Abstract
Background
Fungal infections are an emerging health risk, especially those involving yeast that are resistant to antifungal agents. To understand the range of mechanisms by which yeasts can respond to anti-fungals, we compared gene expression patterns across three evolutionarily distant species - Saccharomyces cerevisiae, Candida glabrata and Kluyveromyces lactis - over time following fluconazole exposure.
Results
Conserved and diverged expression patterns were identified using a novel soft clustering algorithm that concurrently clusters data from all species while incorporating sequence orthology. The analysis suggests complementary strategies for coping with ergosterol depletion by azoles - Saccharomyces imports exogenous ergosterol, Candida exports fluconazole, while Kluyveromyces does neither, leading to extreme sensitivity. In support of this hypothesis we find that only Saccharomyces becomes more azole resistant in ergosterol-supplemented media; that this depends on sterol importers Aus1 and Pdr11; and that transgenic expression of sterol importers in Kluyveromyces alleviates its drug sensitivity.
Conclusions
We have compared the dynamic transcriptional responses of three diverse yeast species to fluconazole treatment using a novel clustering algorithm. This approach revealed significant divergence among regulatory programs associated with fluconazole sensitivity. In future, such approaches might be used to survey a wider range of species, drug concentrations and stimuli to reveal conserved and divergent molecular response pathways.
Background
Mucosal and invasive mycoses are a major world health problem leading to morbidity [1,2] and a mortality rate of up to 70% in immunocompromised hosts [3]. The most common treatment for fungal infections is the family of chemical compounds known as the azoles, which interfere with formation of the cell membrane by inhibiting synthesis of ergosterol [4]. However, the use of azoles to treat a broad spectrum of fungal infections has led to widespread azole resistance [4-9], and resistance is also emerging against the limited number of secondary compounds that are currently available [10,11].
The fungal response to azoles has been most often studied in yeast [5,7,12-17], primarily through analysis of standard laboratory strains of Candida [12,13,18] or Saccharomyces [14,16,17] or their resistant clinical isolates [2,12,15,19]. Other studies have focused on cultures for which drug resistance has been artificially evolved in-vitro [15,18,20,21]. This work has revealed a number of resistance and response mechanisms that can be invoked to protect cells from drugs, including mutations to drug efflux pumps or their regulators [2,12,20,21], mutations to ergosterol synthesis enzymes [20], duplication of the fluconazole target Erg11 [18], and a possible role for Hsp90 [15,22].
Although these represent a wide array of mechanisms, it is likely that the full range of anti-fungal resistance pathways is even greater, for several reasons. The first relates to genetic diversity: the number of clinical isolates that have been studied to-date is relatively modest, and resistant strains produced by artificial evolution are only a few generations removed from the common laboratory strains used as starting material. The second reason relates to the environment: it is very difficult to mirror in the laboratory the range of conditions that must be experienced by yeast in the wild during the evolution of stress response pathways. Thus, an important goal moving forward is to better understand the entire pool of genotypic variation underlying fungal stress responses, particularly as they relate to antifungal agents.
Towards this goal, we performed a comparative study of the transcriptional program activated by fluconazole in three evolutionarily distinct yeasts: Saccharomyces cerevisiae (Sc), Candida glabrata (Cg), and Kluyveromyces lactis (Kl). These species were selected to provide a survey of transcriptional networks at intermediate evolutionary distance, that is, at sufficient distance to observe evolutionary change but sufficiently close to ensure significant conservation. Sc and Cg diverged approximately 100 million years ago, and both harbor evidence of an ancient whole-genome duplication event [23]. Cg is an established human pathogen while Sc has been occasionally found to cause systemic infection in immunocompromised individuals [2]. Kl was selected as an outgroup since its evolutionary history is clearly distinct from Sc (having diverged prior to whole-genome duplication [24]) but its transcriptional network is substantially closer to Sc than, for instance, is the network of Candida albicans [25]. In addition, Sc, Cg, and Kl share functional and phenotypic characteristics (for example, growth as haploids [26], similar codon usage [26]) that make them suitable for comparison.
Earlier efforts to profile expression across different species have been limited to the examination of matched conditions across two organisms [27-29] or curated compendia of microarrays across many conditions [24,30,31]. Such studies have previously identified transcriptional mechanisms leading to large phenotypic divergence among yeasts, often related to the whole-genome duplication event [24,30,31]. Accordingly, we reasoned that matched expression time courses of three yeasts might reveal evolutionary differences in the transcriptional stress response elicited by an anti-fungal drug.
Results and discussion
Kl is dramatically more sensitive to fluconazole than other species
For each of the three species Sc, Cg, and Kl, we obtained standard laboratory strains for which genome sequences were available (Materials and methods). We examined the phenotypic response of these species to a range of concentrations of fluconazole (Additional file 1: Testing Fluconazole Susceptibility), a triazole antifungal drug commonly used in the treatment and prevention of superficial and systemic fungal infections [4]. We found that Kl was approximately 70 times more sensitive to fluconazole than Sc and Cg, with a 50% inhibitory concentration of 0.06 μg/ml versus 4.0 μg/ml for both Sc and Cg (Figure S1 in Additional file 1). Cross-species differences in sensitivity could be due to a variety of factors, including differences in membrane permeability or drug transport, divergence in sequence or regulation of the drug target Erg11, or in any of the pathways previously linked to azole resistance.
