Summary
Background
Homolog pairing, synaptonemal complex (SC) assembly (chromosome synapsis), and crossover recombination are essential for successful meiotic chromosome segregation. A distinguishing feature of meiosis in budding yeast and mammals is that synapsis between homologs depends upon recombination; however, the molecular basis for this contingency is not understood.
Results
We show that the yeast proline isomerase, Fpr3, and the SUMO ligase, Zip3, ensure that SC assembly is dependent upon recombination initiation. When Fpr3 and Zip3 are absent, synapsis occurs even in a mutant that fails to initiate recombination and homolog pairing. Fpr3 and Zip3 appear to specifically prevent synapsis initiation at centromeric sites. This result is consistent with previous observations of SC proteins localizing to centromeres prior to and independent of meiotic recombination initiation. Finally, we show that without Fpr3 and Zip3 activities, the synapsis initiation components, Zip2 and Zip4, are dispensable for chromosome synapsis.
Conclusion
Fpr3 and Zip3 represent parallel pathways that function, in a checkpoint-like manner, to ensure that chromosome synapsis is contingent on the initiation of recombination. We propose that, during normal meiosis, Zip2 and Zip4 act downstream of recombination signals to oppose Fpr3- and Zip3-mediated inhibitions to initiating SC assembly at centromeres. These data suggest a role for centromeres in coordinating major meiotic chromosomal events and draw an interesting parallel between yeast centromeres and C. elegans Pairing Centers.
Introduction
During meiosis, the two members of every homologous chromosome pair segregate from one another in order to generate viable gametes. Homologs must first physically associate in order to ensure that they orient properly on the meiosis I spindle and, subsequently, move toward opposite spindle poles. For most organisms, homologous chromosome alignment (pairing), crossover recombination, and chromosome synapsis are the major chromosomal events that promote stable homolog associations.
Conceptually, stable pairing between homologous chromosomes involves two steps: homolog pairing, followed by reinforcement of paired associations. In some organisms, early steps in recombination are required for initial homologous pairing [1]. Furthermore, meiotic recombination events leading to a crossover outcome result in physical links that stabilize homolog associations. Thus, meiotic recombination appears to contribute to both pairing and reinforcement. On the other hand, synaptonemal complex (SC) assembly (i.e., synapsis) is clearly involved in reinforcement. The SC is formed by the assembly of “central region” proteins at the interface of the proteinaceous cores of lengthwise-aligned chromosomes [2]. The SC appears to bolster and/or maintain initial paired associations, either through a direct, structural role or by promoting a normal level and distribution of crossover events. While normally a hallmark of successful homolog alignment, SC is not a prerequisite for homologous pairing. Moreover, SC can form between nonhomologous chromosomes [3-7].
The fact that SC assembly does not intrinsically require homologous chromosomal associations raises the question of how homology recognition is coordinated with reinforcement of paired associations, such that synapsis does not occur inappropriately between nonhomologus chromosomes. In budding yeast, SC does not form if early recombination and pairing events fail to occur. Zip1 is a major structural component of the SC central region, while Zip2, Zip3, Zip4/Spo22 and Spo16 are Synapsis Initiation Complex proteins that localize to synapsis initiation sites and are required for the initiation and/or progression of SC assembly [8-13]. In the absence of recombination and homolog pairing, the bulk of Zip1 and Synapsis Initiation Complex proteins co-assemble into an extra-chromosomal structure, called a polycomplex, and a limited amount of Zip1 and Zip3 localizes to centromeric chromosomal regions, but SC does not form. How do cells regulate synapsis to ensure that SC does not assemble on chromosomes at the wrong time?
Here, we demonstrate that multiple pathways prevent inappropriate synapsis during meiosis in budding yeast. We show that in the combined absence of a proline isomerase protein, Fpr3, and a SUMO ligase protein, Zip3, SC assembles on chromosomes even in the absence of meiotic recombination initiation and homolog pairing. Our data suggest that Fpr3 and Zip3 regulate Zip1 in mechanistically different ways, to ensure that SC assembly is contingent upon earlier chromosomal events, such as homolog pairing. Moreover, we provide evidence that Zip2 and Zip4 normally oppose the Fpr3 and Zip3 pathways in order to link recombination and/or pairing with synapsis. Further, our data strongly suggest that Fpr3 and Zip3 regulate synapsis specifically at centromeres. This result bolsters the notion that centromeres are a special subset of synapsis initiation sites in budding yeast and suggest a role for centromeres in coordinating homolog pairing with synapsis.
