Abstract
Understanding the origin of cellular life on Earth requires the discovery of plausible pathways for the transition from complex prebiotic chemistry to simple biology, defined as the emergence of chemical assemblies capable of Darwinian evolution. We have proposed that a simple primitive cell, or protocell, would consist of two key components: a protocell membrane that defines a spatially localized compartment, and an informational polymer that allows for the replication and inheritance of functional information. Recent studies of vesicles composed of fatty-acid membranes have shed considerable light on pathways for protocell growth and division, as well as means by which protocells could take up nutrients from their environment. Additional work with genetic polymers has provided insight into the potential for chemical genome replication and compatibility with membrane encapsulation. The integration of a dynamic fatty-acid compartment with robust, generalized genetic polymer replication would yield a laboratory model of a protocell with the potential for classical Darwinian biological evolution, and may help to evaluate potential pathways for the emergence of life on the early Earth. Here we discuss efforts to devise such an integrated protocell model.
Studies of synthetic protocells, in which fatty acids encapsulate genetic polymers, provide clues to how life forms first emerged on Earth and coordinated their growth with division.
The emergence of the first cells on the early Earth was the culmination of a long history of prior chemical and geophysical processes. Although recognizing the many gaps in our knowledge of prebiotic chemistry and the early planetary setting in which life emerged, we will assume for the purpose of this review that the requisite chemical building blocks were available, in appropriate environmental settings. This assumption allows us to focus on the various spontaneous and catalyzed assembly processes that could have led to the formation of primitive membranes and early genetic polymers, their coassembly into membrane-encapsulated nucleic acids, and the chemical and physical processes that allowed for their replication. We will discuss recent progress toward the construction of laboratory models of a protocell (Fig. 1), evaluate the remaining steps that must be achieved before a complete protocell model can be constructed, and consider the prospects for the observation of spontaneous Darwinian evolution in laboratory protocells. Although such laboratory studies may not reflect the specific pathways that led to the origin of life on Earth, they are proving to be invaluable in uncovering surprising and unanticipated physical processes that help us to reconstruct plausible pathways and scenarios for the origin of life.
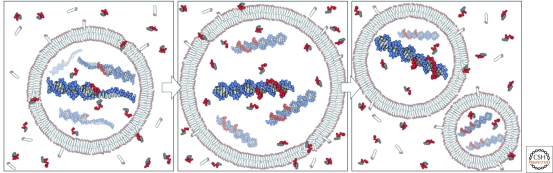
Figure 1.
A simple protocell model based on a replicating vesicle for compartmentalization, and a replicating genome to encode heritable information. A complex environment provides lipids, nucleotides capable of equilibrating across the membrane bilayer, and sources of energy (left), which leads to subsequent replication of the genetic material and growth of the protocell (middle), and finally protocellular division through physical and chemical processes (right). (Reproduced from Mansy et al. 2008 and reprinted with permission from Nature Publishing ©2008.)
The term protocell has been used loosely to refer to primitive cells or to the first cells. Here we will use the term protocell to refer specifically to cell-like structures that are spatially delimited by a growing membrane boundary, and that contain replicating genetic information. A protocell differs from a true cell in that the evolution of genomically encoded advantageous functions has not yet occurred. With a genetic material such as RNA (or perhaps one of many other heteropolymers that could provide both heredity and function) and an appropriate environment, the continued replication of a population of protocells will lead inevitably to the spontaneous emergence of new coded functions by the classical mechanism of evolution through variation and natural selection. Once such genomically encoded and therefore heritable functions have evolved, we would consider the system to be a complete, living biological cell, albeit one much simpler than any modern cell (Szostak et al. 2001).
BACKGROUND
Membranes as compartment boundaries
All biological cells are membrane-bound compartments. The cell membrane fulfills the essential function of creating an internal environment within which genetic materials can reside and metabolic activities can take place without being lost to the environment. Modern cell membranes are composed of complex mixtures of amphiphilic molecules such as phospholipids, sterols, and many other lipids as well as diverse proteins that perform transport and enzymatic functions. Phospholipid membranes are stable under a wide range of temperature, pH, and salt concentration conditions. Such membranes are extremely good permeability barriers, so that modern cells have complete control over the uptake of nutrients and the export of wastes through the specialized channel, pump and pore proteins embedded in their membranes. A great deal of complex biochemical machinery is also required to mediate the growth and division of the cell membrane during the cell cycle. The question of how a structurally simple protocell could accomplish these essential membrane functions is a critical aspect of understanding the origin of cellular life.
Vesicles formed by fatty acids have long been studied as models of protocell membranes (Gebicki and Hicks 1973; Hargreaves and Deamer 1978; Walde et al. 1994a). Fatty acids are attractive as the fundamental building block of prebiotic membranes in that they are chemically simpler than phospholipids. Fatty acids with a saturated acyl chain are extremely stable compounds and therefore might have accumulated to significant levels, even given a relatively slow or episodic synthesis. Moreover, the condensation of fatty acids with glycerol to yield the corresponding glycerol esters provides a highly stabilizing membrane component (Monnard et al., 2002). Finally, phosphorylation and the addition of a second acyl chain yields phosphatidic acid, the simplest phospholipid, thus providing a conceptually simple pathway for the transition from primitive to more modern membranes. The prebiotic chemistry leading to the synthesis of fatty acids and other amphiphilic compounds is treated in more detail in Mansy (2010).
The best reason for considering fatty acids as fundamental to the nature of primitive cell membranes is not, however, their chemical simplicity. Rather, fatty-acid molecules in membranes have dynamic properties that are essential for both membrane growth and permeability. Because fatty acids are single chain amphiphiles with less hydrophobic surface area than phospholipids, they assemble into membranes only at much higher concentrations. This equilibrium property is mirrored in their kinetics: Fatty acids are not as firmly anchored within the membrane as phospholipids; they enter and leave the membrane on a time scale of seconds to minutes (Chen and Szostak 2004). Fatty acids can also exchange between the two leaflets of a bilayer membrane on a subsecond time scale. Rapid flip-flop is essential for membrane growth when new amphiphilic molecules are supplied from the environment. New molecules enter the membrane primarily from the outside leaflet, and flip-flop allows the inner and outer leaflet areas to equilibrate, leading to uniform growth.
Considering that protocells on the early Earth did not, by definition, contain any complex biological machinery, they must have relied on the intrinsic permeability properties of their membranes. Membranes composed of fatty acids are in fact reasonably permeable to small polar molecules and even to charged species such as ions and nucleotides (Mansy et al. 2008). This appears to be largely a result of the ability of fatty acids to form transient defect structures and/or transient complexes with charged solutes, which facilitate transport across the membrane. The subject of the permeability of fatty-acid based membranes is dealt with in greater detail by Mansy (2010).