Comparative expression profiling of Sc, Cg, and Kl
While it is possible that complementary strategies might be observed at different fluconazole dosages [20], we exposed each species to fluconazole at its 50% inhibitory concentration to facilitate direct comparison of the transcriptional response between species. We then monitored global mRNA expression levels at 1/3, 2/3, 1, 2, and 4 population doubling times (Figure 1a). We also found that sampling based on the doubling time of each species, as opposed to absolute time measurements, led to greater coherence in the expression profiles across species (Figure S2 in Additional file 1; Additional file 1: Analysis of Doubling Time Points vs. Absolute Time Points). Selected mRNA measurements were validated using quantitative RT-PCR against six genes (Figure S3 in Additional file 1). We also found significant overlap of the Sc differentially expressed genes with several previous microarray studies and some overlap with gene deletions conferring fluconazole sensitivity (Additional file 1: Microarray Design and Analysis).
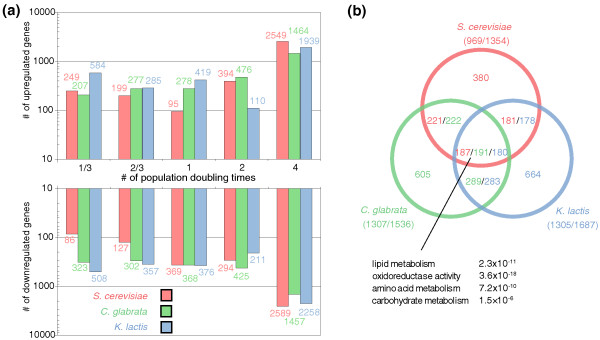
Figure 1.
Differentially expressed genes. (a) Number of differentially expressed (up- and down-regulated) genes by species versus the number of cell doublings. (b) Venn diagram showing the overlap in the sets of differentially expressed genes selected in each species at a false discovery rate of q ≤ 0.1. The number of differentially expressed genes in each region of the Venn diagram is not identical across species, since the number of genes that a species contributes to an orthologous group (that is, number of paralogs) can vary. Ratios in parentheses indicate the number of differentially expressed orthologs by the total number of differentially expressed genes (not all genes possess orthologs).
To compare expression profiles across species, orthologous genes were defined using MultiParanoid [32]. As might be expected based on known phylogenetic distances [23], Cg shared more differentially expressed genes with Sc than with Kl (Figure 1b). We also found some overlap with previously published C. albicans microarray data, especially with the functions of the responsive genes such as those involved in ergosterol biosynthesis and oxido-reductase activity (Additional file 1: Microarray Design and Analysis).
Soft clustering: a novel cross-species clustering algorithm
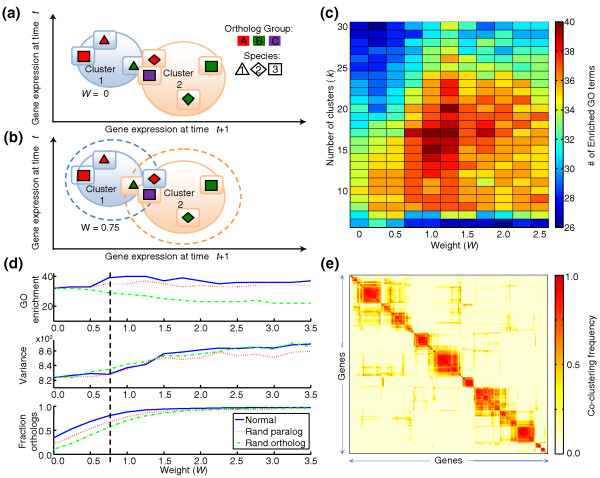
Due to factors such as measurement error and ambiguity of cluster boundaries, we found that the available clustering methods led to situations in which orthologous genes with similar expression patterns could be misplaced into different clusters (Additional file 1: Constrained Clustering Algorithm). Accordingly, we developed a 'soft' clustering approach that integrates expression profiles with gene sequence orthology in a modified k-means model. This algorithm includes an adjustable weight that rewards ortholog co-clustering (Figures 2a, b; Materials and methods; Additional file 1: Constrained Clustering Algorithm). The term 'soft clustering' has also previously been used in other clustering methods to define cases in which a gene can belong to more than one cluster rather than any constraint used to identify clusters [12,13]. Unlike standard clustering methods, which focus solely on cluster coherence, the soft clustering method can simultaneously detect both similar and divergent behavior between orthologs. For instance, when orthologs are not co-clustered despite the addition of a reward, one can be assured that their dynamic profiles truly differ. The weight W and the number of clusters k were scanned over a range of values (Figure 2c). We selected W = 0.75 and k = 17 as choices that approximately optimized the enrichment for Gene Ontology (GO) terms (Additional file 1: Constrained Clustering Algorithm; Additional file 1: Selecting Parameters for the Constrained Clustering Method).
Figure 2.
Soft clustering method. (a) Standard clustering based on expression only: two sets of orthologs are depicted (color represents orthology, shape represents species) where orthologs are split between clusters 1 and 2. For illustrative purposes, only two time points (t and t + 1) are shown. (b) Soft clustering based on expression and orthology: dashed circles denote regions where orthologs will be co-clustered. Since the purple square has no orthologs in cluster 1, it remains assigned to cluster 2. (c) Effect of number of clusters k and orthology weight W on GO term enrichment. (d) The number of enriched GO terms, variance, and fraction of co-clustered orthologs for k = 17 as a function of W in comparison to randomized paralogs/orthologs. Randomization was performed as described in Additional file 1: Randomizing the Orthology Mapping. (e) Since k-means is non-deterministic, to ensure robustness we performed 50 runs of the algorithm recording the fraction of times each gene pair was co-clustered (including all genes from all species). This matrix was hierarchically clustered.