Results
Fpr3 Regulates Zip1 Spatial Distribution in spo11 Meiotic Cells
To identify factors that prevent SC assembly when recombination and chromosome pairing fail, we transposon mutagenized [14] a strain expressing a Zip1-GFP fusion [15] and lacking the Spo11 enzyme, which is required for recombination initiation. We screened for mutations that disrupt the spatial distribution of Zip1-GFP in spo11 meiotic nuclei (see Supplemental Experimental Procedures). At the pachytene stage of meiotic prophase, wild-type cells expressing Zip1-GFP display dynamic, flexible “lines” moving throughout the nucleus, corresponding to full-length SCs [15] (Figure 1A, Supplemental Movies S1 and S4). On the other hand, spo11 meiotic cells producing Zip1-GFP exhibit a single, bright, GFP focus, corresponding to a polycomplex (Figure 1A, Supplemental Movies S2 and S5). We reasoned that meiotic spo11 ZIP1-GFP cells carrying a mutation in a regulator of SC assembly might exhibit moving Zip1-GFP lines instead of a bright focus.
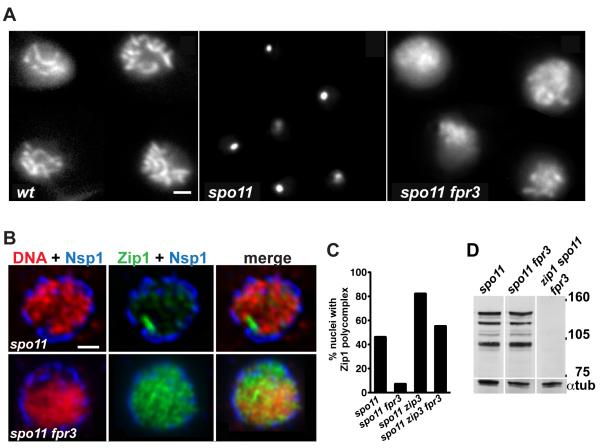
Figure 1.
Fpr3 Regulates Zip1 Spatial Distribution in Meiotic Nuclei of spo11 Mutants
(A, B) Zip1-GFP in live diploid cells at mid-meiotic prophase. Images in (B) show the midsection of fixed, semi-intact, meiotic nuclei. Staining with anti-Nsp1 antibody to mark the nuclear envelope (blue), DAPI to label DNA (red) and anti-Zip1 antibody (green) revealed increased dispersion of Zip1 throughout the nucleoplasm of spo11 fpr3 cells, compared to spo11 cells. (A and B) Bar, 1 micron.
(C) The frequency of polycomplexes was scored in meiotic prophase nuclei (selected based on the presence of the meiosis-specific chromosomal protein, Red1) that were surface-spread, using a standard method that extracts nucleoplasmic material [8], 16 hours after introduction into sporulation medium. The number of nuclei scored was 189, 150, 153 and 113 for spo11, spo11 fpr3, spo11 zip3 and spo11 zip3 fpr3 mutants, respectively. The following pairs of strains showed significant differences (two-sided Fisher’s Exact Test, p < 0.0001): spo11 vs. spo11 fpr3, spo11 vs. spo11 zip3 and spo11 zip3 vs. spo11 zip3 fpr3.
(D) Immunoblot of Zip1 immunoprecipitated from meiotic cell extracts. Each strain carries an ndt80 mutation, which causes pachytene arrest. Thus, after 24 hours of sporulation, when extracts were prepared, the majority of cells are in late prophase. The level of multiple, discrete Zip1 bands running between 90-150 kD markers, is similar between spo11 and spo11 fpr3 cells. Labeling of alpha-tubulin on the same blot confirms that similar quantities of extract were loaded.
Two independent transposon insertions identified a role for the proline isomerase, Fpr3, in regulating Zip1 distribution during meiosis. Live spo11 meiotic cells carrying either insertion in FPR3 showed a reduced frequency and size of polycomplexes, with Zip1-GFP non-uniformly dispersed, in linear segments, throughout the nucleus (Supplemental Movies S3 and S6). We created a spo11 strain carrying an fpr3 deletion and observed the same effect on Zip1-GFP distribution (Figure 1A).
We used the fpr3 deletion in combination with an untagged ZIP1 gene for all subsequent experiments. The presence of Zip1 in meiotic nuclei was further examined using immunofluoresence on cells fixed with a method that preserves most nuclear membrane and nucleoplasmic contents (Figure 1B). In such “semi-squashed” nuclei from spo11 meiotic prophase cells, Zip1 protein is either hardly detectable, or aggregated into a large polycomplex. In contrast, Zip1 is dispersed throughout the nuclei of spo11 fpr3 meiotic cells. To assess whether spo11 fpr3 nuclei assemble Zip1 on chromosomes, lysed meiotic nuclei were fixed and spread on glass slides using a technique that removes nucleoplasmic and cytoplasmic structures but preserves chromatin and associated proteins, including SCs and polycomplexes. Zip1 linear stretches were not detectable on spo11 fpr3 chromosomes at any stage in meiotic prophase, indicating that abnormally distributed Zip1-GFP in spo11 fpr3 cells corresponds to nucleoplasmic Zip1, not Zip1 assembled on chromosomes.