Prebiotic vesicles were almost certainly composed of complex mixtures of amphiphiles. Amphiphilic molecules isolated from meteorites (Deamer 1985; Deamer and Pashley 1989) as well as those synthesized under simulated prebiotic conditions (McCollum et al. 1999; Dworkin et al. 2001; Rushdi and Simoneit 2001) are highly heterogeneous, both in terms of acyl chain length and head group chemistry. Membranes composed of mixtures of amphiphiles often have superior properties to those composed of single pure species. For example, mixtures of fatty acids together with the corresponding alcohols and/or glycerol esters generate vesicles that are stable over a wider range of pH and ionic conditions (Monnard et al. 2002), and are more permeable to nutrient molecules including ions, sugars and nucleotides (Chen et al. 2004; Sacerdote and Szostak 2005; Mansy et al. 2008). This is in striking contrast to the apparent requirement for homogeneity in the nucleic acids, where even low levels of modified nucleotides can be destabilizing or can block replication.
RECENT RESULTS
Pathways for Vesicle Growth
Fatty-acid vesicle growth has been shown to occur through at least two distinct pathways: growth through the incorporation of fatty acids from added micelles, and growth through fatty-acid exchange between vesicles. The growth of membrane vesicles from micelles has been observed following the addition of micelles or fatty-acid precursors to pre-formed vesicles (Walde et al. 1994a, 1994b; Berclaz et al. 2001). When initially alkaline fatty-acid micelles are mixed with a buffered solution at a lower pH, the micelles become thermodynamically unstable. As a consequence, the fatty-acid molecules can either be incorporated into pre-existing membranes, leading to growth (Berclaz et al. 2001), or can self-assemble into new vesicles (Blochliger et al. 1998; Luisi et al. 2004). These pioneering studies were done by cryo-TEM, which does not allow growth to be followed in real time, and by light scattering, which is difficult to interpret in the case of samples with heterogeneous size distributions. We therefore adapted a fluorescence assay based on FRET (Förster resonance energy transfer) to measure changes in membrane area in real time. This assay is based on the distance dependence of energy transfer between donor and acceptor fluorescent dyes; thus when a membrane grows in area by incorporating additional fatty-acid molecules, the dyes are diluted and the efficiency of FRET decreases. Studies on small (typically 100 nm in diameter) unilamellar fatty-acid vesicles using this assay showed that the slow addition of fatty-acid micelles led to vesicle growth with an efficiency of ∼90% (Hanczyc et al. 2003).
The real-time FRET assay allowed for a kinetic dissection of the growth process, revealing a surprisingly complex series of events after the rapid addition of micelles (Chen and Szostak 2004). Two major processes were observed. The first fast phase resulted in membrane area growth that was limited to ∼40% increase in area, independent of the amount of added micelles. A second much slower phase led to a further increase in membrane area that varied with the amount of added micelles. We interpreted the fast phase as reflecting the rapid assembly of a layer of adhering micelles around the pre-formed vesicles, with rapid monomer exchange resulting in the efficient incorporation of this material into the pre-formed membrane. We interpreted the slow phase as the consequence of micelle–micelle interactions leading to the assembly of intermediate structures that could partition between two pathways—with some monomers dissociating and contributing to membrane growth and the remainder ultimately assembling into new membrane vesicles. Although these interpretations are consistent with our data, the experiments are rather indirect, and further exploration of the mechanism of membrane growth is certainly desirable.
A second, distinct pathway for vesicle growth involves fatty-acid exchange between vesicles. Under certain conditions this exchange can lead to growth of a subpopulation of vesicles at the expense of their surrounding neighbors. Within populations of osmotically relaxed vesicles, such exchange processes do not result in significant changes in size distribution with time. Similarly, a population of uniformly osmotically swollen vesicles does not change in size distribution, but such vesicles are in equilibrium with a lower solution concentration of fatty acids because the tension in the membrane of the swollen vesicles makes it more energetically favorable for fatty-acid molecules to reside in membrane. When osmotically swollen vesicles are mixed with osmotically relaxed (isotonic) vesicles, rapid fatty-acid exchange processes result in growth of the swollen vesicles and corresponding shrinkage of the relaxed vesicles (Chen et al. 2004). Because vesicles can be osmotically swollen as a result of the encapsulation of high concentrations of nucleic acids such as RNA, this process allows for the growth of vesicles containing genetic polymers at the expense of empty vesicles (or vesicles that contain less internal nucleic acid). Because faster replication would increase the internal nucleic acid concentration, this pathway of competitive vesicle growth provides the potential for a direct physical link between the rate of replication of an encapsulated genetic polymer and the rate of growth of the protocell as a whole.
Assuming that the division of osmotically swollen vesicles could occur either stochastically or at some threshold size, protocells that developed some heritable means of faster replication and growth would have a shorter cell cycle, on average, and would therefore gradually take over the population. This simple physical mechanism might therefore lead to the emergence of Darwinian evolution by competition at the cellular level. However, if replication is limited by the rapid reannealing of complementary strands (see later discussion), it may be difficult to reach osmotically significant concentrations. Furthermore, osmotically swollen vesicles are intrinsically difficult to divide owing to the energetic cost of reducing the volume of a spherical vesicle to that of two daughter vesicles of the same total surface area. One possibility is that osmotically driven competitive growth might alternate with the faster membrane growth that follows micelle addition. If new fatty-acid material was only available sporadically, rapid membrane growth might follow an influx of fresh fatty acids, facilitating division (see following discussion).
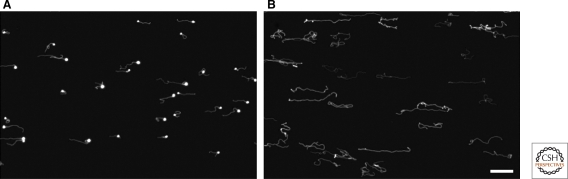
All of the experiments discussed earlier were done with small unilamellar vesicles prepared by extrusion through 100 nm pores in filters. In contrast, fatty-acid vesicles that form spontaneously by rehydrating dry fatty-acid films tend to be several microns in diameter and multilamellar (Hargreaves and Deamer 1978; Hanczyc et al. 2003). Such large multilamellar fatty-acid vesicles are so heterogeneous that quantitative studies of growth and division are difficult. We have recently developed a simple procedure for the preparation of micron-sized, monodisperse (homogeneous in size) multilamellar vesicles by large-pore dialysis (Zhu and Szostak 2009b). The preparation of large monodisperse multilamellar vesicles has allowed us to directly observe an unusual mode of vesicle growth (Zhu and Szostak 2009a). We showed that feeding a micron-sized multilamellar fatty-acid vesicle with fatty-acid micelles results in the formation of a thin membranous protrusion which extends from the side of the initially spherical parental vesicle. Over time, this thin membrane tubule elongates and thickens, gradually incorporating more and more of the parental vesicle, until eventually the entire vesicle is transformed into a long, hollow threadlike vesicle (Fig. 2). This pathway occurs with vesicles ranging in size from 1 to at least 10 µm in diameter, composed of a variety of different fatty acids and related amphiphiles. Only multilamellar vesicles grow in this manner and only when vesicle volume increases slowly (relative to surface area growth) because of a relatively impermeable buffer solute. Confocal microscopy has provided insight into the mechanism of this mode of growth: The outermost membrane layer grows first, and because there is little volume between it and the next membrane layer, and that volume cannot increase on the same time scale, the extra membrane area is forced into the form of a thin tubule. Over time, this tubule grows, and as a result of poorly understood exchange processes, the entire original vesicle is ultimately transformed into a long thread-like hollow vesicle (Fig. 3).
