. More recent research incorporating genetic approaches has added to the complexity of transcription, involving literally hundreds of factors, cofactors, remodeling complexes, histone modifiers, and elongation-, splicing- and termination-factors required for or associated with a single transcriptional event. Furthermore the discovery of, sometimes distant, sequence elements required for regulated transcription of some genes has added to the intricacy of the transcriptional process that occurs in vivo. Though there is still much to learn, the difficult task of integrating this information and placing it in the context of the nucleus is gathering momentum.
The widely held view of transcriptional mechanics, of the RNA polymerase complex sliding along a template to generate a transcript is also dominated by biochemistry. Textbooks are full of descriptions of promoter bound factors recruiting RNA polymerase, which initiates transcription before sliding along the transcription unit. Indeed, single molecules of prokaryotic RNA polymerase have been visualized in vitro, sliding along a fixed DNA template during a one-dimensional diffusional search for a promoter (Kabata et al. 1993; Guthold et al. 1999; Harada et al. 1999) or during transcription (Schafer et al. 1991; Wang et al. 1998; Guthold et al. 1999; Davenport et al. 2000). However, which molecule actually moves, the polymerase or the DNA depends on which is fixed (Iborra et al. 1996b). Several studies have shown that an anchored polymerase generates considerable pulling force on a DNA template, rotating the double helix in a clockwise manner as it threads the strand through the protein during transcription (Kabata et al. 1993; Wang et al. 1998; Guthold et al. 1999). It is impossible to know from these in vitro studies what actually happens in vivo. Although the answer to this question may seem trivial, it has profound implications for our understanding of transcription and genome function, and can only be answered by examining evidence of transcription in the nucleus.
Fakan and colleagues have studied nascent transcripts at high resolution for decades (Fakan and Bernhard 1971; Fakan et al. 1976; Fakan 2004). They found that nascent RNA is located in perichromatin fibrils (PF). PF are structures observed using electron microscopy by specific contrasting methods, and most often located in the perichromatin region, the boundaries between condensed and decondensed chromatin (Fakan and Bernhard 1971). Early autoradiographic studies combined with later findings indicated that PF are the in situ form of nascent RNA complexed with processing factors (Nash et al. 1975; Fakan et al. 1976; Cremer et al. 2004).
Seminal studies on transcription in mammalian nuclei were carried out by Jackson and Cook (Jackson et al. 1981; Jackson and Cook 1985). They uniformly labeled DNA of HeLa cells with 14C in vivo, and encapsulated them in agarose beads before a short incubation with [3H]uridine to label nascent RNA. The cells were then lysed in an isotonic solution and chromatin was digested with a restriction enzyme or DNase followed by electrophoresis to remove the digested chromatin. They found that over 90% of nascent RNA is retained in the beads after as much as 98% of the DNA/chromatin had been removed. The chromatin released from the beads by electrophoresis was assayed further and found to be considerably larger than an RNAPII holocomplex, suggesting that nonattached transcribing complexes should have been released with the chromatin. However, they found that 60% of the original RNA polymerase activity was retained in the beads after loss of 75% of the chromatin. These results suggested a model whereby newly synthesized RNA and the transcriptional machinery are retained in the nucleus by a structure that is resistant to nuclease digestion. Indeed analysis of the DNA retained in the beads indicated that it was enriched in active genes. Jackson and Cook proposed the involvement of a “nucleoskeleton” in the process of transcription, suggesting attachment of the polymerase complex, and arguing against the concept of free RNA polymerase complexes tracking DNA templates. These were controversial studies in their time, and the merits and conclusions are still debated today. Though Jackson and Cook were careful to maintain physiological conditions during their experiments (Jackson and Cook 1985), critics argued that the system employed may have created an artificial network of proteins that retained active gene sequences.
TRANSCRIPTION OCCURS IN FACTORIES
These studies were expanded and advanced by Jackson et al. (1993), and particularly Wansink et al. (1993) in which transcription sites were visualized by indirect immunofluorescence after pulse-labeling or microinjecting cells with halogenated ribonucleoside precursors. The use of in vivo labeling methods excluded potential artifacts, which may have been introduced by the earlier run-on procedures. These studies revealed that nascent transcripts were not equally distributed throughout the nucleoplasm, but that transcription occurred in a limited number of discrete sites or foci that were sensitive to transcriptional inhibitors. An important observation was that increased labeling times did not result in detection of more foci, only increased intensity suggesting that all sites of transcription were being detected. Estimates of the number of transcription foci vary between several hundred and several thousand per nucleus depending on the cell type (Jackson et al. 1993; Iborra et al. 1996a; Pombo et al. 1999; Osborne et al. 2004). The total number of sites appeared to be several times lower than the number of active transcription units suggesting that each site contained multiple genes (Jackson et al. 1993; Jackson et al. 1998). The term “transcription factories” was coined (Iborra et al. 1996a) to reflect the potential gathering of several transcription units to each factory in similarity to replication factories, which form during the S phase of the cell cycle, each accommodating several replicons at once. Fakan and colleagues have often observed that immunogold detection of nascent RNAs after short labeling is most often represented by individual gold particles associated with PF (Cmarko et al. 1999). This was taken as evidence that transcription takes place in individual localized sites rather than in factories. However quantitative estimates based on immunogold labeling on the surface of EM sections is tenuous. Though PF and transcription factories have not been linked directly, probably because with the various imaging techniques used one can only see what one attempts to detect, it seems likely that PFs do represent nascent transcripts (Fakan 1994) produced at focal RNAPII factories (Cmarko et al. 1999).