Immunofluoresence analysis of spread nuclei confirmed that polycomplex formation is reduced in cells containing an fpr3 mutation. At 16 hours after introduction into sporulation medium, about half of spo11 meiotic nuclei exhibit a Zip1 polycomplex (Figure 1C). In contrast, significantly fewer spo11 fpr3 cells (7%) exhibit polycomplexes (P < 0.0001) (Figure 1C). The Synapsis Initiation Complex protein, Zip3, must normally act to discourage polycomplex formation in spo11 cells, as a significantly greater fraction of spo11 zip3 meiotic cells (82%) exhibit a polycomplex (P < 0.0001) (Figure 1C). The fpr3 mutation significantly reduces the frequency of polycomplex formation in spo11 zip3 cells (P < 0.0001) (Figure 1C).
Total levels of Zip1 protein are similar between spo11 and spo11 fpr3 cells at mid-meiotic prophase (Figure 1D). Thus, instead of regulating overall levels of Zip1, the Fpr3 protein appears to promote the aggregation of Zip1 protein into polycomplexes, under circumstances that prevent SC assembly on chromosomes.
Fpr3 Localizes to Polycomplexes
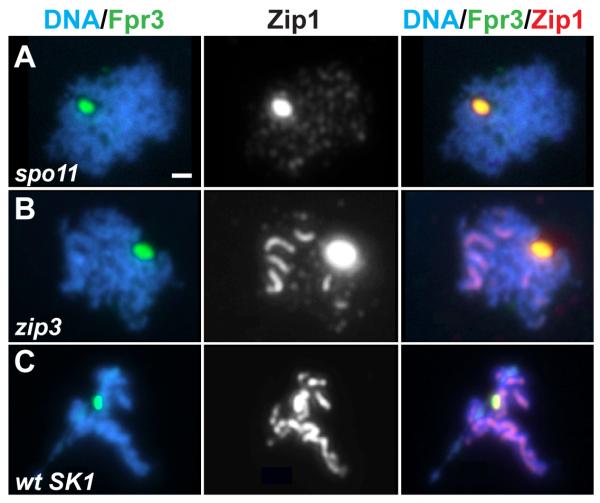
Fpr3 localizes predominantly to nucleoli during mitotic growth, but disperses throughout the nucleoplasm upon progression into meiosis [16, 17]. Fpr3 is present in the nucleoplasm of spo11 meiotic cells (data not shown), indicating that the redistribution of Fpr3 during meiosis is independent of recombination. Interestingly, we further observed that Fpr3 co-localizes with Zip1 in polycomplexes in spo11 and zip3 mutants (Figure 2A, B), and in meiotic cells of the SK1 wild-type strain (which, unlike cells of the BR strain used in most of our experiments, contain polycomplexes) (Figure 2C).
Figure 2.
Fpr3 Localizes to Polycomplexes
Meiotic prophase nuclei from genetic backgrounds known to form polycomplexes were labeled with DAPI to mark DNA (blue), anti-Fpr3 antiserum (green) and anti-Zip1 antibodies (white in middle panels, red in right panels). Fpr3 and Zip1 colocalize as a large aggregate, the polycomplex, in nuclei from spo11 (A), zip3 (B), or wild-type (C) cells of the SK1 strain. Bar, 1 micron.
Fpr3 and Zip3 Together Prevent SC Assembly on Unpaired Chromosomes in spo11 Mutants
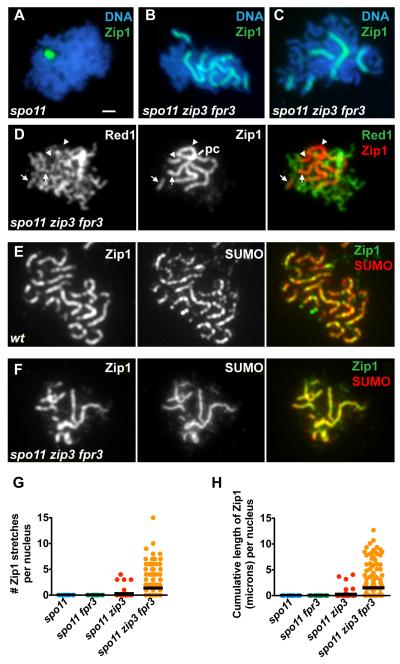
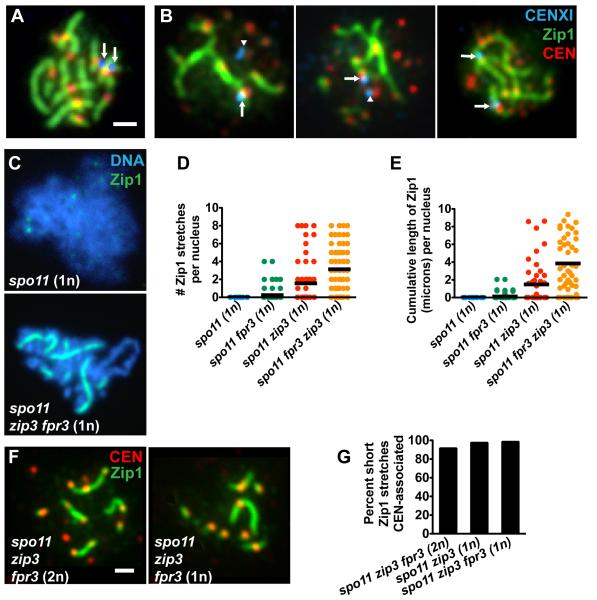
Our analysis of Fpr3 led us to an unexpected role for the Synapsis Initiation Complex protein, Zip3, in preventing synapsis. While meiotic nuclei from spo11 fpr3 cells never assemble Zip1 on chromosomes (>100 nuclei scored), 34% (n = 198) of nuclei from the spo11 zip3 fpr3 triple mutant display robust, sometimes extremely long and continuous, linear stretches of Zip1 on meiotic chromosomes (Figure 3B-D and F-H). spo11 zip3 fpr3 triple mutants exhibit between one and 15 Zip1 linear stretches, corresponding to up to 13 microns of Zip1 length per nucleus (Figure 3G, H) (in wild type, Zip1 cumulative length measures up to 24 microns (Figure 5C)). Nuclei in spo11 zip3 double mutants exhibit one or two Zip1 linear stretches only very rarely (Figure 3G).