Figure 2.
Vesicle shape transformations during growth. All vesicles are labeled with 2 mM encapsulated HPTS, a water-soluble fluorescent dye, in their internal aqueous space. (A) 10 min and (B) 30 min after the addition of five equivalents of oleate micelles to oleate vesicles (in 0.2 M Na-bicine, pH 8.5). Scale bar: 50 µm. (Reproduced from Zhu and Szostak 2009a and reprinted with permission from ACS Publications ©2009.)
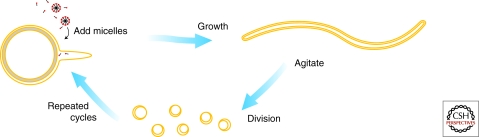
Figure 3.
Schematic diagram of coupled vesicle growth and division. (Reproduced from Zhu and Szostak 2009a and reprinted with permission from the Journal of the American Chemical Society ©2009.)
Pathways for Vesicle Division
Vesicle division by the extrusion of large vesicles through small pores is a way in which mechanical energy can be used to drive division (Hanczyc et al. 2003). Vesicle growth by micelle feeding followed by division by extrusion can be performed repetitively, resulting in cycles of growth and division in which both membrane material and vesicle contents are distributed to daughter vesicles in each cycle. However, division by extrusion results in the loss of 30%–40% of the encapsulated vesicle contents to the environment during each cycle (Hanczyc et al. 2003; Hanczyc and Szostak 2004). Most of this loss is a result of the unavoidable geometric constraint of dividing a spherical vesicle into two spherical (or subspherical) daughter vesicles with conservation of surface area; some additional loss may occur as a result of pressure-induced membrane rupture. Although extrusion is a useful laboratory model for vesicle division, an analogous extrusion process appears unlikely to occur in a prebiotic scenario on the early Earth because vesicle extrusion from the flow of suspended vesicles through a porous rock would require both the absence of any large pores or channels and a very high pressure gradient (Zhu and Szostak 2009a).
The above problems stimulated a search for a more realistic pathway for vesicle division. The possible spontaneous division of small unilamellar vesicles after micelle addition has been reported (Luisi et al. 2004; Luisi 2006), and electron microscopy has revealed structures that are possible intermediates in growth and division, notably pairs of vesicles joined by a shared wall (Stano et al. 2006). However, the mechanism of the proposed division as well as the nature of the energetic driving force remain unclear. Additional studies are required to clearly distinguish between the vesicle-stimulated assembly of new vesicles, and the more biologically relevant processes of growth and division.
We have recently found that the growth of large multilamellar vesicles into long threadlike vesicles, described above, provides a pathway for coupled vesicle growth and division (Zhu and Szostak 2009a). The long threadlike vesicles are extremely fragile, and divide spontaneously into multiple daughter vesicles in response to modest shear forces. In an environment of gentle shear, growth and division become coupled processes because only the filamentous vesicles can divide (Fig. 3). If the initial parental vesicle contains encapsulated genetic polymers such as RNA, these molecules are distributed randomly to the daughter vesicles and are thus inherited. The robustness and simplicity of this pathway suggests that similar processes might have occurred under prebiotic conditions. The mechanistic details of this mode of division remain unclear. One possibility, supported by some microscopic observations, is that the long thin membrane tubules are subject to the “pearling instability” (Bar-Ziv and Moses 1994), and minimize their surface energy by spontaneously transforming from a cylindrical shape to a string of beads morphology. The very thin tether joining adjacent spherical beads may be a weak point that can be easily disrupted by shear forces.
RNA-Catalyzed RNA Replication on the Early Earth and the Modern Laboratory
A core assumption of the RNA world hypothesis is that the RNA genomes of primitive cells were replicated by a ribozyme RNA polymerase (Gilbert 1986). The idea of RNA-catalyzed RNA replication provides a solution to the apparent paradox of DNA replication catalyzed by proteins that are encoded by DNA. This simplification of early biochemistry gained instant plausibility from the discovery of catalytic RNAs almost 30 years ago (Kruger et al. 1982; Guerrier-Takada et al. 1983). In the time after the discovery of the first ribozymes, the RNA World hypothesis has continued to gain support, most dramatically from the discovery that the ribosome is a ribozyme (Nissen et al. 2000), and that all proteins are assembled through the catalytic activity of the ribosomal RNA. Support has also come from the in vitro evolution of a wide range of new ribozymes, including bona fide RNA polymerases made of RNA (Johnston et al. 2001). On the other hand, no ribozyme polymerase yet comes close to being a self-replicating RNA, like the replicase envisaged as the core of the RNA World biochemistry.
Why has the in vitro evolution of an RNA replicase been so much more difficult than originally expected? It is clear that the problem is not with catalysis of the chemical step, even with catalytically demanding triphosphate substrates. Evolutionarily optimized versions of the Class I ligase carry out multiple-turnover ligation reactions at >1 s−1, with over 50,000 turnovers overnight (Ekland et al. 1995), and optimized versions of the smaller, simpler DSL ligase carry out sustained multiple turnover ligation reactions at rates >1/min (Voytek and Joyce 2007). These catalytic rates are more than sufficient to carry out the replication of a 100–200 nt ribozyme in minutes to hours, if these rates could be maintained in the context of a polymerase reaction using monomer substrates. However, even the best available polymerase ribozyme requires 1–2 days to copy 10–20 nucleotides of a template strand, apparently as a consequence of poor binding to both the ribonucleoside triphosphate (NTP) monomers and the primer-template substrate. The need to overcome the electrostatic repulsion between negatively charged NTP and RNA substrates and ribozyme is thought to contribute to the very high Mg++ requirement for the polymerase ribozyme (Glasner et al. 2000). Such high levels of Mg++ lead to hydrolytic degradation of the ribozyme, and are also not compatible with known fatty-acid based membranes because of crystallization of the fatty acid-magnesium salt. In addition, fatty-acid membranes are almost impermeable to NTPs (Mansy et al. 2008).