Both groups (Jackson et al. 1993; Wansink et al. 1993), investigated the localization of splicing components relative to nascent transcription sites. Large, intensely labeled SC-35 domains localized close to some nascent transcription sites but did not overlap with them, whereas some colocalization was observed between transcription sites and weak SC-35 foci (Wansink et al. 1993). These observations are consistent with suggestions that large SC-35 domains are storage sites containing no transcriptional activity (Iborra et al. 1996a; Pombo and Cook 1996; Fay et al. 1997), whereas small SC-35 foci may result from recruitment of splicing components to transcription sites for cotranscriptional splicing (Huang and Spector 1996; Misteli et al. 1997; Lamond and Spector 2003).
Building on previous work, Cook and colleagues employed electron microscopy (EM) to study nascent transcripts in pulse labeled cells (Iborra et al. 1996a) using indirect labeling with immuno-gold particles. They found that gold particles marking nascent transcripts appeared as clusters, which increased in particle number with increased labeling times suggesting that the clusters marked synthetic sites. Once again, the total number of clusters did not change with increasing labeling times, which they interpreted as evidence that the technique was sensitive enough to detect all sites. Importantly, they found that the diameter of nascent transcript clusters remained fairly constant (at ∼75 nm) regardless of the labeling time and increased elongation, even during extended chase periods. Iborra et al. (1996a) suggest that this result is contrary to models in which the polymerase slides along the template during transcription. They argue that such a polymerase tracking mechanism would result in nascent transcripts occupying an increased volume with increasing elongation. Instead they proposed their data to be consistent with a model in which the DNA template slides through a polymerase that is immobilized in a factory. This has been one of the most difficult concepts for many in the transcription field to accept, and perhaps rightly so because it contradicts the textbook view that holds the gene as the central scaffold to which the transcription complex is recruited. Interestingly, in the case of DNA replication, it is widely accepted that DNA is drawn through and extruded from discrete replication factories containing many active polymerases (Berezney et al. 2000). Though this is not proof that a similar mechanism occurs during transcription, it appears to be the most plausible given the available evidence in favor of this concept and relative lack of evidence to the contrary.
Iborra et al. (1996a) also examined the relationship between nascent transcript sites and RNAPII by combining detection of nascent RNA and RNAPII with immno-gold particles of different sizes. Gold particles marking RNAPII were also found in discrete clusters throughout the nucleus and averaged 56 nm in diameter. They showed that nascent transcript gold clusters were intimately associated with RNAPII clusters but did not overlap exactly. The centers of nascent transcript clusters and RNAPII clusters were shifted relative to each other by an average of 24 nm, consistent with two overlapping zones, one rich in transcripts, and the other rich in RNA polymerase II. Simultaneous immunofluorescent detection of RNAPII and nascent transcripts combined with confocal microscopy confirmed this association between focal RNAPII sites and transcriptional activity (Grande et al. 1997). RNAPII-containing foci colocalized with sites of nascent transcript labeling and vice versa, however, there were also many sites strongly labeled for RNAPII that contained little or no BrUTP label, and vice versa. With hindsight, this might be taken as one of the first hints of specialized polymerase factories and the possibility that not all factories are equally active (see below).
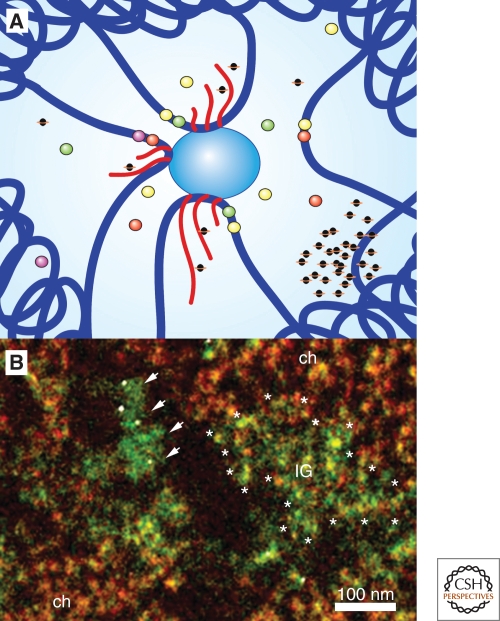
What actually constitutes the factory is a question open to interpretation. Nascent transcripts are undoubtedly the product but the enzymatic machinery is in essence the factory. The most recent and probably most accurate measurements of transcription factories size employed electron spectroscopic imaging (ESI) (Eskiw et al. 2008). Rather than measure the size of a cluster of gold particles, ESI records atomic signatures (Fig. 1). Transcription factories appear as large proteinacious (nitrogen rich) structures with an average diameter of 87 nm. Chromatin and nascent transcripts with associated RNPs appear as fibrous, relatively phosphorus-rich structures located at the periphery of factories.
Figure 1.
(A) Schematic diagram of transcription of multiple genes at a nuclear RNAPII transcription factory. RNAPII factory shown as central blue circle with three transcribing genes and their associated transcription factors (small colored circles). Nascent transcripts are shown in red, chromatin is dark blue, and splicing components are depicted as small black circles with orange halo. (B) Electron spectroscopic imaging of HeLa cell nucleus. Phosphorous-rich structures are colored red and nitrogen green. Arrows point to nitrogen-rich transcription factory. White dots are immunogold detection of BrdU pulse-labeled nascent transcripts. Asterisks outline a small region of interchromatin granules (IG) and ch denotes regions of relatively compact chromatin. Image courtesy of Dr. Christopher Eskiw.