Figure 3.
Zip3 and Fpr3 Together Regulate Synapsis in spo11 Mutant Cells
(A-F) The molecules labeled are indicated, along with their corresponding colors, in the upper right of each panel. Bar, 1 micron.
(A-D, F) Linear stretches of Zip1 on meiotic chromosomes in spo11 mutants. Surface-spread nuclei were labeled with antibodies to Zip1, DAPI (to mark DNA), anti-Red1 antibodies (to label chromosome cores [33]) and anti-Smt3 antibodies (to mark SUMO).
(D) Examples of Zip1 linear stretches that colocalize with Red1 are indicated by arrows; the few stretches that do not colocalize with Red1 (arrowheads) (but usually colocalize with DAPI (not shown)) appear to emanate from the polycomplex (pc).
(E, F) SUMO decorates Zip1 linear stretches in mid-prophase nuclei from (E) wild-type cells and from (F) spo11 zip3 fpr3 mutant cells.
(G and H) The extent of Zip1 polymerization in nuclei from various strains is displayed on scatterplots. In (G) each nucleus is plotted along the Y axis according to the total number of distinct Zip1 stretches it contained. In (H) each nucleus is plotted along the Y axis according to the total, cumulative length of Zip1 it contained. A horizontal black bar depicts the mean for each strain. The fraction of nuclei exhibiting one or more Zip1 linear segments was 6/57 for spo11 zip3 and 67/198 for spo11 zip3 fpr3 mutants. (100 nuclei were plotted for spo11 and spo11 fpr3). Polycomplexes, which sometimes are linear in shape, but stain more intensely than stretches of SC, were not included in these counts.
Figure 5.
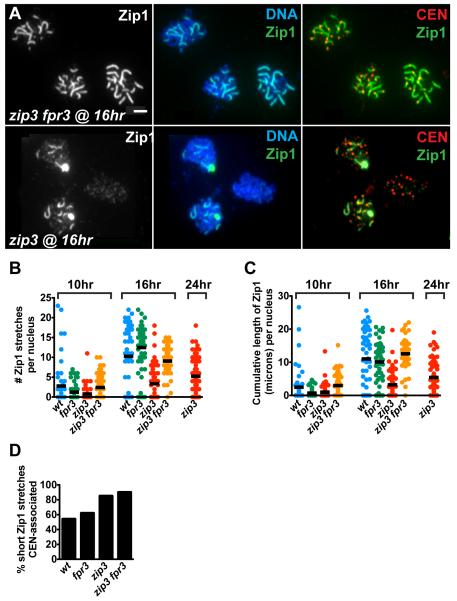
fpr3 Mutation Allows Increased Synapsis from Centromeres in Cells Missing Zip3
(A) The average extent of Zip1 polymerization on chromosomes is higher in nuclei from zip3 fpr3 (top) than from zip3 (bottom) cells at a similar time point in meiotic prophase. Labeled molecules, and their respective colors, are indicated at the upper right of each panel. Bar, 1 micron.
(B, C), The extent of Zip1 polymerization was measured at multiple time points for the zip3 fpr3 double mutant and control strains. Nuclei were plotted on each graph as described in Figure 3G and H. At least 50 nuclei were plotted for each strain. At 16 hours, both the number of Zip1 stretches (B) and their cumulative length (C) are significantly reduced in zip3 mutants compared to the other strains (Mann-Whitney two-tailed p < 0.0001); these values are not significantly different between wild-type and zip3 fpr3 strains.
(D) Frequency of centromere-associated, short Zip1 stretches (0.35-0.65 microns in length) from nuclei at 10- and 16-hour time points. Because synapsis is delayed in the zip3 mutant, nuclei at 24 hours after induction of sporulation were also included. The number of short Zip1 stretches analyzed was 521, 643, 290 and 195 for wild type, fpr3, zip3, and zip3 fpr3 cells, respectively. Significant differences were observed between the following pairs of strains (two sided p <0.0001): zip3 vs. wild-type, zip3 vs. fpr3, zip3 fpr3 vs. wild type, and zip3 fpr3 vs. fpr3. The values exhibited by wild type and fpr3 are not significantly different.