We compared our soft clustering approach to additional standard clustering methods (Figure S4a in Additional file 1). In comparison to classical k-means (equivalent to W = 0), the fraction of co-clustered orthologs increased from approximately 35% to 70%, with a negligible increase in within-cluster variance (Figure 2d). For W > 0.75, we saw no improvement in the number of enriched GO terms, a marked increase in total cluster variance, and little improvement in the fraction of co-clustered orthologs (Additional file 1: Constrained Clustering Algorithm). Since k-means is non-deterministic, to ensure robustness the results of 50 runs of the algorithm were used to populate a matrix recording the fraction of times each gene pair was co-clustered. This matrix was used as a similarity matrix for subsequent hierarchical clustering (Figure 2e; Additional file 1: Co-clustering Matrix). The resulting 17 cross-species gene expression clusters are shown in Figure 3a, b, Figure S7 in Additional file 1, and Table S1 in Additional file 2.
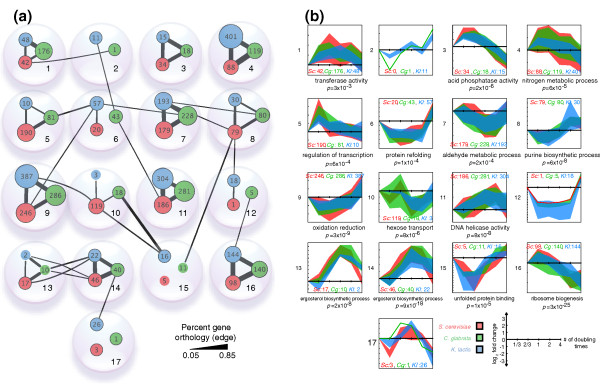
Figure 3.
Cluster structure and dynamics. (a) Each of the 17 clusters appears as a bubble containing up to three colored nodes whose sizes represent the number of genes contributed by each species. Edge thickness denotes the percent of gene orthology shared within or between clusters, measured using the size of the intersection divided by the size of the union of the sample sets. Only significant edges (P < 0.01) are shown. Several clusters show conserved orthology but not dynamics (for example, cluster 10 Sc, Cg with cluster 15 Kl). Note that clusters were ordered to minimize orthology edge crossings. (b) Expression dynamics of the 17 soft clusters over time following fluconazole exposure. Separate plots for each species can be found in Additional file 1. The width of each band corresponds to ± one standard deviation about the mean. A selection of enriched GO terms are shown for different clusters; see Figure S11 in Additional file 1 for full GO enrichment results. The number of genes for each species in each cluster is also shown.
Conservation of cis-regulatory motifs across clusters
We found that two cross-species clusters (13 and 14) were highly enriched for ergosterol biosynthetic genes (P ≤ 10-8) and were coherently up-regulated in all three species - likely in response to ergosterol depletion. Both clusters were also enriched for the upstream DNA-binding motif of the sterol biosynthesis regulators Ecm22 and Upc2 [33]. Interestingly, Upc2 has also been implicated in increased fluconazole resistance in the fungal pathogen C. albicans [34]. Rox1 motifs were enriched in Sc and Cg but not Kl. A likely explanation for this divergence is that Rox1 is a repressor of hypoxia-induced genes, and Kl both lacks a Rox1 ortholog and the capacity for anaerobic growth.
Beyond the clusters representing ergosterol biosynthesis, we found two additional clusters (9 and 16) in which high conservation of expression patterns, sequence orthology, and cis-motif conservation were observed across species. Cluster 9 was regulated by the general stress-response transcription factors Msn2p and Msn4p (q < 10-5; Additional file 1: Expression Conservation of the General Stress Response) and showed GO enrichment for oxido-reductase activity (q <10-8) and carbohydrate metabolism (q <10-7). Cluster 16 was enriched for ribosomal biogenesis and assembly (q <10-13) with upstream PAC [35] and RRPE motifs previously implicated in regulating genes involved in the general stress response and ribosomal regulation (Additional file 1: Expression Conservation of the General Stress Response) [28,31,35,36].
For other clusters, conserved motifs were absent, suggesting divergence across species. This lack of motif conservation was particularly surprising for clusters 3, 4, 7, and 11, which contained large numbers of co-expressed orthologous genes. On the other hand, this finding is consistent with previous studies finding low motif conservation [24,28,30,31]. We also found no significant enrichment for binding sites of orthologous transcription factors (Tac1, Mrr1, Crz1) known to mediate fluconazole-resistance in the evolutionarily diverged pathogen C. albicans [37].
Despite application of the soft-clustering algorithm, some clusters nevertheless shared significant gene orthology (but not expression) with other clusters, such as clusters 10 and 15 in Figure 3a. In these cases, we also found no conserved motifs between these clusters, indicating both promoter and expression divergence among orthologs in addition to species-specific motifs (Additional file 1: Species-specific Motifs).
Co-clustering implicates both highly conserved and divergent pathways
Next, we analyzed the soft clusters to identify pathways for which the fluconazole response is either highly conserved or strikingly divergent. For this purpose, differentially expressed pathways were identified using the GO Biological Process database [38] (Materials and methods). For each pathway, we computed the number of orthologous gene groups for which: 1, all three species were in the same cluster (full co-clustering); 2, two species were in the same cluster (partial co-clustering); or 3, no two species were in the same cluster (no co-clustering). The pathways with the highest percentage of orthologs with full co-clustering are shown in Figure 4a. The pathways with the highest percentage of orthologs that do not co-cluster are shown in Figure 4b. Clustering results for all pathways are given in Table S2 in Additional file 3.
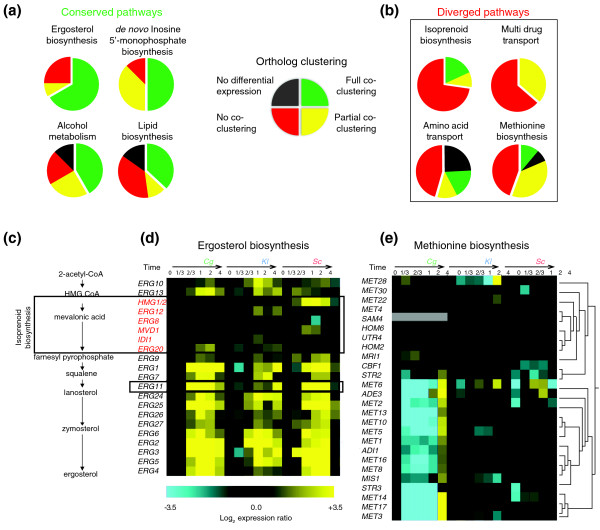
Figure 4.