A GFP-tagged form of the human catalytic subunit of RNAPII (Sugaya et al. 2000) was used to monitor RNAPII kinetics in living cells by fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) (Kimura et al. 2002; Hieda et al. 2005). GFP-RNAPII dynamics were consistent with the existence of at least two populations of RNAPII in nuclei. Most of the tagged RNAPII (75–80% of the pool) had a very short recovery time on the order of seconds characteristic of rapid free diffusion as has been seen for other nuclear factors. The transcriptionally engaged fraction (20–25%) had a considerably longer recovery time (t1/2 approx. 20 min) consistent with relative immobilization. The latter fraction could be further subdivided into initiating and elongating subfractions of decreasing turnover rates and differential sensitivity to transcriptional inhibitors of initiation and elongation. These results suggested that RNAPII and most likely other factory components undergo continual but relatively slow dynamic exchange with freely diffusing components, but that the position of the factories themselves may be more stable. Under conditions in which new initiation is specifically and globally inhibited (Allen et al. 2004; Espinoza et al. 2004; Mariner et al. 2008), it was found that expressed genes disengage from transcription factories but focal sites of Ser5-RNAPII were still present after 30 min (Mitchell and Fraser 2008). These findings suggest that factories are not simply aggregates of RNAPII on active genes but appear to be genuine subnuclear compartments. Consistent with this model, inhibition with drugs which freeze or slow the elongating polymerase (Palstra et al. 2008), did not lead to genome reorganization.
HOW MANY FACTORIES ARE THERE?
The number of transcription sites per cell nucleus has been determined in a number of ways in many different cell and tissue types. The earliest methods used nascent transcript labeling in commonly used cultured cell lines such as HeLa and other fibroblastic cells and determined that there were 100–500 sites of nascent RNA per nucleus (Jackson et al. 1993; Wansink et al. 1993). In those days, the most reliable estimates put the number of eukaryotic genes at over 100,000 with the number of expressed or active genes in a given cell type around 10,000–30,000, far greater than the number of RNA synthesis sites. Wansink et al. (1993) suggested two possible reasons to explain this discrepancy. Because few eukaryotic genes are transcribed at relatively high rates and most are transcribed at low rates, the small number of nascent RNA sites could be caused by detection of only the most highly transcribed genes, and a failure to detect the bulk of active genes making few nascent transcripts. In this scenario there would be no evidence in favor of a factory model because each gene could be transcribing in isolation. Another possibility, particularly favored by Jackson and Cook was that each transcription site was occupied by a number of actively transcribed genes. Over the subsequent decade, refinements in nascent transcript labeling and detection have led to improved estimates of the number of factories per nucleus (Martin and Pombo 2003). Approximately 2100 (Iborra et al. 1996a) and 2400 (Jackson et al. 1998) nascent transcript sites were detected respectively in HeLa nuclei and an equivalent number of RNAPII sites were observed (Iborra et al. 1996a). Fay et al. (1997) had similar results but noted considerable variations in the number of nascent transcript sites (849–3888) in individual human fibroblasts. Pombo et al. (1999) using cryosectioning found evidence for 10,000 nucleoplasmic factories in HeLa cells with separate factories for RNAPII and RNAPIII; approximately 8000 RNAPII and 2000 RNAPIII factories. As the number of transcription sites increased, estimates of the number of eukaryotic genes in the genome and the number of active genes in a particular cell type decreased. However, the new appreciation of the extent of nongenic noncoding transcription has again increased the total number of transcription units and may now in fact exceed even the early estimates of gene numbers. So we are left with the realization that the number of active transcription units greatly outweighs the number of transcription sites per cell nucleus even with the highest estimates of factory numbers from HeLa cells.
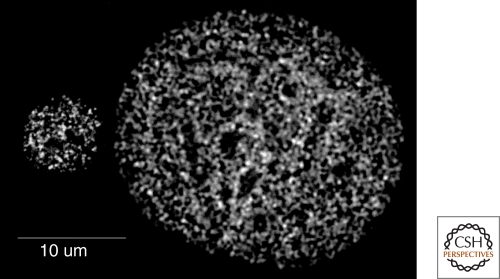
Some of the reports detailing the number of factories per nucleus make claims of highly sensitive techniques enabling the detection of all nascent transcription sites. Part of the variation in factory numbers observed may be because of differences in the ability to detect weak nascent RNA sites, but it also seems that there may be substantial variation within a cell type and between different cell types. In many cases HeLa or fibroblastic cells, which flatten out in culture were used, resulting in a substantial increase in nuclear diameter and nuclear volume when compared to cells with spherical nuclei. Osborne et al. (2004) used immunofluorescence to detect nuclear sites with high concentrations of the active form of RNAPII phosphorylated on Serine 5 (Ser5-RNAPII). They found approximately 2000 Ser5-RNAPII sites in the extended and flattened nuclei of mouse embryo fibroblast, consistent with previous nascent transcript labeling studies in fibroblastic cells (Fig. 2). In contrast, they found that erythroblasts, B-cells, T-cells, and fetal brain cells, which have spherical nuclei with significantly smaller radii and nuclear volumes, have dramatically fewer Ser5-RNAPII sites (100–300 per nucleus). Though the claim of detecting all RNAPII transcription sites was not made, the RNAPII detection threshold used by two different groups was sufficient to show that 90–99% of nascent RNA FISH signals for a variety of different genes overlapped with RNAPII factories (Osborne et al. 2004; Ragoczy et al. 2006; Osborne et al. 2007; Schoenfelder et al. 2010). Thus, although some RNAPII sites may have been missed because of the RNAPII detection threshold, the fact that nearly all nascent gene transcript signals colocalize with the RNAPII sites detected suggest that the majority of sites were detected. These results support the concept that all gene transcription occurs in RNAPII factories, consistent with conclusions from nascent transcript labeling studies. The large differences in factory numbers seen in nuclei from tissues versus cells grown on a surface appear genuine and may be a consequence of a reduced potential for intrachromosomal and especially interchromosomal sharing of factories in flattened cell nuclei. More factories may be required as an adaptation to service the same number of genes in flattened nuclei. Interestingly, changes in cell and nuclear morphology have been demonstrated to result in widespread changes in gene expression (Dalby et al. 2007; Chang and Hughes-Fulford 2009).