The small ubiquitin-like modifier protein, SUMO, colocalizes with Zip1 assembled in the context of SC, and is dependent on Zip1 assembly within the SC central region for its localization to synapsed chromosomes [18, 19]. If the Zip1 stretches exhibited by spo11 zip3 fpr3 cells reflect normal SC central region assembly, SUMO is expected to colocalize with Zip1 stretches in the triple mutant. Indeed, SUMO labels the Zip1 stretches exhibited by spo11 zip3 fpr3 meiotic nuclei (Figure 3F).
Like spo11 single mutants, spo11 zip3 fpr3 meiotic cells show no induction of gene conversion above mitotic levels. Thus, the zip3 and fpr3 mutations do not permit Zip1 polymerization by suppressing the spo11 defect in DSB formation (Table 1).
Table 1.
Meiotic Induction of Gene Conversion
| Genotype | Hours of Sporulation |
His+ Spores (X 10−5) |
Arg+ Spores (X 10−5) |
|---|---|---|---|
| spo11 | 0 | 7.8 (+/− 1.1) | 1.6 (+/− 2.5) |
| spo11 | 80 | 6.4 (+/− 1.7) | 0.5 (+/− 0.5) |
| spo11 zip3 fpr3 | 0 | 6.5 (+/− 1.2) | 1.7 (+/− 0.2) |
| spo11 zip3 fpr3 | 80 | 4.8 (+/− 1.1) | 0.6 (+/− 0.2) |
Meiotic gene conversion was measured at 0, and 80 hours following introduction of spo11 or spo11 zip3 fpr3 mutant cells into sporulation medium. While gene conversion is induced 100-1000 fold at HIS4 and ARG4 during meiosis in wild-type cells of the BR1919 background [34], spo11 cells experience no induction above mitotic levels. Frequencies of prototrophs are the averages of three independent cultures; standard errors are indicated in parentheses.
Two types of analyses indicate that the absence of Fpr3 and Zip3 allows synapsis to occur in the absence of homologous pairing. First, pairing between homologous centromeres (CENXI) was monitored using LacO arrays in combination with LacI-GFP expressed in trans [20] (Figure 4A). Immunofluoresence on chromosome spreads from the spo11 zip3 fpr3 triple mutant revealed that Zip1 assembles on unpaired CENXI chromosomal regions (Figure 4B). Second, SC assembly was examined in haploid cells, where chromosomes are devoid of pairing partners. Haploid cells forced to undergo sporulation can progress through meiotic prophase and assemble SC [21]. SC assembly in haploids is dependent on Spo11 activity: no Zip1 linear stretches were detected in nuclei from spo11 haploid cells at mid-prophase (n = 57) (Figure 4C, D, E). In contrast, 74% (n = 57) of mid-prophase, haploid nuclei from the spo11 zip3 fpr3 triple mutant exhibited between one and eight distinct Zip1 linear stretches per nucleus, and up to 12 microns of linear Zip1 per nucleus (Figure 4C, D, E). Thus, Spo11-independent SC assembly induced by the combined absence of Zip3 and Fpr3 does not require the presence of homologs.
Figure 4.
Zip1 Assembly Occurs on Unpaired Chromosomes and Initiates at Centromeres in the spo11 zip3 fpr3 Triple Mutant
(A, B) Meiotic nuclei carrying a LacO array inserted at centromere XI (CENXI) were labeled to assess the colocalization of Zip1 (green) with homologous CENXI regions (blue, arrows and arrowheads). All centromeres (CEN) were labeled using Ctf19-myc (red). Arrows indicate cases where CENXI is incorporated into a Zip1 linear segment, while arrowheads indicate centromeres unassociated with Zip1.
(A) Two CENXI foci associate with a Zip1 stretch in wild type, indicating homologous synapsis. Homologously synapsed CENXI signals were sometimes fused, and sometimes unfused (as in this example).
(B) Meiotic nuclei from spo11 zip3 fpr3 triple mutants frequently exhibited a Zip1 stretch that encompassed an unpaired CENXI focus.
(C) Haploid meiotic nuclei from (MATa/MATα) spo11 or spo11 zip3 fpr3 cells were labeled with DAPI (blue) and anti-Zip1 (green) antibodies 20 hours after induction of meiosis.
(D, E) Extent of Zip1 polymerization in haploid meiosis is depicted on the two scatterplots. Nuclei for each strain were plotted as described in Figure 3G and H. The fraction of nuclei exhibiting one or more Zip1 linear segments was 0/57 for spo11, 9/79 for spo11 fpr3, 19/53 for spo11 zip3, and 42/57 for spo11 zip3 fpr3 haploid cells.
(F) Zip1 (green) and CENs (Ctf19, red) were labeled on meiotic chromosome spreads from spo11 zip3 fpr3 diploids and haploids. Bar, 1 micron.