The incompatibility between currently available ribozyme polymerases and fatty-acid based vesicles suggests either that early replicases were quite different, or that RNA replication in early cells proceeded in a very different manner. For example, less charged and more activated nucleotides might be easier for a ribozyme polymerase to bind, with little or no Mg++. Many potential leaving groups, such as imidazole, adenine or 1-Me-adenine have been examined in template-directed and nontemplated polymerization reactions, but have not yet been tested as substrates for ribozyme polymerases (Prabahar and Ferris 1997; Huang and Ferris 2006). The tethering of ribozyme and primer-template to hydrophobic aggregates has been examined as a way of increasing local substrate concentration (Müller and Bartel 2008). However, this approach did not lead to a dramatic improvement in the extent of template copying, apparently as a result of ribozyme inhibition at high effective RNA concentrations. It may be possible to evolve polymerases that operate well at high RNA concentrations, but membrane localization by chemical derivatization adds further complexity to a replication pathway because of the need for a specific catalyst for the derivatization step. Alternative means of facilitating the interaction of a ribozyme with a primer-template substrate, such as the presence of basic peptides or other cofactors, might overcome this problem. Finally it is noteworthy that very small self-aminoacylating ribozymes have been obtained using RNA libraries that have an unconstrained sequence at their 3′-end (Chumachenko et al. 2009). This strategy may also be fruitful in selections for ribozyme polymerases.
Given the above constraints and uncertainties, what can we say about the emergence of RNA-catalyzed RNA replication in the origin of life? It is still possible that under the proper conditions, and using the right substrates, a small simple ribozyme could effectively catalyze RNA replication. However, a replicase must do more than catalyze a simple phospho-transfer reaction. Binding in a nonsequence-specific manner to a primer-template complex, facilitating binding of the proper incoming monomer, catalyzing primer extension, and repeating this process until the end of the template (or set of templates for a multi-component replicase) is reached might require a complex replicase structure. Such a replicase would presumably be rare in collections of random RNA sequences. If life required a very special sequence to get started, then the origin of life on earth could have been a low probability event and life on other earth-like planets might be very rare. If, on the other hand, RNA-catalyzed RNA replication could have emerged gradually in a series of simpler steps, it might have been easier and thus more likely for life to begin, and life elsewhere might be common. For this reason, we now turn to a consideration of nonenzymatic template-directed replication chemistry.
Chemical Template Replication Revisited
The nonenzymatic template-directed polymerization of activated ribonucleotides was studied in depth by Leslie Orgel, together with his students and colleagues, over a period of several decades (Orgel 2004). Here, the template itself acts as a catalyst by helping to align and orient the monomers so that they are pre-organized for polymerization. The main lesson from this work is that spontaneous chemical copying of RNA sequences is indeed possible, but is subject to several important constraints and limitations. The constraints make template-directed RNA copying incompatible with currently available membrane vesicle systems, and the limitations have, so far at least, made it impossible to obtain repeated cycles of RNA replication through chemical copying.
To obtain reasonable reaction rates, Orgel made use of nucleoside monophosphates activated with a good leaving group such as imidazole. The ribonucleotide 5′-phosphorimidazolides spontaneously assemble on a template oligonucleotide and polymerize over several days, generating a complementary strand. However, monomer binding to RNA templates is weak and concentrations on the order of 0.1 M were required for optimal copying. In addition, a Mg++ concentration of ∼0.1 M, which as noted above is incompatible with the presence of fatty-acid based membranes, is required for optimal polymerization. Even under these rather extreme conditions, polymerization proceeds at only 1–2 nucleotides per day, and is therefore limited by monomer hydrolysis. Beyond these constraints, three aspects of the copying reaction present major hurdles to multiple rounds of replication. First, the chemical structure of RNA results in a problem of regiospecificity, because new linkages can be either 3–5′ or 2′–5′ phosphodiester bonds. Surprisingly, under most conditions, it is the 2′–5′ phosphodiester bonds that are most common in polymerization products. This problem can be ameliorated by the choice of ions and leaving groups; for unknown reasons, Zn2+ ions and the 2-methylimidazole leaving group favor the synthesis of 3′–5′ linkages (Lohrmann et al. 1980; Inoue and Orgel 1982). Studies of oligonucleotide ligation showed that the helical context of an extended RNA duplex also favors the formation of 3′–5′ linkages (Rohatgi et al. 1996). However, it remains difficult to obtain a homogeneous RNA backbone without enzymatic catalysis. Second, adenine (A) residues in the template are difficult to copy, and two or more As in succession block chain growth (Inoue and Orgel 1983), presumably as a result of the poor base-stacking propensity of the incoming U monomers. It has recently been found that template copying at subzero temperatures can proceed past multiple A residues in the template (Vogel and Richert 2007), but this required sequential additions of oxyazabenzotriazole-activated monomer together with a series of helper oligos. Third, there is the issue of fidelity. The copying of G and C residues is remarkably accurate, with error rates estimated at 0.5% or less. However, the addition of A and U residues causes problems, most significantly the formation of G:U wobble base-pairs (Wu and Orgel 1992), which would lead to significant error rates in a four base system. Might an alternative genetic polymer, perhaps even a close relative of RNA, overcome these problems and enable chemical replication? The identification of such a system might ultimately lead to the discovery of plausible progenitors of RNA, or, alternatively, to the discovery of new replication strategies that allow for the chemical replication of RNA itself.
Phosphoramidate Nucleic Acids
Early studies of template copying using more reactive nucleotide derivatives were performed by Orgel et al., who examined both 2′- and 3′-amino ribonucleotide 5′-phosphorimidazolides (Lohrmann and Orgel 1976; Zielinksi and Orgel 1985; Tohidi et al. 1987). Polymerization of these nucleotides yields phosphoramidate nucleic acids, which are generally similar to standard phosphodiester linked nucleic acids except that the phosphoramidate linkage is more acid labile. As expected, replacing a sugar hydroxyl in the monomer with a more nucleophilic amino group resulted in a large increase in monomer reactivity. The activated 3′-amino ribonucleotides participated in rapid copying of short oligonucleotide templates 5-13 nucleotides in length, yielding N3′→P5′ linked complementary oligonucleotides (Tohidi et al. 1987). The increased reactivity also led to faster intramolecular monomer cyclization, which depleted the template copying reactions of activated substrate molecules (Hill et al. 1988).
More recently, the Richert group has begun to explore the potential of 3′-amino- nucleotide analogs in template-directed condensation reactions. Deoxyribonucleotide monomers, activated with an oxyazabenzotriazole leaving group, completed a template-directed reaction with a 3′-amino-terminated primer in seconds (Rothlingshofer et al. 2008). The fidelity of this reaction is sufficient to allow for sequencing by nonenzymatic primer extension. However, the monomer is rapidly consumed by internal cyclization.