Figure 2.
Maximum intensity projections of Ser5-RNAPII factories in splenic B cell (left) and primary mouse embryo fibroblast (right) nuclei. From Osborne et al., 2004.
HIGHER-ORDER CHROMATIN FOLDING, TRANSCRIPTION AND SHARED RNAPII SITES
Since the discovery of transcriptional enhancers, which function to increase gene expression from remote genomic positions and independent of orientation to gene promoters, debate has centered on their mechanism. Most now agree that long-range gene control by remote enhancers and locus control regions involves direct interaction between chromatin at the enhancer sites and gene local regulatory elements (Carter et al. 2002; Tolhuis et al. 2002). It appears that DNA binding factors, transcription factors, and associated factors are required to either create an appropriate chromatin structure for distal elements to engage in interactions or mediate interactions by acting as bridging molecules (Drissen et al. 2004; Vakoc et al. 2005; Kurukuti et al. 2006; Splinter et al. 2006; Song et al. 2007; Majumder et al. 2008; Hadjur et al. 2009). Though we are still in the dark as to what specific transcriptional advantage such folding endows to a particular gene locus, there is little doubt that higher-order chromatin folding plays a critical role in transcriptional regulation over genomic distances of up to a megabase or more (Chakalova et al. 2005a; Kleinjan and van Heyningen 2005). One line of reasoning suggests that the increased local concentration of transcription factors brought into proximity of the promoter by the distal enhancer results in increased recruitment of (or to) the transcriptional machinery. A logical extension then leads to the question of what effect associations or clustering of distal actively transcribed genes and their complexed enhancers at transcription factories would have?
A critical tenet of the transcription factory model is that each site has the potential to be occupied by a number of actively transcribed genes or transcription units. Cook (2002) proposed that transcription factories might form by the aggregation of a local cluster of transcriptionally active genes and their associated polymerases. Logic suggested that active genes in proximity in the primary DNA sequence would be more likely to engage in the same factory than active genes separated by long stretches of inactive genomic sequence. This may be true, but the original demonstration that actively transcribed genes could indeed share the same factory greatly exceeded this expectation. Osborne et al. (2004) showed using primary transcript RNA FISH and immunofluorescence (RNA FISH) for Ser5-RNAPII that genes separated by 25–40 megabases of chromosomal DNA could share factories at remarkably high frequencies in mouse erythroid progenitors. Chromosome conformation capture (3C) (Dekker et al. 2002) was used to verify the spatial association between distal transcribed sequences. It was also apparent that active genes on separate chromosomes could share factories but appeared to do so at reduced frequency compared to linked transcribed genes. These results, coupled with a realization of the transcriptional behavior of “active” genes in vivo lead to some important conclusions.
Several lines of evidence suggest that transcription of “active” genes in a particular cell or tissue type is not continuous. Genes appear to be transcribed in bursts or pulses of transcriptional activity separated by variable periods of inactivity (Chubb et al. 2006; Raj et al. 2006). Similarly, RNA FISH for most “active” genes results in signals for only a portion of alleles across a population of expressing cell, with individual cells displaying none, one, or two actively transcribed alleles (Wijgerde et al. 1995; Levsky et al. 2002; Osborne et al. 2004; Osborne et al. 2007), There are a handful of so called “super genes,” which appear to defy this general rule (Fraser 2006). For example, the α- and β-globin genes (Hba and Hbb) in erythroid cells (Wijgerde et al. 1995) and the immunoglobulin genes (Igh, Igk, and Igl) in B-cells (Bolland et al. 2004; Osborne et al. 2007) tend to be continually or constitutively active with nearly all alleles in an interphase population associated with transcription factories (Osborne et al. 2004; Osborne et al. 2007). For other “expressed genes,” which appear to be less frequently transcribed, inactive alleles were located away from RNAPII factories. Thus the high degree of co-localization between transcribed genes suggested that upon activation a significantly high proportion of newly transcribed genes join pre-established transcription sites containing other active genes rather than assemble their own transcription site de novo. This was further investigated by studying the dynamics of the immediate early genes Fos and Myc in resting splenic mouse B-cells (Osborne et al. 2007). Most alleles for these genes are transcriptionally inactive and located away from factories in resting cells. However, in cells induced for 5 min, the opposite is found, most Fos and Myc alleles are associated with factories and transcriptionally active. A high percentage of the newly active Fos alleles associated with the same factory as the constitutively transcribed Igh locus located approximately 30 Mb away on chromosome 12. Most interestingly, 25% of the newly active Myc alleles from chromosome 15 were also found to be associated with the same factory as the Igh locus. This appeared to be a highly preferential interchromosomal association because RNA FISH analyses of many other genes in trans had significantly lower interchromosomal association frequencies with Igh. Previous studies had shown that chromosomes 12 and 15 are preferred neighbors in mouse B-cells (Roix et al. 2003; Parada et al. 2004) and that a significant degree of intermingling occurs between chromosome territories (Branco and Pombo 2006). That MYC and IGH come into proximity in B-cells is without doubt, because the two genes are the most common translocation partners in Burkitt's Lymphoma in humans and plasmacytomas in mouse (Hecht and Aster 2000; Potter 2003). Osborne's work showed that induction of MYC correlated with a measurable shift in position of MYC alleles towards IGH alleles, indicating that activation of transcription involves short-range chromatin movements over distances of 0.5–1.5 microns to access a factory, and suggesting that preferential interchromosomal transcription factory coassociations could be involved in the mechanism of chromosomal translocations. Others have also documented long-range intra- and interchromosomal interactions between loci or coassociations with specific nuclear subcompartments (Spilianakis et al. 2005; Bacher et al. 2006; Brown et al. 2006; Ling et al. 2006; Lomvardas et al. 2006; Simonis et al. 2006; Wurtele and Chartrand 2006; Xu et al. 2006; Zhao et al. 2006; Apostolou and Thanos 2008; Brown et al. 2008; Hu et al. 2008) and in many cases correlated clustering or proximity with effects on gene expression. This remains a very controversial area, and even those who promote such concepts, in many cases cannot agree on how, where and to what effect proximity in nuclear space has a functional role.