Pathway expression conservation and divergence. (a) Top conserved and (b) diverged pathway responses as revealed by the soft clustering approach. Each pathway is represented by a pie with four slices - green, yellow, red, and black - denoting the percentage of orthologs in that pathway for which all three species co-clustered, two species co-clustered, no two species co-clustered, and no species' orthologs were differentially expressed, respectively. Pathways were defined using GO biological process annotations. (c) Schematic of ergosterol biosynthesis, the most conserved pathway response. Interestingly, this pathway includes isoprenoid biosynthesis, for which the response was one of the most divergent. (d) mRNA expression responses of ergosterol pathway genes are shown in order of occurrence in the pathway. Expression levels of genes 3 to 8 (boxed, and red) corresponding to isoprenoid biosynthesis are strikingly divergent. The fluconazole target Erg11 is boxed. (e) Hierarchically clustered mRNA expression responses of methionine biosynthesis genes show extensive divergence across species. Grey expression values denote a gene for which the species lacks an ortholog.
By this analysis, the most conserved pathway was ergosterol biosynthesis, which is consistent with our study of conserved motifs (above). Fluconazole directly inhibits ergosterol synthesis by targeting of Erg11, and all species appear to respond strongly to this reduction in ergosterol by up-regulating the enzymes required for its novel biosynthesis. ERG11 was up-regulated early in both Sc and Cg and later in Kl. Since ERG11 over-expression is one mechanism by which yeast can overcome fluconazole-induced growth inhibition [18], delays in its induction could contribute to Kl's greater fluconazole sensitivity.
The first stages of ergosterol biosynthesis are carried out by a subset of enzymes of the isoprenoid pathway. While most ergosterol genes were coordinately up-regulated in all three species, the expression levels of isoprenoid biosynthesis genes were strikingly divergent (Figures 4b, d). In all eukaryotes, regulation of isoprenoid biosynthesis is known to be complex with multiple levels of feedback inhibition [39]. Thus, the extensive divergence in isoprenoid biosynthesis expression suggests that the regulation of this pathway has also diverged between species.
Extensive expression divergence was also observed in methionine biosynthesis and amino acid transport (Figure 4b). Curiously, many Cg methionine biosynthesis orthologs were strongly down-regulated early in the time-course (Figure 4e). This strong down-regulation was not mirrored in Sc and Kl, which displayed divergent expression responses that were not co-clustered. Interestingly, it has been previously suggested that differences in methionine biosynthesis may alter azole susceptibility in C. neoformans [40] and C. albicans [41].
Major divergence in mRNA expression of transporters
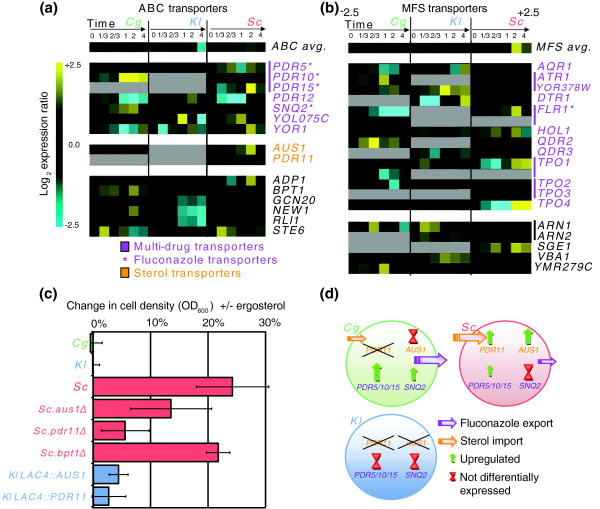
A final pathway for which we observed striking expression divergence was multi-drug transport (Figure 4b; Additional file 1: Transport). Most genes in this pathway were covered by clusters 8, 11, 16 (Figure 5a, b). Multi-drug transporters are divided into two classes: ATP-binding cassette (ABC) and major facilitator superfamily (MFS) transporters [5]. We examined the expression patterns of these transporters and found at least two types of divergent behaviors. First, the fraction of differentially expressed Sc MFS transporters was low compared to Cg and Kl (Fisher exact test, one-tailed P = 0.025 and 0.020, respectively). Second, the timing of MFS gene expression differed, with Sc up-regulated late and Cg up-regulated early (Figure 5b). In SC, several ABC and MFS transporters have been shown to bind fluconazole as a substrate [20,42,43]. Of these, we found that the PDR5/10/15 family of ABC transporters was up-regulated in Cg and Sc but not Kl. Another fluconazole transporter, SNQ2, was up-regulated in Cg only.
Figure 5.
Divergence in transporter usage. Cross-species expression profiles of (a) ATP-binding cassette (ABC) and (b) major facilitator superfamily (MFS) transporters are shown. Grey expression values denote a gene for which the species lacks an ortholog. (c) Change in cell density with addition of exogenous ergosterol at the fluconazole 50% inhibitory concentration across different mutant backgrounds. Sc.bpt1Δ is a gene knockout unrelated to fluconazole response and is included as a control. Error bars indicate one standard deviation. (d) Model for differential usage of transporters among Sc, Cg, and Kl.
We also found strong differences in the expression of other multi-drug transporters that have not been previously linked to fluconazole: PDR12 was strongly down-regulated in Sc and Cg but up-regulated in Kl; ATR1 and YOR378W were up-regulated in Cg and Kl but not Sc; HOL1 was up-regulated in Sc and Kl but not Cg. Some transporters also showed differences in expression timing (YOR1, PDR12).