(G) The fraction of short Zip1 stretches (0.35-0.65 microns in length, representing the most recent synapsis initiations) that are centromere-associated. 71, 60 and 64 short Zip1 stretches were analyzed for spo11 zip3 fpr3 (2n), spo11 zip3 (1n), and spo11 zip3 fpr3 (1n), respectively.
Taken together, these observations suggest that Fpr3 and Zip3 function in parallel to prevent inappropriate SC assembly on chromosomes.
Synapsis Initiates Predominantly at Centromeres in spo11 zip3 fpr3 Meiotic Cells
More than half of SC initiations occur at centromere regions [20, 22]. In a spo11 mutant, the Zip1 protein localizes specifically to centromeres, raising the possibility that the inappropriate synapsis exhibited by spo11 zip3 fpr3 meiotic nuclei initiates preferentially at centromeric locations. To address this question, we co-labeled Zip1 and the centromere protein, Ctf19, on spread meiotic chromosomes (Figure 4F). We then selected Zip1 stretches that had, based on their length, recently initiated synapsis (see Supplemental Experimental Procedures) and measured the fraction associated with a centromere. We found that over 90% of short Zip1 stretches in spo11 zip3 fpr3 meiotic nuclei (diploid or haploid) are centromere associated (Figure 4G). Thus, in the absence of recombination initiation, Zip3 and Fpr3 prevent SC assembly at centromeric synapsis initiation sites.
Zip3 and Fpr3 Prevent Synapsis Initiation at Centromeres in Recombination-Proficient Cells
Since Fpr3 and Zip3 prevent SC assembly from initiating at centromeres in spo11 meiotic cells, we wondered whether they also negatively regulate synapsis in recombination-proficient (SPO11+) cells. We quantified synapsis in wild type, fpr3, zip3 and fpr3 zip3 mutants by measuring the cumulative length of Zip1 and by counting the number of Zip1 stretches exhibited by meiotic nuclei at all stages of synapsis. As reported previously [22], the cumulative length of Zip1 is significantly lower, and synapsis initiations are enriched at centromeres, in nuclei from the zip3 mutant compared to wild type (Figure 5A, C). Analysis of the fpr3 single mutant indicated that Fpr3 does not regulate either the kinetics or centromere association of synapsis events in otherwise wild-type meiotic nuclei (Figure 5B-D). On the other hand, fpr3 zip3 double mutant cells exhibit cumulative lengths of Zip1 per nucleus that are not significantly different from wild-type nuclei scored at similar time points (Figure 5C), as if Fpr3 mediates the synapsis delay observed in the zip3 mutant. Although fpr3 suppresses the zip3 defect in synapsis, it does not suppress the defects in meiotic progression and spore viability (Figure S2).
Closer analysis revealed that the fpr3 mutation does not precisely rescue synapsis in zip3 mutants: fpr3 zip3 double mutant cells maintain a deficit in non-centromeric synapsis initiations. While the number of individual Zip1 stretches per nucleus in fpr3 zip3 double mutants is higher, on average, than in zip3 single mutants, it nevertheless rarely exceeds 16, as expected for a single synapsis initiation event per chromosome pair (Figure 5B), and 90% of short Zip1 stretches are centromere-associated (Figure 5D). These observations indicate that Fpr3 activity prevents a large fraction of centromeric synapsis initiation events in zip3 mutants, and are consistent with the idea that Fpr3 and Zip3, in parallel, regulate SC assembly at centromeres in recombination-proficient cells.
Does Fpr3 influence SC assembly in other mutants that exhibit a synapsis delay? DMC1 encodes a recombinase required for strand exchange during meiosis [23, 24]. In dmc1 mutants, meiotic recombination is initiated, but not completed properly; synapsis is initiated, but accumulation of chromosome-length Zip1 stretches is delayed [23, 24]. Consistent with published results [16], we found no role for Fpr3 in preventing SC assembly in dmc1 meiotic nuclei; furthermore, fpr3 does not change the fraction of SC initiation events that are centromere associated (Supplementary Data, Figure S1, compare dmc1 to dmc1 fpr3).