CHROMATIN DYNAMICS AND TRANSCRIPTION
Constrained Brownian motion of chromatin could in theory account for the rapid, short-range chromatin movement (Chakalova et al. 2005a) of induced Myc alleles toward the Igh factory mentioned earlier. The Myc gene in resting B-cells is not transcriptionally naïve. Myc expression is involved in stimulating proliferation of immature B-cells and would have been induced at various points in B-cell differentiation concurrent with Igh transcription. However, that is not to say that active or directed processes are not involved in reorganization of chromatin and transcription in vivo. More dramatic chromatin movements may be involved, and have been suggested in the initial activation of a locus from a completely silent state. Ground-breaking live cell studies (Chuang et al. 2006) reported dramatic vectorial movements of chromatin in response to targeting a transcriptional activator to a silent transgene array. Migration occurred 1–2 h after targeting at speeds of up to 0.9 microns/min over distances of 1–5 microns. Rapid vectorial movements appeared to be punctuated by brief periods of random diffusional motion. Chuang et al. (2006) showed that repositioning did not require transcription and did not appear to involve extensive decondensation of the transgene array structure. Actin and nuclear myosin were required for directed movements and the analysis of an actin point mutant defective in actin polymerization suggested that filamentous, F-actin may be involved. Dundr et al. (2007) also showed actin-dependent, long-range repositioning using a tagged array of inducible U2 snRNA genes. Again, hours rather than minutes after induction, they observed vectorial movement of the transgene array toward relatively stably positioned Cajal bodies that are normally found in association with snRNAs and histone gene loci. Expression of a dominant negative mutant of b-actin markedly inhibited repositioning, supporting a role for nuclear actin in long-range chromatin movements.
Large-scale chromatin movement has not been seen in every instance of transcriptional activation in live cells. Kumaran and Spector (Kumaran and Spector 2008) targeted a transgene array to the peripheral nuclear lamina. Transcriptional induction resulted in activation at the periphery with a clear increase in local RNAPII concentration. It was not clear whether the transgenes used local factories, which increased in size to accommodate the 200-copy array, or whether RNAPII was recruited to each promoter in the array de novo. However long-range movements were not observed. Similarly, Yao et al. (Yao et al. 2007) observed recruitment of polymerase components to heat shock loci in live cells on Drosophila polytene chromosomes creating characteristic puffs. Apart from local swelling, movement of the heat shock loci on the giant polytene chromosomes was not observed upon activation of transcription. Clearly live cell imaging holds the most potential for understanding the dynamic relationships between genes, transcriptional components and transcription factories. The studies of Kumaran and Spector (2008) and Yao et al. (2007) are probably the best available evidence in favor of the textbook model of transcription, in which RNAPII is recruited to gene promoters. However, aside from their obvious use, it remains to be seen whether the hundreds of gene copies in transgene arrays or giant polytene chromosomes are good models that accurately reflect the behavior of an isolated single copy gene in a diploid nucleus. It is possible that these localized multi-gene assemblies are in themselves super factories that alter the dynamic exchange of polymerase components (Kimura et al. 2002) to create a steady state structure that is several times larger than the average transcription factory.
TRANSCRIPTIONAL INTERACTOME AND TRANSCRIPTION HOTSPOTS
The number of transcription units sharing a factory at any one time has been estimated to be between two and 30 in HeLa cell nuclei (Jackson et al. 1998; Pombo et al. 1999). Mouse erythroid precursors, which have a few hundred factories, approximately 13,000 expressed alleles (6500 active genes assuming that most are bi-allelically expressed) with approximately half actively transcribing at any given moment are estimated to hold about 10–30 genes per factory (Schoenfelder et al. 2010). Obviously the large amount of nongenic transcription must be factored into this calculation, but most studies suggest that a substantial fraction of these transcripts occur in the vicinity of active genes (Kapranov et al. 2007) and in the case of the globin genes are found in a relatively small fraction of the erythroid cells concomitant with gene transcription (Ashe et al. 1997; Gribnau et al. 2000; Chakalova et al. 2005b; Miles et al. 2007). The observation that preferential interchromosomal gene associations at factories can occur raises the possibility that the combination of genes at a particular factory could be meaningful in terms of transcriptional output of the assembled genes. Recent studies examining the intranuclear localization of transcriptionally active mini-chromosomes suggested that similarly regulated genes cluster at a limited number of factories. Xu and Cook (2008) used expression constructs carrying transcription units driven by different promoters. The multicopy constructs were assembled into nucleosomes in cells and the resulting mini-chromosomes underwent rounds of transcription and replication. RNA immuno-FISH experiments showed that transcriptionally active copies of mini-chromosomes clustered at a subset of the available transcription factories. Moreover, the mini-chromosome appeared to share transcription sites with endogenous genes suggesting the introduced constructs were nonrandomly accommodated by host sites. The authors went on to cotransfect pairs of different constructs to test whether plasmids containing functionally similar genes and regulatory elements would share nuclear factories more often than functionally unrelated transcription units. Firstly, mini-chromosomes carrying RNAPI, II, or III transcription units separated into different factories, specialized in RNAPI, II, or III transcription, respectively. RNAPI genes acquired nucleolar localization, whereas RNAPII and III plasmids were spread in nonoverlapping foci throughout the nucleoplasm. These results were in agreement with earlier findings showing separate RNAPII and RNAPIII nucleoplasmic transcription sites (Pombo et al. 1999), and supported the view that the mini-chromosomes mimicked the behavior of endogenous chromosomal loci. Secondly, evidence for specialization was found within the RNAPII factory subclass. Mini-chromosomes carrying different genes driven by identical RNAPII promoters co-occupied the same foci at high frequencies. Conversely, plasmids carrying different promoters were mostly transcribed in separate factories. These preferences extended to endogenous genes too, because an edogenous gene was more likely to be in close proximity to its plasmid-borne counterpart than an unrelated gene. Taken together, these results suggested that factories are not functionally equal, and may be specialized to transcribe similarly regulated genes.