Additionally, two ABC transporters, AUS1 and PDR11, which uptake sterol under anaerobic conditions [44], were up-regulated in Sc but were not differentially expressed in Cg (Cg does not possess a PDR11 ortholog). This suggests that Sc but not Cg increases sterol transport during fluconazole exposure. Intriguingly, since the direct effect of fluconazole is to inhibit sterol synthesis, increased sterol transport could be a mechanism for increased fluconazole tolerance. In support of this hypothesis, we found that the normally repressed cell wall mannoprotein DAN1, whose expression is required for sterol uptake [45], was up-regulated in Sc but not Cg. Since Kl lacks sterol transporters, it cannot import sterol and only grows aerobically [46,47] (Additional file 1: Analysis of Sterol Import Machinery in Fungal Genomes). As a possible explanation for this divergent behavior, we found that the promoter regions of ScAUS1, ScPDR11, and ScDAN1 contain binding motifs for ergosterol biosynthesis and/or sterol transport regulators Ecm22p, Rox1p and Sut1p, all of which were absent upstream of CgAUS1 and CgDAN1.
Therefore, the striking divergence in expression of fluconazole export and sterol import pathways suggests differing strategies in the azole response: following fluconazole exposure, Sc appears to activate sterol influx through up-regulation of PDR11 and AUS1; in contrast, Cg may activate fluconazole efflux through strong up-regulation of SNQ2 and a PDR5/10/15 ortholog (Figure 5a).
Sterol import increases fluconazole tolerance in Sc, but not Cg or Kl
To investigate these hypotheses, we grew wild-type Sc and Cg along with deletion mutants Sc.aus1Δ and Sc.pdr11Δ under fluconazole treatment in the presence or absence of exogenous ergosterol (4 μg/ml). As shown in Figure 5c, we found that addition of ergosterol had no effect on growth of Cg but led to an increase in growth of Sc (P = 0.018). This increase was attenuated in Sc.aus1Δ and Sc.pdr11Δ (P = 0.033), which lack sterol import genes, but not in an unrelated control knockout, Sc.bpt1Δ. Thus, Sc but not Cg is aided by adding ergosterol to the environment, and this process is likely dependent on AUS1 and/or PDR11.
Three additional lines of evidence support the hypothesis that Sc prefers sterol import while Cg prefers fluconazole export in response to fluconazole treatment. A retrospective analysis of deletion mutant fitness in Sc [48] revealed that a greater proportion of gene deletions involved in the sterol pathway lead to fluconazole sensitivity than deletion of fluconazole transporters themselves (Fisher exact test, one-tailed P = 0.043). This suggests a role for sterol transporters in the Sc fluconazole response. Second, fluconazole tolerance in Cg has been shown to be unaffected when constitutively expressing CgAUS1 in the presence of exogenous free cholesterol (though not in the presence of serum) [49]. Third, deletion of the Cg orthologs of fluconazole transporters PDR5 (CgCDR1) [50] or SNQ2 [51] both resulted in increased fluconazole sensitivity.
Expression of sterol importers in Kl increases fluconazole tolerance
Since Kl neither up-regulates drug exporters nor encodes sterol importers, we considered that this lack of a transport response might be responsible for the higher drug sensitivity we observed for Kl in relation to the other species. Consistent with this hypothesis, we found that Kl growth was unaffected by addition of exogenous ergosterol (Figure 5c), similar to Cg but in sharp contrast to Sc. We also predicted that transgenic expression of sterol importers ScAus1 or ScPdr11 in Kl might increase fluconazole tolerance in the presence of exogenous ergosterol. To test this prediction, we chromosomally integrated ScAUS1 and ScPDR11 into Kl non-disruptively at the KlLAC4 locus under control of the strong constitutive Kl PLAC4-PBI promoter (Materials and methods). Transformed Kl strains were grown under fluconazole treatment with and without exogenous ergosterol (4 μg/ml). We observed that transgenic expression of sterol importer AUS1 in Kl significantly increased fluconazole tolerance (P = 0.012; Figure 5c) in an ergosterol-dependent manner. Thus, it appears that differences in sterol import and drug export are responsible for a component of the anti-fungal response, and of the observed functional divergence across the three yeast species.
Conclusions
In this study, we have compared the dynamic transcriptional responses of three diverse yeast species to fluconazole treatment, revealing significant divergence in their regulatory programs. The data suggest several different mechanisms of azole tolerance, depending on the species (Figure 5d). The Sc response depends on sterol influx, through up-regulation of PDR11 and AUS1. In contrast, the Cg response relies on fluconazole efflux through strong up-regulation of SNQ2 and a PDR5/10/15 ortholog. Neither of these responses have evolved in Kl, leading to its severe drug sensitivity. These conclusions are supported by follow-up experiments demonstrating that growth in ergosterol increases the fluconazole tolerance of Sc, but not other species, in a PDR11- and AUS1-dependent fashion. They are also supported by the finding that transgenic expression of AUS1 in Kl increases the fluconazole tolerance of this species.
To arrive at these conclusions, we employed a novel 'soft clustering' approach that is of general use in the fields of comparative and systems biology. This approach is distinct from other methods for cross-species expression analysis [27,28,30,52] in several important ways. Chief among these, it integrates sequence orthology with gene expression patterns to produce accurate orthologous clusters. This integration is accomplished by a symmetric process that does not require the designation of one species as a reference. In addition, soft clustering handles data from more than two species and can, in principle, analyze any number of species simultaneously. In future, such approaches might be used to survey a wider range of species, drug concentrations and stimuli to reveal conserved and divergent molecular response pathways.