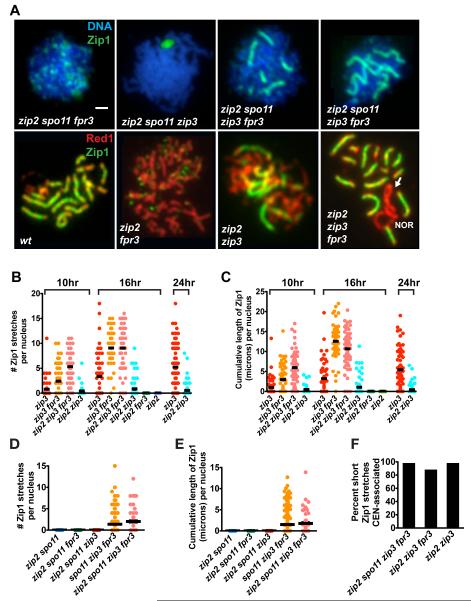
Zip2 and Zip4 are Dispensable for Synapsis in the zip3 fpr3 Mutant
In contrast to Zip3, Zip2 and Zip4 functions are normally essential for all synapsis initiations [9, 13]. It was therefore surprising to find that SC assembly occurs independently of Zip2 (Figure 6A-E) and Zip4 (not shown) activities in zip3 fpr3 mutants. Strikingly, the fraction of meiotic nuclei exhibiting Zip1 linear stretches and the overall extent of Zip1 polymerization is indistinguishable between zip3 fpr3 and zip2 zip3 fpr3 cells (Figure 6B, C). Moreover, the extent of Zip1 polymerization is similar between spo11 zip3 fpr3 cells with, or without, Zip2 (Figure 6D, E). Synapsis initiations in both spo11 zip3 fpr3 and zip3 fpr3 meiotic cells missing Zip2 activity are almost exclusively centromere-associated (Figure 6F). Thus, while Zip2 (and Zip4) functions are essential for synapsis initiation during “normal” meiosis, the synapsis that occurs when Zip3 and Fpr3 are both absent does not require Zip2 or Zip4. Even though Zip2 is not required for synapsis in zip3 fpr3, the Zip2 protein does localize to chromosomes to an extent similar to that observed in zip3 (Figure S3).
Figure 6.
Synapsis from Centromeres in zip3 fpr3 Cells is Zip2-Independent
(A) Upper panels show DNA (blue) and Zip1 (green) on spread meiotic nuclei from spo11 mutants. Lower panels show meiotic cores (anti-Red1, red) and anti-Zip1 (green) in spread meiotic nuclei from SPO11+ strains. Arrow points to a zip2 zip3 fpr3 chromosome XII pair with a Zip1-deficient (unsynapsed) region, adjacent to the nucleolar organizing region (NOR). As SC does not span the NOR, synapsis on one side of this chromosomal region would require synapsis initiation at a non-centromeric site. Bar, 1 micron.
(B-E) Scatterplots show the rate and extent of synapsis in zip2 zip3 fpr3 combinations in SPO11+(B and C), or spo11 (D and E) backgrounds. Nuclei were plotted as described in Figure 3G and H; at least 50 nuclei were plotted for each strain. Data shown in (B and C) belongs to a large time course data set, which also includes the data shown in Figure 5; note that wild-type and fpr3 data (shown in Figure 5) are omitted here. At 16 hours, zip2 zip3 fpr3 cells exhibit significantly higher cumulative levels of Zip1 as compared with zip3 cells (two-tailed Mann Whitney p < 0.0001); no significant differences in the cumulative length of Zip1 are apparent between zip2 zip3 fpr3 and either zip3 fpr3 or wild-type strains.
(F) Short Zip1stretches (0.35-0.65 microns in length) exhibited by zip2 zip3 fpr3 nuclei are almost exclusively centromere-associated. 42, 207, and 30 short Zip1 stretches were analyzed for zip2 spo11 zip3 fpr3, zip2 zip3 fpr3, and zip2 zip3 cells, respectively.
Discussion
Separate Pathways Prevent Inappropriate Synapsis at Centromeres
We began this study with the speculation that yeast cells have a mechanism that prevents SC assembly in the absence of meiotic recombination. Our characterization of two genes involved in regulating Zip1 polymerization indicates that yeast cells, indeed, utilize multiple pathways to suppress inappropriate synapsis.
Fpr3 and Zip3 together prevent Zip1 polymerization from centromeric chromosomal regions in spo11 mutants. In fact, the two genes appear to function redundantly in the sense that the absence of either gene alone has little effect on synapsis, while missing both functions leads to quite dramatic SC assembly. However, a couple of observations suggest that Fpr3 and Zip3 play separate molecular roles in regulating synapsis. First, the spatial distributions of Fpr3 and Zip3 suggest that these factors regulate Zip1 polymerization at different sub-cellular locations. Fpr3 localizes predominantly to the meiotic nucleoplasm, whereas Zip3 localizes to centromeres early in meiotic prophase [22]. Second, the fpr3 and zip3 mutations have opposing effects on polycomplex formation, with fpr3 mutation reducing polycomplex formation and zip3 mutation enhancing polycomplex assembly. Finally, the fact that the fpr3 mutation modulates the synapsis phenotype of zip3 mutants suggests that Fpr3 controls Zip1, at least in part, independently of Zip3 activity.
We speculate that, prior to recombination initiation, nucleoplasmic Fpr3 forces Zip1 to be in an “inactive” state, while Zip3 acts at centromeric sites, perhaps on chromatin proteins themselves or alternatively on Zip1, to prevent SC assembly (Figure S3). Signals downstream of recombination initiation then oppose Fpr3 activity, to increase the pool of “active” Zip1. Spo11 signaling, directly or indirectly, also triggers SIC proteins, such as Zip2 and Zip4, to localize to centromeres and counteract the barriers to synapsis previously established by Fpr3 and Zip3.