However, important questions remain: Does the tendency for factory preference or specialization result in a non-random organization of native or chromosomally integrated transcription units? Schoenfelder et al. (2010) answered these questions in two ways. First they observed the nuclear localization of ectopically integrated, transcriptionally active human HBB transgenes in erythroid cells from several different lines of transgenic mice. They found that the HBB transgene locus had a three- to 15-fold preference to be transcribed in the same factory as the endogenous mouse Hbb genes compared to the endogenous Hba locus. The fact that the ectopically integrated transgene in all six lines tested showed preferential interchromosomal associations, in one case occurring in up to 35% of cells, suggested that preferential factory co-associations had the potential to reorganize the genome in the nucleus. They next developed an enhanced 4C assay to screen the entire genome for genes sharing transcription factories with the Hbb or Hba genes. Supported by extensive FISH and 3C analyses they revealed two overlapping transcription networks, each involving hundreds of preferred transcription partners in cis and trans. Genes regulated by the erythroid-specific transcription factor, Klf1 (erythroid Kruppel-like factor) were overrepresented in the globin transcription networks suggesting that coregulated genes preferentially transcribed in shared factories. Klf1 was previously shown to be required for high-level transcription of the Hbb gene in definitive erythroid cells (Nuez et al. 1995; Perkins et al. 1995) and also appears to contribute to Hba regulation (Shyu et al. 2006; Vernimmen et al. 2007). Schoenfelder et al. (2010) used immunofluorescence to show that, Klf1 localized to 30-40 small foci in erythroid nuclei and that nearly all overlapped with Ser5-RNAPII foci. These results suggested the possibility that a subset of factories were specialized because of an increased concentration of a particular transcription factor; a phenomena seen before for other factors (Grande et al. 1997). Actively transcribed alleles of Klf1-regulated genes were more often found in association with factories containing high levels of Klf1 suggesting that they had a higher probability of transcription when associated with a specialized factory. Furthermore, they found that many Klf1-regulated genes were preferentially clustered at these sites. This data provided strong evidence for functional specialization of transcription factories, and suggested that individual factories could become hotspots for optimal transcription of a subset of coregulated genes. There seemed to be no specific requirement for the globin genes to transcribe in the same factory with any other particular gene, but that in most, if not all, erythroid cells the globin genes associated with a varied subset of other Klf1 regulated genes (Schoenfelder et al. 2010).
PULLING SOME STRINGS
The emerging evidence provides strong support for the transcription factory model where distant genes in cis and trans nonrandomly associate during transcription. A key feature of this model as originally proposed by Cook is that transcribed genes are reeled through factories by the relatively immobilized polymerase while extruding nascent RNA, rather than the polymerase tracking along the template. Cook and colleagues recently tested this prediction by assessing nuclear proximity of two distal, rapidly induced genes, one long and one short. SAMD4A is 221 kb long and located approximately 50 Mb from TNFAIP2—a relatively short gene of 11 kb. Both genes are activated simultaneously by tumor necrosis factor α(TNFα). Induction of SAMD4A has been shown to result in a fairly synchronous wave of transcription and RNAPII traversing the transcription unit over a 70-min period (Wada et al. 2009). The 3C assay was used to investigate long-range associations over this time period between the short, repeatedly transcribed TNFAIP2 gene and several regions along the length of SAMD4A. Before induction there was no evidence for long-range association between these genes, but upon activation they found that proximity between TNFAIP2 and the SAMD4A promoter was rapidly induced. As the wave of RNAPII moved along SAMD4A, contacts between the promoter and TNFAIP2 were lost in favor of new contacts between TNFAIP2 and successive downstream regions of SAMD4A. In other words, it appeared that the long gene was sliding past the distal short gene with a temporal pattern that matched the progress of the transcribing polymerase on the long gene. These results showing that different parts of transcription units are brought into proximity are consistent with the idea that transcribed genes are reeled through factories and very difficult to reconcile with the textbook model of RNAPII sliding along a relatively immobile template.
CONCLUDING REMARKS
The realization that the active form of RNAPII is compartmentalized in the nucleus into a finite number of discrete transcription factories and that actively transcribed genes are non-randomly organized around these sites has widespread implications in understanding nuclear organization of the genome and genome function. This turns the classical view of gene transcription on its head, and with this new understanding come new insights and new questions. Rather than the requirement for near simultaneous recruitment and assembly of hundreds of factors required for regulated activation of transcription at the site of individual genes, this model suggests that many transcriptional components are preassembled into active sites which genes migrate to for transcription. There is still of course ample scope for regulatory factors and transcription components being recruited to genes while outside factories. For example, transcription factors, cofactors, chromatin remodeling complexes, and various histone modifying activities may act as licensing factors that affect genes before factory entry to prepare them for productive engagements with an RNAPII complex. The hypophosphorylated or initial binding form of RNAPII may also engage genes outside factories. The chance or directed coassociation of coregulated genes at a factory may favor re-initiation of those genes through sharing of dynamically binding or interacting factors, which will in turn stabilize the presence of those genes at the factory for as long as it takes to transcribe them. This may then become a favorable place for other coregulated genes to alight leading to a dynamically self-organizing hotspot of transcription for a specific subgroup or network of genes. Within the factory, epigenetic modifications may also be laid down or re-affirmed in transcribed regions, which may further promote re-initiation and/or serve as a memory for the activation state. Obviously chromatin mobility is a potentially important component of this new understanding. Factors or activities that compact chromatin or tether them to “repressive” nuclear domains may serve to restrict mobility and access to transcription factories. Genes or alleles not able to reach one of their hotspots may disengage after relative few rounds of transcription at a generic factory because of a relatively low local concentration of specific factors required for re-initiation and stabilization at a factory.