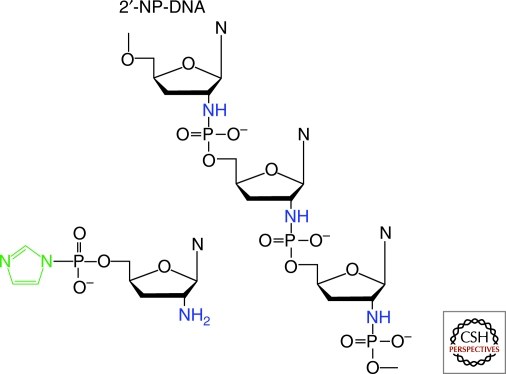
The high intrinsic reactivity of the amino-sugar modified nucleotides suggested to us that an alternative phosphoramidate nucleic acid might act as a good platform for chemical self-replication. Our group has therefore started to study a series of phosphoramidate nucleic acids with sugar phosphate backbones that vary in their degree of conformational flexibility or constraint. We are currently focusing on the 2′-5′ linked phosphoramidate analog of DNA, and the corresponding monomers, the 2′-amino dideoxyribonucleotide 5′-phosphorimidazolides (Fig. 4). The 2′-amino ImpddNs are advantageous as monomers because they cannot undergo intramolecular cyclization because of the steric constraint of the ribose ring; they are only depleted during polymerization reactions by competing hydrolysis.
Figure 4.
Structures of 2′-5′ phosphoramidate DNA, and the corresponding activated monomers.
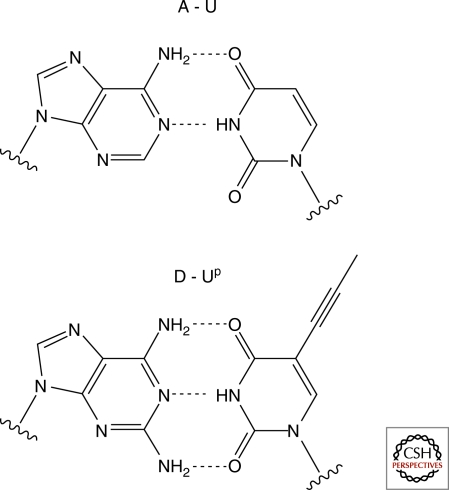
Our first experiments with 2′-amino ImpddG led to rapid and efficient primer-extension across a dC15 template, generating full-length product in ∼6 hour (Mansy et al. 2008). Encouraged by the rapid copying of oligo-dC templates by 2′-amino ImpddG, we performed a more extensive study of the copying of templates with differing sugar-phosphate backbones, lengths and sequences (Schrum et al. 2009). The most important property contributing to good template activity appears to be preorganization in the form of an A-type helix. Thus, RNA templates were uniformly superior to DNA templates of the same sequence, and LNA (locked nucleic acid) templates, which are chemically locked in a C3′-endo sugar conformation, were superior to RNA templates. This result is consistent with previous observations that under most conditions RNA-template directed polymerization of ribonucleotides leads to a majority of 2′-5′ linkages; it appears that an A-type helical geometry generally favors polymerization through attack by a 2′ nucleophile. A second key factor is that enhanced monomer affinity for the template increases the reaction efficiency. Thus G:C base-pairs lead to efficient copying, whereas A:U base-pairs were very poorly copied. Replacing A with D (diaminopurine) results in a D:U base-pair with three hydrogen bonds, and slightly improved primer-extension. However, the poor base stacking of U residues must also be improved to obtain efficient template copying. When we replaced U with C5-propynyl-U, the resulting A:Up base-pairs led to improved copying, but the D:Up combination had a clearly synergistic effect (Fig. 5). In the context of a DNA duplex, the D:Up base-pair has previously been shown to be energetically almost equivalent to a C:G base-pair (Chaput et al. 2002). When we used G, C, D, and Up as the four monomer and template bases, we were able to copy mixed-sequence RNA templates over 15 nts in length in about a day. This represents a significant step toward developing a robust, generalized chemical replication system.
Figure 5.
Watson Crick base-pairs. Top: Standard A:U base-pair. Bottom: Alternative diaminopurine (D): C5-propynyl-uracil (Up) base-pair.
This system is, however, far from ideal, and there are strong indications that the fidelity of template copying becomes an issue when all four nucleotides are present, largely because of the formation of G:U wobble base-pairs. Given that primer-extension on 2′–5′ linked DNA templates is approximately similar to that on the corresponding RNA templates, we are currently synthesizing 2′–5′ linked phosphoramidate DNA templates to assess self-replication in this system. In light of the efficient templating of LNA oligonucleotides, we are also interested in exploring prebiotically plausible nucleic acids that are more conformationally constrained than RNA. A particularly interesting candidate is TNA (threose nucleic acid) (Schöning et al. 2000) and its 2′-amino substituted phosphoramidate version, NP-TNA (Wu et al. 2002). These are both base-pairing systems that form standard Watson-Crick duplexes, despite having only five atoms per backbone repeat unit (vs. six for RNA and DNA). It will be of great interest to see if the resulting decrease in flexibility leads to increased fidelity in template copying reactions.
Protocell Assembly
In principle, protocell-like objects could form spontaneously as new membranes self-assemble and encapsulate genetic molecules in solution. Recently, simple physical processes that would enhance the efficiency of the coassembly of nucleic acids and membrane vesicles have been proposed. One such alternative scenario is based on the fact that the clay mineral montmorillonite is not only a catalyst of RNA polymerization (Ferris et al. 1996; Huang and Ferris 2006) but also catalyzes membrane assembly (Hanczyc et al. 2003). Experiments with clay particles containing surface-adsorbed RNA showed that such particles stimulated vesicle assembly, and frequently became trapped inside the vesicles whose assembly they had catalyzed. Thus, a common mineral can catalyze both the assembly of a genetic polymer and the assembly of a membrane vesicle, and bring these two components together to generate a protocell-like structure consisting of a genetic polymer trapped within a membrane compartment. Although the effectiveness of this process is attractive, some means of releasing at least some of the bound nucleic acid from the mineral surface, and/or replicating it on the surface, would be necessary for subsequent replication to occur.