Materials and methods
Strains and growth conditions
Standard laboratory strains with known genomic sequence [53] were used: Sc BY4741, Cg CBS138 (ATCC 2001), and Kl NRRL Y-1140 (ATCC 8585). Cultures were grown in rich media (YPD) from OD600 of 0.05 to 0.2 at 30°C and 225 rpm. Cells were treated with fluconazole at species-specific sub-inhibitory concentrations (Figure S1 in Additional file 1), and harvested at 0, 1/3, 2/3, 1, 2 or 4 doubling times as measured for untreated cells.
Microarray expression profiling
RNA was isolated by hot phenol/chloroform extraction and enriched for mRNA via poly-A selection (Ambion 1916, Austin, TX, USA). mRNA from untreated cells was combined in equal amounts from all time points to form a species-specific reference sample. Six replicates per time point were dUTP labeled (three biological replicates by two technical replicates) with Cy3 and Cy5 dyes (Invitrogen SKU11904-018, Carlsbad, CA, USA) creating a dye-swapped reference design. Samples were hybridized to Agilent expression arrays using the protocol recommended by Agilent. Differential expression was called using the VERA error model [54] and false discovery rate multiple-test correction [55]. Additional description of both the microarray platform and analysis can be found in Additional file 1.
Soft clustering algorithm
We developed a constrained clustering method based on the k-means algorithm, but using a revised objective function (Additional file 1). Like regular k-means, the objective function considers the similarity of each gene's expression profile to the center of its assigned class. However, it also rewards class assignments in which orthologs are co-clustered. The reward (W) is a user-defined parameter that serves as a tradeoff between cluster expression coherence and percentage of co-clustered orthologs: each gene, x ∈ X, is assigned to cluster h* such as to minimize the objective function:
where ∑(D(x, Ch) - W) refers to all possible partitions of genes in the same orthology group, D() refers to a user defined distance function, and Ch denotes the center of cluster h. As discussed in the main text and in Additional file 1, the appropriate value of the reward, W, can be determined using complementary information. Here, it was tuned to maximize the GO enrichment of the clusters.
The new objective function also leads to changes in the search algorithm for determining the optimal cluster assignments: for each group of orthologs across the three species, we search for the partitions that result in the minimum total distance between all pairs of group members. Since there are 2m possible subgroups, where m is the size of the orthology group (here, most orthology groups are of size m = 3), and each subgroup is checked for all possible k clusters, the search complexity for each group is O(2m * k). Since m is small, the running time of the algorithm is typically very fast. Detailed methods, including algorithm pseudo-code, are presented in Additional file 1.
Identifying highly conserved and divergent pathways
We first ranked GO processes categories [38] based on their significance of overlap with differentially expressed orthologous groups [32]. An orthologous group was considered differentially expressed if at least one member was differentially expressed. We used the top 20 ranked GO processes for identifying conserved and divergent pathways. Conserved pathways were defined as those with the highest 'full co-clustering' fraction of genes known to be involved in the process and divergent pathways were defined as those with the highest 'no co-clustering' fractions.
Insertion of ScAUS1/ScPDR11 into Kl
To facilitate insertion of ScAUS1 and ScPDR11 into Kl, open reading frames were placed under control of the strong PLAC4-PBI promoter by cloning into plasmid pKLAC2 (NEB N3742S), which possesses approximately 2-kb homology to the Kl.LAC4 locus. Open reading frames were amplified with a SacI restriction site (3' end), which was used to ligate a kanamycin marker from pCR-Blunt (Invitrogen K-2800-20). XhoI (5' end) and SbfI (3' end) restriction sites were added by PCR for ligation into pKLAC2. Modified plasmids were transformed into Escherichia coli and screened on Luria-Bertani media containing ampicillin and kanamycin. Plasmids were mini-prepped (GE Healthcare #US79220-50RXNS, Piscataway, NJ, USA) and verified by PCR and SacII digestion. All restriction enzymes were obtained from New England Biolabs (Ipswich, MA, USA).
SacII-linearized plasmids were transformed into Kl NRRL Y-1140 by electroporation, thereby inserting ScAUS1 and ScPDR11 non-disruptively at the Kl.LAC4 locus. Colonies were selected on YCB + 5 mM acetamide (New England Biolabs N3742 S and verified by PCR. mRNA expression of ScAUS1 and ScPDR11 was validated by quantitative RT-PCR.
Data
The data reported in this paper have been deposited in the Gene Expression Omnibus database, accession number [GEO:GSE15710].
Abbreviations
ABC: ATP-binding cassette; CG: Candida glabrata; GO: Gene Ontology; Kl: Kluyveromyces lactis; MFS: major facilitator superfamily; SC: Saccharomyces cerevisiae.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DK, KT, TR and TI designed the study. DK performed all experimental work. ZBJ and GZ developed the soft-constraint clustering approach. DK, KT, and GZ analyzed the data. DK and TI wrote the manuscript. ZBJ and TI supervised the work.
Supplementary Material
Supplementary Methods, Results, and Discussion.
Supplementary Table S1.
Supplementary Table S2.
Contributor Information
Dwight Kuo, Email: dkuo@bioeng.ucsd.edu.
Kai Tan, Email: kai-tan@uiowa.edu.
Guy Zinman, Email: zinman@cs.cmu.edu.
Timothy Ravasi, Email: travasi@bioeng.ucsd.edu.
Ziv Bar-Joseph, Email: zbj@cs.cmu.edu.
Trey Ideker, Email: tideker@ucsd.edu.
Acknowledgements
We thank Katherine Licon, Justin Catalana and Kevin Thai for technical assistance. DK was supported by the National Science and Engineering Research Council of Canada. KT and TI were supported by a David and Lucille Packard Foundation Award and NIH Grant #R01 ES014811 to TI. GZ and ZBJ were supported by NIH grant #RO1 GM085022 and NSF CAREER award 0448453 to ZBJ.