There is still much to do and many of the important questions will best be answered by live cell experiments on single copy or endogenous genes. For example, the transcriptional period of individual genes needs to be assessed. Are factory encounters transient and highly dynamic, potentially reshuffling at every transcriptional cycle, or once established, do active alleles/genes remain associated with factories throughout a cell cycle. It is likely the answers will vary widely for different genes and may encompass the entire range, potentially influenced by the character of long-range regulatory elements that individual active genes are complexed with as well as other transcription units they may encounter in factories. Also, how does the genome end up in such highly-organized, tissue-specific conformations that allow specific subgroups of genes the opportunity to cluster at specialized factories? There is controversy surrounding evidence suggesting chromosome positions may be partially inherited through mitosis (Gerlich et al. 2003; Walter et al. 2003; Cvackova et al. 2009), and that in very early G1 phase genome reorganization is more dramatic than in the rest of interphase (Dimitrova and Gilbert 1999). It appears likely that mitosis offers an essential opportunity for large-scale, though imperfect, genome reorganization which may be further refined in early G1. If the organized associations of co-regulated genes are beneficial then we may expect considerable pressure to be exerted on the primary organization of the genome, such that individual genes are optimally placed to take advantage of their eventual folding in 3 dimensions in the various tissues in which they will be expressed. These are exciting times and the advent of new technologies coupled with deep sequencing along with advanced microscopy hold great promise for further exciting advances in understanding the relationships between genome organization and transcriptional control as well as other genome functions. And hopefully, future textbooks will reflect these important aspects of the transcriptional mechanism so that the next generation of genome investigators will be more adequately prepared to advance the understanding of transcription inside the complex world of the nucleus.
ACKNOWLEDGMENTS
We would like to thank in particular C. Eskiw and other members of the Laboratory of Chromatin and Gene Expression for many helpful discussions. We are also grateful to A. Papantonis and P. Cook for generously sharing unpublished data.
Footnotes
Editors: Tom Misteli and David L. Spector
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Allen TA, Von Kaenel S, Goodrich JA, Kugel JF 2004. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol 11: 816–821 [DOI] [PubMed] [Google Scholar]
- Apostolou E, Thanos D 2008. Virus Infection Induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell 134: 85–96 [DOI] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ 1997. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev 11: 2494–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol 8: 293–299 [DOI] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE 2004. Antisense intergenic transcription in V(D)J recombination. Nat Immunol 5: 630–637 [DOI] [PubMed] [Google Scholar]
- Branco MR, Pombo A 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biology 4: e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, et al. 2008. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol 182: 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ 2006. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol 172: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P 2002. Long-range chromatin regulatory interactions in vivo. Nat Genet 32: 623–626 [DOI] [PubMed] [Google Scholar]
- Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P 2005a. Replication and transcription: Shaping the landscape of the genome. Nat Rev Genet 6: 669–677 [DOI] [PubMed] [Google Scholar]
- Chakalova L, Osborne CS, Dai YF, Goyenechea B, Metaxotou-Mavromati A, Kattamis A, Kattamis C, Fraser P 2005b. The Corfu δβ thalassemia deletion disrupts γ-globin gene silencing and reveals post-transcriptional regulation of HbF expression. Blood 105: 2154–2160 [DOI] [PubMed] [Google Scholar]
- Chang TT, Hughes-Fulford M 2009. Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue Eng Part A 15: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH 2006. Transcriptional pulsing of a developmental gene. Curr Biol 16: 1018–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, van Driel R, Fakan S 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell 10: 211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR 2002. Predicting three-dimensional genome structure from transcriptional activity. Nat Genet 32: 347–352 [DOI] [PubMed] [Google Scholar]
- Cremer T, Kupper K, Dietzel S, Fakan S 2004. Higher order chromatin architecture in the cell nucleus: On the way from structure to function. Biol Cell 96: 555–567 [DOI] [PubMed] [Google Scholar]
- Cvacková Z, Masata M, Stanĕk D, Fidlerová H, Raska I 2009. Chromatin position in human HepG2 cells: Although being non-random, significantly changed in daughter cells. J Struct Biol. 165:107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby MJ, Gadegaard N, Herzyk P, Sutherland D, Agheli H, Wilkinson CD, Curtis AS 2007. Nanomechanotransduction and interphase nuclear organization influence on genomic control. J Cell Biochem 102: 1234–1244 [DOI] [PubMed] [Google Scholar]
- Davenport RJ, Wuite GJ, Landick R, Bustamante C 2000. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science 287: 2497–2500 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993 [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W 2004. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179: 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw CH, Rapp A, Carter DR, Cook PR 2008. RNA polymerase II activity is located on the surface of protein-rich transcription factories. J Cell Sci 121: 1999–2007 [DOI] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA 2004. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol 11: 822–829 [DOI] [PubMed] [Google Scholar]
- Fakan S 1994. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol 4: 86–90 [DOI] [PubMed] [Google Scholar]
- Fakan S 2004. The functional architecture of the nucleus as analysed by ultrastructural cytochemistry. Histochem Cell Biol 122: 83–93 [DOI] [PubMed] [Google Scholar]
- Fakan S, Bernhard W 1971. Localisation of rapidly and slowly labelled nuclear RNA as visualized by high resolution autoradiography. Exp Cell Res 67: 129–141 [DOI] [PubMed] [Google Scholar]
- Fakan S, Puvion E, Sphor G 1976. Localization and characterization of newly synthesized nuclear RNA in isolate rat hepatocytes. Exp Cell Res 99: 155–164 [DOI] [PubMed] [Google Scholar]
- Fay FS, Taneja KL, Shenoy S, Lifshitz L, Singer RH 1997. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A). Exp Cell Res 231: 27–37 [DOI] [PubMed] [Google Scholar]