More recent experiments suggest a very different geochemical scenario leading to the assembly of similar protocell-like structures. The hollow channels within the rocks of the alkaline off-axis hydrothermal vents provide a protected compartmentalized environment where it has been suggested that primitive metabolic activities might have originated (Martin and Russell 2003). Recent theoretical studies suggested that the strong thermal gradients present in hydrothermal vents, together with the thin channels produced by mineral precipitation, could greatly concentrate small organic molecules such as nucleotides as well as larger nucleic acids from a very dilute external reservoir (Baaske et al. 2007). Work from our laboratory (Budin et al. 2009) has confirmed the predicted concentration effect, and has also shown that subcritical concentrations of fatty acids can be concentrated to the extent that they self-assemble into vesicles at the bottom of the capillary channels. Moreover, DNA oligonucleotides can also be greatly concentrated and can become encapsulated within the vesicles, resulting in the spontaneous assembly of protocell-like structures.
Encapsulated Template Replication: Emergence of a Protocell
The experiments discussed earlier suggest that the assembly of protocell-like structures is not that difficult, because it appears to be possible through multiple distinct mechanisms. The more challenging question is, how could such a structure replicate? We have already considered the replication of the protocell membrane and the genetic material as separate entities. To address the question of their replication as a combined structure we must consider in more detail the molecular constituents of the protocell membrane and the molecular nature of the encapsulated genetic material.
Genome replication within a protocell can only occur if the building blocks used to copy template strands are able to enter the fatty-acid vesicle compartment. Early work using phospholipid-based vesicles and protein enzymes showed the feasibility of constructing primitive cell-like compartments (Chakrabarti et al. 1994; Luisi et al 1994). More recent permeability studies (Mansy et al. 2008; see also Mansy 2010) showed that nucleotides could spontaneously diffuse across simple fatty-acid membranes, but that net negative charge is a critical determinant of permeability. Thus, nucleotides that are chemically activated, e.g., by conversion of the 5′-phosphate to a 5′-phosphorimidazolide, equilibrate across vesicle membranes much more rapidly on account of the reduction of the net negative charge. In addition, mixtures of fatty acids with their glycerol esters generate membranes that are more permeable to polar and charged molecules. By combining these observations, we were able to show that activated 2′-amino-2′,3′-dideoxyguanosine-5′-phosphorimidazolide, the same nucleotide previously shown to rapidly copy oligo-dC templates, could be added to the outside of fatty-acid vesicles containing an encapsulated primer-template complex, and copy the internal dC15 template. Copying of the encapsulated dC15 template by primer-extension reached >95% completion in 12–24 hour (Mansy et al. 2008), compared with 6–12 hour in free solution. The longer time required for copying encapsulated templates reflects the time required for entry of external nucleotides to the interior of the vesicles. Importantly, the presence of high concentrations (5–10 mM) of highly reactive activated nucleotides did not have any disruptive effects on the integrity of the vesicle membrane, as no leakage of encapsulated primer-template complexes was observed. It is also important to note that control experiments with phospholipid membranes showed no copying of internal template, because the activated nucleotides could not enter the vesicle; similarly, “modern” activated nucleotides such as nucleoside triphosphates cannot cross fatty-acid based membranes. Successful copying of encapsulated templates therefore requires both “primitive” nucleotides with reduced charge, and “primitive” membranes composed of single chain amphiphiles.
The copying of a genetic polymer inside a membrane compartment is an important step toward the realization of a self-replicating system capable of Darwinian evolution. What, then, are the remaining barriers to the assembly of such a system? The copying of a single-stranded template produces a double-stranded product; these strands would have to separate before a second cycle of genome replication could begin. Separate follow-up experiments by our group showed that some fatty-acid based vesicles are able to retain encapsulated DNA and RNA oligonucleotides over a temperature range of 0 °C to 100 °C (Mansy and Szostak 2008). As with permeability, mixtures of amphiphiles lead to improved thermostability, with glycerol esters being particularly stabilizing, possibly because of the additional hydrogen bond donors and acceptors provided by the glycerol head group. Furthermore, we found that encapsulated double-stranded DNA could be denatured at elevated temperatures, with the strands reannealing once the temperature was lowered (Mansy and Szostak 2008). This implies that thermal fluctuations could provide a mechanism for strand separation that is compatible with the integrity of fatty-acid vesicles, potentially allowing for complete cycles of replication of encapsulated genetic polymers. The mutual compatibility of nucleic acid replication and fatty-acid compartment growth is very encouraging because it alleviates concerns related to the permeability and stability of membrane vesicles. Vesicles therefore do seem to be a physically plausible way to segregate and spatially localize genomes, keep emergent catalytic polynucleotides physically close to their encoding genome, and protect the nascent evolving system from parasitic polymers.
CHALLENGES AND FUTURE RESEARCH DIRECTIONS
Prospects for a Complete Protocell Model
Although considerable progress has been made toward the assembly of model protocells, several remaining issues must be solved before multiple cycles of protocell replication can be achieved in the laboratory. These factors are also relevant to protocell replication on the early Earth. The most important factor at this time appears to be the competition between strand reannealing and strand copying, after thermal strand separation. PCR reactions generally plateau at about 1-µM DNA strand concentration, which is the concentration at which strand reannealing and strand copying occur on a similar time scale of about 1 minute However, nonenzymatic template copying requires on the order of a day for completion, which implies that either template copying must be much faster, or reannealing must be much slower. One way to make reannealing sufficiently slow is to keep strand concentrations subnanomolar. Low strand concentrations are possible in large vesicles, but it is hard to see how a few molecules of a genetic polymer could have any significant phenotypic effect on a large vesicle composed of millions of amphiphilic molecules. The emergence of metabolic ribozymes would be more plausible if nucleic acid strand concentrations were much higher, so that a catalyst of modest efficiency could generate enough product to influence cell properties.
What other factors might affect the rate of strand reannealing? Perhaps the most obvious possibility is that secondary structure, which can form extremely rapidly because the interactions are intramolecular, could greatly slow down strand annealing. This phenomenon is essential to the viability of single-stranded RNA phage such as Qβ• (Axelrod et al. 1991). Significant intrastrand secondary structure would also be an expected consequence of selection for sequences that fold into functional shapes with catalytic activity. On the other hand, chemical replication through dense secondary structure would probably be much slower than replication of an unfolded, open template. The outcome of simultaneous selection for an open, accessible template sequence, and a folded functional structure remains unclear. An alternative but even more speculative solution might result from the rapid binding by base-pairing of short oligonucleotides or even monomers to freshly separated strands. If a template strand was largely occupied by monomers or short oligomers, even if these were in rapid exchange, the strand might be prevented from annealing to a complementary strand. This possibility has the advantage that it need not block or slow the copying reaction, however its effectiveness remains to be tested experimentally.