References
- Paulitsch A, Weger W, Ginter-Hanselmayer G, Marth E, Buzina W. A 5-year (2000-2004) epidemiological survey of Candida and non-Candida yeast species causing vulvovaginal candidiasis in Graz, Austria. Mycoses. 2006;49:471–475. doi: 10.1111/j.1439-0507.2006.01284.x. [DOI] [PubMed] [Google Scholar]
- Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother. 2005;49:668–679. doi: 10.1128/AAC.49.2.668-679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson LS, Reyes CM, Stolpman M, Speckman J, Allen K, Beney J. The direct cost and incidence of systemic fungal infections. Value Health. 2002;5:26–34. doi: 10.1046/j.1524-4733.2002.51108.x. [DOI] [PubMed] [Google Scholar]
- Maertens JA. History of the development of azole derivatives. Clin Microbiol Infect. 2004;10(Suppl 1):1–10. doi: 10.1111/j.1470-9465.2004.00841.x. [DOI] [PubMed] [Google Scholar]
- Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat Rev Microbiol. 2005;3:547–556. doi: 10.1038/nrmicro1179. [DOI] [PubMed] [Google Scholar]
- Carrillo-Munoz AJ, Giusiano G, Ezkurra PA, Quindos G. Antifungal agents: mode of action in yeast cells. Rev Esp Quimioter. 2006;19:130–139. [PubMed] [Google Scholar]
- Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. Molecular basis of resistance to azole antifungals. Trends Mol Med. 2002;8:76–81. doi: 10.1016/S1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- Sanglard D. Resistance of human fungal pathogens to antifungal drugs. Curr Opin Microbiol. 2002;5:379–385. doi: 10.1016/S1369-5274(02)00344-2. [DOI] [PubMed] [Google Scholar]
- Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- Cowen LE, Steinbach WJ. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot Cell. 2008;7:747–764. doi: 10.1128/EC.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Lee AY, Epp E, Fredette A, Surprenant J, Harcus D, Scott M, Tan E, Nishimura T, Whiteway M, Hallett M, Thomas DY. Chemogenomic profiling predicts antifungal synergies. Mol Syst Biol. 2009;5:338. doi: 10.1038/msb.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol. 2006;61:704–722. doi: 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- Lepak A, Nett J, Lincoln L, Marchillo K, Andes D. Time course of microbiologic outcome and gene expression in Candida albicans during and following in vitro and in vivo exposure to fluconazole. Antimicrob Agents Chemother. 2006;50:1311–1319. doi: 10.1128/AAC.50.4.1311-1319.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammert GF, Fostel JM. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob Agents Chemother. 2000;44:1255–1265. doi: 10.1128/AAC.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist S. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot Cell. 2006;5:2184–2188. doi: 10.1128/EC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis DP. Efflux-mediated resistance to fluconazole could be modulated by sterol homeostasis in Saccharomyces cerevisiae. J Antimicrob Chemother. 2000;46:199–203. doi: 10.1093/jac/46.2.199. [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J Biol Chem. 2003;278:34998–35015. doi: 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin Microbiol Infect. 2004;10(Suppl 1):11–23. doi: 10.1111/j.1470-9465.2004.t01-1-00844.x. [DOI] [PubMed] [Google Scholar]
- Anderson JB, Sirjusingh C, Parsons AB, Boone C, Wickens C, Cowen LE, Kohn LM. Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics. 2003;163:1287–1298. doi: 10.1093/genetics/163.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermitsky JP, Edlind TD. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother. 2004;48:3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Lelandais G, Shukla S, Mukhopadhyay G, Jacq C, Devaux F, Prasad R. Responses of pathogenic and nonpathogenic yeast species to steroids reveal the functioning and evolution of multidrug resistance transcriptional networks. Eukaryot Cell. 2008;7:68–77. doi: 10.1128/EC.00256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais G, Tanty V, Geneix C, Etchebest C, Jacq C, Devaux F. Genome adaptation to chemical stress: clues from comparative transcriptomics in Saccharomyces cerevisiae and Candida glabrata. Genome Biol. 2008;9:R164. doi: 10.1186/gb-2008-9-11-r164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Xie D, Fan Z, Chipperfield H, Marden J, Wong WH, Zhong S. Modeling co-expression across species for complex traits: insights to the difference of human and mouse embryonic stem cells. PLoS Comput Biol. 2010;6:e1000707. doi: 10.1371/journal.pcbi.1000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Berman J, Barkai N. Comparative gene expression analysis by differential clustering approach: application to the Candida albicans transcription program. PLoS Genet. 2005;1:e39. doi: 10.1371/journal.pgen.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyenko A, Tamas I, Liu G, Sonnhammer ELL. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;22:e9–15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- Davies BS, Rine J. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics. 2006;174:191–201. doi: 10.1534/genetics.106.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PM, Oliver BG, White TC. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell. 2004;3:1391–1397. doi: 10.1128/EC.3.6.1391-1397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, Philippakis AA, Hu Y, De Masi F, Pacek M, Rolfs A, Murthy T, Labaer J, Bulyk ML. High-resolution DNA binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DH, D'Haeseleer P. Deciphering principles of transcription regulation in eukaryotic genomes. Mol Syst Biol. 2006;2:2006.0012. doi: 10.1038/msb4100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhauser J. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet Biol. 2009;47:94–106. doi: 10.1016/j.fgb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Consortium GO. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimster-Denk D, Rine J, Phillips J, Scherer S, Cundiff P, DeBord K, Gilliland D, Hickman S, Jarvis A, Tong L, Ashby M. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix(TM). J Lipid Res. 1999;40:850–860. [PubMed] [Google Scholar]