- Fraser P 2006. Transcriptional control thrown for a loop. Curr Opin Genet Dev 16: 490–495 [DOI] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112: 751–764 [DOI] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, de Jong L, van Driel R 1997. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci 110: 1781–1791 [DOI] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell 5: 377–386 [DOI] [PubMed] [Google Scholar]
- Guthold M, Zhu X, Rivetti C, Yang G, Thomson NH, Kasas S, Hansma HG, Smith B, Hansma PK, Bustamante C 1999. Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys J 77: 2284–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, Yanagida T 1999. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys J 76: 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht JL, Aster JC 2000. Molecular biology of Burkitt's lymphoma. J Clin Oncol 18: 3707–3721 [DOI] [PubMed] [Google Scholar]
- Hieda M, Winstanley H, Maini P, Iborra FJ, Cook PR 2005. Different populations of RNA polymerase II in living mammalian cells. Chromosome Res 13: 135–144 [DOI] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. 2008. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci 105: 19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL 1996. Dynamic organization of pre-mRNA splicing factors. J Cell Biochem 62: 191–197 [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR 1996a. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci 109: 1427–1436 [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, McManus J, Jackson DA, Cook PR 1996b. The topology of transcription by immobilized polymerases. Exp Cell Res 229: 167–173 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Cook PR 1985. Transcription occurs at a nucleoskeleton. EMBO J 4: 919–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, McCready SJ, Cook PR 1981. RNA is synthesized at the nuclear cage. Nature 292: 552–555 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR 1993. Visualization of focal sites of transcription within human nuclei. Embo J 12: 1059–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR 1998. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell 9: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, Shimamoto N 1993. Visualization of single molecules of RNA polymerase sliding along DNA. Science 262: 1561–1563 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR 2007. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Kimura H, Sugaya K, Cook PR 2002. The transcription cycle of RNA polymerase II in living cells. J Cell Biol 159: 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V 2005. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am J Hum Genet 76: 8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL 2003. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo RC, Singer RH 2002. Single-cell gene expression profiling. Science 297: 836–840 [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272 [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413 [DOI] [PubMed] [Google Scholar]
- Majumder P, Gomez JA, Chadwick BP, Boss JM 2008. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med 205: 785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29: 499–509 [DOI] [PubMed] [Google Scholar]
- Martin S, Pombo A 2003. Transcription factories: Quantitative studies of nanostructures in the mammalian nucleus. Chromosome Res 11: 461–470 [DOI] [PubMed] [Google Scholar]
- Miles J, Mitchell JA, Chakalova L, Goyenechea B, Osborne CS, O'Neill L, Tanimoto K, Engel JD, Fraser P 2007. Intergenic transcription, cell-cycle and the developmentally regulated epigenetic profile of the human β-globin locus. PLoS One 2: e630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387: 523–527 [DOI] [PubMed] [Google Scholar]
- Mitchell JA, Fraser P 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev 22: 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash RE, Puvion E, Bernhard W 1975. Perichromatin fibrils as components of rapidly labeled extranucleolar RNA. J Ultrastruct Res 53: 395–405 [DOI] [PubMed] [Google Scholar]
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375: 316–318 [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by igh. PLoS Biol 5: e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W 2008. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One 3: e1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T 2004. Tissue-specific spatial organization of genomes. Genome Biol 5: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH 1995. Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375: 318–322 [DOI] [PubMed] [Google Scholar]
- Pombo A, Cook PR 1996. The localization of sites containing nascent RNA and splicing factors. Exp Cell Res 229: 201–203 [DOI] [PubMed] [Google Scholar]
- Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR 1999. Regional specialization in human nuclei: Visualization of discrete sites of transcription by RNA polymerase III. EMBO J 18: 2241–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M 2003. Neoplastic development in plasma cells. Immunol Rev 194: 177–195 [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M 2006. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 20: 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 34: 287–291 [DOI] [PubMed] [Google Scholar]
- Schafer DA, Gelles J, Sheetz MP, Landick R 1991. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature 352: 444–448 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42:53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu YC, Wen SC, Lee TL, Chen X, Hsu CT, Chen H, Chen RL, Hwang JL, Shen CK 2006. Chromatin-binding in vivo of the erythroid kruppel-like factor, EKLF, in the murine globin loci. Cell Res 16: 347–355 [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Song SH, Hou C, Dean A 2007. A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol Cell 28: 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya K, Vigneron M, Cook PR 2000. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J Cell Sci 113: 2679–2683 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. Embo J 26: 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, et al. 2009. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci 106:18357–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160:685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM 1998. Force and velocity measured for single molecules of RNA polymerase. Science 282: 902–907 [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L 1993. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol 122: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M, Grosveld F, Fraser P 1995. Transcription complex stability and chromatin dynamics in vivo. Nature 377: 209–213 [DOI] [PubMed] [Google Scholar]
- Wurtele H, Chartrand P 2006. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res 14: 477–495 [DOI] [PubMed] [Google Scholar]
- Xu M, Cook PR 2008. Similar active genes cluster in specialized transcription factories. J Cell Biol 181: 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT 2007. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28: 978–990 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]