Another challenge faced by replicating protocells, whether on the early earth or in the modern laboratory, is the continuous dilution of protocells through the competing formation of new empty vesicles. When new fatty acids are supplied as micelles, the efficiency of incorporation into pre-formed vesicles (or protocells) can be quite high, but some new vesicles are always formed. Thus over time, the descendants of a given protocell will gradually be diluted out by the continuous formation of these new vesicles. To avoid extinction by dilution, the protocells must out-compete other vesicles either by having a more rapid cell cycle, thereby generating more progeny during division, or by surviving destructive processes more efficiently. We have previously proposed (Chen et al. 2004) that faster growth, driven by the osmotic pressure of encapsulated nucleic acid, could lead to an effectively shorter cell cycle for protocells that contain high copy numbers of their replicating genome. However, in light of the problems associated with this approach, it is of considerable interest to explore new ways in which a protocell genome could lead to faster growth or growth that occurs at the expense of empty vesicles. An alternative strategy for surviving dilution would be for a protocell genome to colonize empty vesicles. This could occur through a low level of stochastic vesicle–vesicle fusion events, possibly catalyzed by low levels of divalent cations such as Ca2+. Systematic efforts to measure vesicle fusion frequencies under different environmental conditions could therefore be quite useful. Finally, it is possible that this problem could be circumvented entirely if early life was discontinuous (Budin et al. 2009). For example, protocells could be occasionally disrupted by drying, or simply dissolve as a result of dilution with water to a level below the critical aggregate concentration; subsequent re-hydration or concentration would result in reformation of vesicles encapsulating genomic nucleic acids, thus generating a new “randomized” set of protocells. As long as such events were fairly uncommon, and assuming that genomic replication had kept ahead of vesicle replication so that each vesicle contained multiple genome copies prior to disruption, this process of disruption and reformation would lead to the spread of evolving genomic sequences through the “new” vesicles.
Laboratory models of protocell systems should be helpful in modeling many of the above scenarios. Assuming that protocell reproduction can be achieved, and made efficient enough to continue through many generations, it should then be possible to observe the spontaneous evolution of adaptive innovations in this relatively simple chemical system. The nature of such adaptations may provide clues as to how modern cells evolved from their earliest ancestors. Ultimately this line of research may also tell us whether the conserved biochemistry of life is driven by chemical necessity, or whether biochemically very different forms of life are also possible.
ACKNOWLEDGMENTS
We thank Itay Budin, Matt Powner, and other members of the Szostak lab for helpful discussions.
Footnotes
Editors: David Deamer and Jack W. Szostak
Additional Perspectives on The Origins of Life available at www.cshperspectives.org
REFERENCES
- Axelrod VD, Brown E, Priano C, Mills DR 1991. Coliphage Q β RNA replication: RNA catalytic for single-strand release. Virology 184: 595–608 [DOI] [PubMed] [Google Scholar]
- Baaske P, Weinert FM, Duhr S, Lemke KH, Russell MJ, Braun D 2007. Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proc Natl Acad Sci 104: 9346–9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ziv R, Moses E 1994. Instability and “pearling” states produced in tubular membranes by competition of curvature and tension. Phys Rev Lett 73: 1392–1395 [DOI] [PubMed] [Google Scholar]
- Berclaz N, Muller M, Walde P, Luisi PL 2001. Growth and transformation of vesicles studied by ferritin labeling and cryotransmission electron microscopy. J Phys Chem B 105: 1056–1064 [Google Scholar]
- Blochliger E, Blocher M, Walde P, Luisi PL 1998. Matrix effect in the size distribution of fatty acid vesicles. J Phys Chem B 102: 10383–10390 [Google Scholar]
- Budin I, Bruckner R, Szostak JW 2009. Formation of protocell-like vesicles in a thermal diffusion column. J Am Chem Soc 131: 9628–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti AC, Breaker RR, Joyce GF, Deamer DW 1994. Production of RNA by a polymerase protein encapsulated within phospholipid vesicles. J Mol Evol 39: 555–9 [DOI] [PubMed] [Google Scholar]
- Chaput JC, Sinha S, Switzer C 2002. 5-propynyluracil.diaminopurine: An efficient base-pair for non-enzymatic transcription of DNA. Chem Commun 15: 1568–9 [DOI] [PubMed] [Google Scholar]
- Chen IA, Roberts RW, Szostak JW 2004. The emergence of competition between model protocells. Science 305: 1474–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IA, Szostak JW 2004. A kinetic study of the growth of fatty acid vesicles. Biophys J 87: 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumachenko NV, Novikov Y, Yarus M 2009. Rapid and simple ribozymic aminoacylation using three conserved nucleotides. J Am Chem Soc 131: 5257–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer DW 1985. Boundary structures are formed by organic-components of the Murchison carbonaceous chondrite. Nature 317: 792–794 [Google Scholar]
- Deamer DW, Pashley RM 1989. Amphiphilic components of the Murchison carbonaceous chondrite: Surface properties and membrane formation. Orig Life Evol Biosph 19: 21–38 [DOI] [PubMed] [Google Scholar]
- Dworkin J, Deamer D, Sandford S, Allamandola L 2001. Self-assembling amphiphilic molecules: Synthesis in simulated interstellar/precometary ices. Proc Natl Acad Sci 98: 815–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland EH, Szostak JW, Bartel DP 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 1319–25 [DOI] [PubMed] [Google Scholar]
- Ferris JP, Hill AR Jr, Liu R, Orgel LE 1996. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 381: 59–61 [DOI] [PubMed] [Google Scholar]
- Gebicki JM, Hicks M 1973. Ufasomes are stable particles surrounded by unsaturated fatty acid membranes. Nature 243: 232–234 [DOI] [PubMed] [Google Scholar]
- Gilbert W 1986. The RNA World. Nature 319: 618 [Google Scholar]
- Glasner ME, Yen CC, Ekland EH, Bartel DP 2000. Recognition of nucleoside triphosphates during RNA-catalyzed primer extension. Biochemistry 39: 15556–62 [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–57 [DOI] [PubMed] [Google Scholar]
- Hanczyc MM, Szostak JW 2004. Replicating vesicles as models of primitive cell growth and division. Curr Opin Chem Biol 8: 660–664 [DOI] [PubMed] [Google Scholar]