- Pascon RC, Ganous TM, Kingsbury JM, Cox GM, McCusker JH. Cryptococcus neoformans methionine synthase: expression analysis and requirement for virulence. Microbiology. 2004;150:3013–3023. doi: 10.1099/mic.0.27235-0. [DOI] [PubMed] [Google Scholar]
- Ha KC, White TC. Effects of azole antifungal drugs on the transition from yeast cells to hyphae in susceptible and resistant isolates of the pathogenic yeast Candida albicans. Antimicrob Agents Chemother. 1999;43:763–768. doi: 10.1128/aac.43.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenreiro S, Rosa PC, Viegas CA, Sa-Correia I. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast. 2000;16:1469–1481. doi: 10.1002/1097-0061(200012)16:16<1469::AID-YEA640>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Broco N, Tenreiro S, Viegas CA, Sa-Correia I. FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on pdr3 transcriptional regulator. Yeast. 1999;15:1595–1608. doi: 10.1002/(SICI)1097-0061(199911)15:15<1595::AID-YEA484>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Wilcox LJ, Balderes DA, Wharton B, Tinkelenberg AH, Rao G, Sturley SL. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem. 2002;277:32466–32472. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- Alimardani P, Regnacq M, Moreau-Vauzelle C, Ferreira T, Rossignol T, Blondin B, Berges T. SUT1-promoted sterol uptake involves the ABC transporter Aus1 and the mannoprotein Dan1 whose synergistic action is sufficient for this process. Biochem J. 2004;381:195–202. doi: 10.1042/BJ20040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussereau F, Casaregola S, Lafay JF, Bolotin-Fukuhara M. The Kluyveromyces lactis repertoire of transcriptional regulators. FEMS Yeast Res. 2006;6:325–335. doi: 10.1111/j.1567-1364.2006.00028.x. [DOI] [PubMed] [Google Scholar]
- Snoek IS, Steensma HY. Why does Kluyveromyces lactis not grow under anaerobic conditions? Comparison of essential anaerobic genes of Saccharomyces cerevisiae with the Kluyveromyces lactis genome. FEMS Yeast Res. 2006;6:393–403. doi: 10.1111/j.1567-1364.2005.00007.x. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Nakayama H, Tanabe K, Bard M, Hodgson W, Wu S, Takemori D, Aoyama T, Kumaraswami NS, Metzler L, Takano Y, Chibana H, Niimi M. The Candida glabrata putative sterol transporter gene CgAUS1 protects cells against azoles in the presence of serum. J Antimicrob Chemother. 2007;60:1264–1272. doi: 10.1093/jac/dkm321. [DOI] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother. 1999;43:2753–2765. doi: 10.1128/aac.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli R, Posteraro B, Ferrari S, La Sorda M, Fadda G, Sanglard D, Sanguinetti M. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol Microbiol. 2008;68:186–201. doi: 10.1111/j.1365-2958.2008.06143.x. [DOI] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Generalized singular value decomposition for comparative analysis of genome-scale expression data sets of two different organisms. Proc Natl Acad Sci USA. 2003;100:3351–3356. doi: 10.1073/pnas.0530258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DJ, Martin T, Nikolski M, Cayla C, Souciet JL, Durrens P. Genolevures: protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 2009;37:D550–554. doi: 10.1093/nar/gkn859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, Feizi H, Ideker T. Correcting for gene-specific dye bias in DNA microarrays using the method of maximum likelihood. Bioinformatics. 2008;24:71–77. doi: 10.1093/bioinformatics/btm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci USA. 2002;99:8778–8783. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- Barker KS, Pearson MM, Rogers PD. Identification of genes differentially expressed in association with reduced azole susceptibility in Saccharomyces cerevisiae. J Antimicrob Chemother. 2003;51:1131–1140. doi: 10.1093/jac/dkg217. [DOI] [PubMed] [Google Scholar]
- Ernst J, Nau GJ, Bar-Joseph Z. Clustering short time series gene expression data. Bioinformatics. 2005;21(Suppl 1):i159–168. doi: 10.1093/bioinformatics/bti1022. [DOI] [PubMed] [Google Scholar]
- Lu Y, Huggins P, Bar-Joseph Z. Cross species analysis of microarray expression data. Bioinformatics. 2009;25:1476–1483. doi: 10.1093/bioinformatics/btp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Banerjee A, Mooney RJ. Active semi-supervision for pairwise constrained clustering. Proceedings of the 2004 SIAM International Conference on Data Mining, (SDM-2004); June 13-16, 2004: Nashville, TN. 2004. pp. 333–344.
- Wagstaff KL. In: Knowledge Discovery in Inductive Databases: 5th International Workshop, KDID 2006; Berlin, Germany: September 18th, 2006. Dzeroski S, Struyf J, editor. Springer; 2007. Value, cost, and sharing: issues in constrained clustering; pp. 1–10. [Lecture Notes in Computer Science, volume 4747.] [Google Scholar]
- Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/S0219720005001375. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Eisen MB. Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol. 2002;3:RESEARCH0059. doi: 10.1186/gb-2002-3-11-research0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, Pesole G. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics. 2001;17(Suppl 1):S207–214. doi: 10.1093/bioinformatics/17.suppl_1.s207. [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Hendrix D, Golden A, Smith TJ, Rokhsar DS. Transcription factor binding site identification using the self-organizing map. Bioinformatics. 2005;21:1807–1814. doi: 10.1093/bioinformatics/bti256. [DOI] [PubMed] [Google Scholar]
- Merico A, Sulo P, Piskur J, Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. Febs J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M. In: Biodiversity and Ecophysiology of Yeasts. Rosa C, Péter G, editor. Springer; 2006. Genomics and biodiversity in yeasts. [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Results, and Discussion.
Supplementary Table S1.
Supplementary Table S2.