- Hanczyc MM, Fujikawa SM, Szostak JW 2003. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science 302: 618–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves WR, Deamer DW 1978. Liposomes from ionic, single-chain amphiphiles. Biochemistry 17: 3759–3768 [DOI] [PubMed] [Google Scholar]
- Hill AR Jr, Nord LD, Orgel LE, Robins RK 1988. Cyclization of nucleotide analogues as an obstacle to polymerization. J Mol Evol 28: 170–1 [DOI] [PubMed] [Google Scholar]
- Huang W, Ferris JP 2006. One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J Am Chem Soc 128: 8914–9 [DOI] [PubMed] [Google Scholar]
- Inoue T, Orgel LE 1982. Oligomerization of (guanosine 5′-phosphor)-2-methylimidazolide on poly(C). An RNA polymerase model. J Mol Biol 162: 201–17 [DOI] [PubMed] [Google Scholar]
- Inoue T, Orgel LE 1983. A nonenzymatic RNA polymerase model. Science 219: 859–62 [DOI] [PubMed] [Google Scholar]
- Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 1319–25 [DOI] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR 1982. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147–57 [DOI] [PubMed] [Google Scholar]
- Lohrmann R, Orgel LE 1976. Template-directed synthesis of high molecular weight polynucleotide analogues. Nature 261: 342–344 [DOI] [PubMed] [Google Scholar]
- Lohrmann R, Bridson PK, Orgel LE 1980. Efficient metal-ion catalyzed template-directed oligonucleotide synthesis. Science 208: 1464–5 [DOI] [PubMed] [Google Scholar]
- Luisi PL 2006. The emergence of life: From chemical origins to synthetic biology, Cambridge University Press, Cambridge [Google Scholar]
- Luisi PL, Walde P, Oberholzer T 1994. Enzymatic RNA synthesis in self-reproducing vesicles: An approach to the construction of a minimal synthetic cell. Ber Bunsenges Phys Chem 98: 1160–5 [Google Scholar]
- Luisi PL, Stano P, Rasi S, Mavelli F 2004. A possible route to prebiotic vesicle reproduction. Artif Life 10: 297–308 [DOI] [PubMed] [Google Scholar]
- Mansy SS 2010. Membrane transport in primitive cells. Cold Spring Harb Perspect Biol 2: a002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy SS, Szostak JW 2008. Thermostability of model protocell membranes. Proc Natl Acad Sci 105: 13351–13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy SS, Schrum JP, Krishnamurthy M, Tobé S, Treco DA, Szostak JW 2008. Template-directed synthesis of a genetic polymer in a model protocell. Nature 454: 122–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Russell MJ 2003. On the origins of cells: A hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci 358: 59–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollom TM, Ritter G, Simoneit BR 1999. Lipid synthesis under hydrothermal conditions by Fischer-Tropsch-type reactions. Orig Life Evol Biosph 29: 153–166 [DOI] [PubMed] [Google Scholar]
- Monnard PA, Apel CL, Kanavarioti A, Deamer DW 2002. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: Implications for a prebiotic aqueous medium. Astrobiology 2: 139–52 [DOI] [PubMed] [Google Scholar]
- Müller UF, Bartel DP 2008. Improved polymerase ribozyme efficiency on hydrophobic assemblies. RNA 14: 552–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–30 [DOI] [PubMed] [Google Scholar]
- Orgel LE 2004. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol 39: 99–123 [DOI] [PubMed] [Google Scholar]
- Prabahar KJ, Ferris JP 1997. Adenine derivatives as phosphate-activating groups for the regioselective formation of 3’,5’-linked oligoadenylates on montmorillonite: Possible phosphate-activating groups for the prebiotic synthesis of RNA. J Am Chem Soc 119: 4330–7 [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Bartel DP, Szostak JW 1996. Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′-5′ phosphodiester bonds. J Am Chem Soc 118: 3340–4 [DOI] [PubMed] [Google Scholar]
- Röthlingshöfer M, Kervio E, Lommel T, Plutowski U, Hochgesand A, Richert C 2008. Chemical primer extension in seconds. Angew Chem Int Ed Engl 47: 6065–8 [DOI] [PubMed] [Google Scholar]
- Rushdi AI, Simoneit BR 2001. Lipid formation by aqueous Fischer-Tropsch-type synthesis over a temperature range of 100 to 400 degrees C. Orig Life Evol Biosph 31: 103–118 [DOI] [PubMed] [Google Scholar]
- Sacerdote MG, Szostak JW 2005. Semi-permeable lipid bilayers exhibit diastereoselectivity favoring ribose; implications for the origins of life. Proc Natl Acad Sci 102: 6004–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning K, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A 2000. Chemical etiology of nucleic acid structure: The α-threofuranosyl-(3′→2′) oligonucleotide system. Science 290: 1347–51 [DOI] [PubMed] [Google Scholar]
- Schrum JP, Ricardo A, Krishnamurthy K, Blain JC, Szostak JW 2009. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′, 3′-dideoxyribonucleoside-5′-phosphorimidazolide monomers. J Am Chem Soc 31: 14560–14570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stano P, Wehrli E, Luisi PL 2006. Insights into the self-reproduction of oleate vesicles. J Phys: Condens Matter 18: S2231–S2238 [Google Scholar]
- Szostak JW, Bartel DP, Luisi PL 2001. Synthesizing life. Nature 409: 387–390 [DOI] [PubMed] [Google Scholar]
- Tohidi M, Zielinski WS, Chen CH, Orgel LE 1987. Oligomerization of 3′-amino-3′deoxyguanosine-5′phosphorimidazolidate on a d(CpCpCpCpC) template. J Mol Evol 25: 97–99 [DOI] [PubMed] [Google Scholar]
- Vogel SR, Richert C 2007. Adenosine residues in the template do not block spontaneous replication steps of RNA. Chem Commun 19: 1896–8 [DOI] [PubMed] [Google Scholar]
- Voytek SB, Joyce GF 2007. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc Natl Acad Sci 104: 15288–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walde P, Wick R, Fresta M, Mangone A, Luisi PL 1994a. Autopoietic self-reproduction of fatty acid vesicles. J Am Chem Soc 116: 11649–11654 [Google Scholar]
- Walde P, Goto A, Monnard P-A, Wessicken M, Luisi PL 1994b. Oparin’s reactions revisited: Enzymatic synthesis of poly(adenylic acid) in micelles and self-reproducing vesicles. J Am Chem Soc 116: 7541–7547 [Google Scholar]
- Wu T, Orgel LE 1992. Nonenzymatic template-directed synthesis on hairpin oligonucleotides. 3. Incorporation of adenosine and uridine residues. J Am Chem Soc 114: 7963–9 [DOI] [PubMed] [Google Scholar]
- Wu X, Guntha S, Ferencic M, Krishnamurthy R, Eschenmoser A 2002. Base-pairing systems related to TNA: α-Threofuranosyl oligonucleotides containing phosphoramidate linkages. Org Lett 4: 1279–82 [DOI] [PubMed] [Google Scholar]
- Zhu TF, Szostak JW 2009a. Coupled growth and division of model protocell membranes. J Am Chem Soc 131: 5705–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu TF, Szostak JW 2009b. Preparation of large monodisperse vesicles. PLoS ONE 4: e5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski WS, Orgel LE 1985. Oligomerization of activated derivatives of 3′-amino-3′-deoxyguanosine on poly(C) and poly(dC) templates. Nucleic Acids Res 13: 2469–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]