The Role of Fpr3 in Regulating Zip1 Polymerization
The Fpr3 proline isomerase has already been implicated in a role downstream of Spo11 signaling. Hochwagen et al. [16] showed that Fpr3 is required to maintain a checkpoint response triggered by a meiotic recombination defect. Here, we have identified a novel meiotic role for Fpr3 in regulating SC assembly when recombination initiation has failed. In such a regulatory capacity, Fpr3 does not control a true cellular checkpoint, because spo11 cells do not exhibit cell cycle arrest. However, Fpr3 may act in a checkpoint-like manner, to ensure that certain chromosomal events (i.e., SC assembly) are contingent upon the successful completion of earlier events (i.e., early recombination events).
Proline isomerases have been shown to have chaperone-like activity that can affect the enzymatic activity of target proteins [16, 25-27]. Hochwagen et al. [16] demonstrated that Fpr3 mediates a component of the meiotic recombination checkpoint via inhibition of Glc7 phosphatase activity. These authors showed that Glc7 overexpression bypasses the cell cycle delay in dmc1 cells, similar to the bypass elicited by the fpr3 mutation [16]. In contrast, a transgene overexpressing GLC7 during meiosis does not phenocopy the fpr3 mutation with respect to SC assembly on chromosomes (data not shown), suggesting that Glc7 does not promote Zip1 polymerization. Nevertheless, we imagine that Fpr3 targets an enzyme that, in turn, regulates a post-translational modification of Zip1 or another SC component. Alternatively, perhaps Fpr3 acts directly on Zip1 protein folding, to influence the capacity of Zip1 to assemble SC central region.
Zip3 Has Opposing Roles in SC Assembly
The fact that Zip3 plays a negative role in the regulation of Zip1 assembly is surprising, since it was previously implicated in promoting synapsis [8]. One way to reconcile these opposing roles is by postulating that Zip3 inhibits Zip1 polymerization specifically at centromeres, where it is known to be dispensable for promoting synapsis [22].
Zip3 protein has been found to have SUMO E3 ligase activity [18] (although it is not the only E3 ligase active in meiosis [18, 19]). As sumoylation has a broad range of protein regulatory roles [28], one can imagine that Zip3 carries out its positive and negative roles through controlling the sumoylation of different protein targets. Zip3 might regulate sumoylation of SC proteins, chromosomal axis components, or centromeric proteins to either encourage or discourage Zip1 stability and/or Zip1’s capacity to polymerize on meiotic chromatin.
A Central Role for Centromeres in Homologous Synapsis?
In C. elegans, where recombination is dispensable for homolog pairing and synapsis, a single cis-acting site on each chromosome, called the Pairing Center, appears to coordinate homologous pairing with SC assembly [6]. In yeast, recombination may be mechanistically required for homologous pairing, but our observations suggest that it is not required for synapsis. At least early on in meiosis, most synapsis initiates at centromeric sites, where recombination is not likely to occur [22, 29, 30]. Moreover, here, we demonstrate that recombination need not occur anywhere in the genome in order for synapsis to initiate at centromeres. Nevertheless, the earliest synapsis events in yeast, which serve to reinforce chromosome partner choice, overwhelmingly occur at centromeres. Thus, we wonder whether a singular site on each yeast chromosome (the centromere) is a specialized locale, analogous to C. elegans Pairing Center sites, where homologous recognition (pairing) and the reinforcement of paired associations (synapsis) are coordinated. If so, it appears that this coordination involves inhibitory roles of Fpr3 and Zip3 at centromeres, together with counteracting, positive signals downstream of recombination/pairing (presumably involving the Zip2 and Zip4 proteins).
To date, a role for yeast centromeres in ensuring homologous synapsis has not been demonstrated. We have found little evidence for nonhomologous synapsis in the zip3 fpr3 double mutant (Supplementary Data, Figure S5), which, according to our model, is missing a significant portion of the machinery needed to prevent inappropriate synapsis at centromeres. Perhaps yeast and worms share a similar mechanism for homologous synapsis, but yeast cells have evolved to rely on it to a lesser degree. It follows from this argument that meiotic contexts in which alternative potential pairing strategies, such as recombination, are disrupted might result in an increased reliance on the centromeric synapsis machinery in yeast.
Experimental Procedures
Strains
Yeast genetic manipulations were carried out using standard procedures. Except for the wild-type SK1 strain [31], all strains used in this study are diploids in which both haploid parents are isogenic with BR1919-8B [24]. Genetic crosses were used to construct strains carrying multiple mutations. See Supplemental Experimental Procedures for additional strain information.
Western Blot Analysis
Meiotic cell extracts were prepared as described previously [8]. Beads carrying immunoprecipitated Zip1 protein (see Supplemental Experimental Procedures) were boiled in NuPAGE LDS sample buffer (Invitrogen) and eluates were separated in 4-12% Bis-Tris polyacrylamide gels (Invitrogen), using 1X MOPS running buffer (Invitrogen). After transfer, PVDF membranes were incubated with polyclonal rabbit anti-Zip1 [12] and rat anti-tubulin (Sera-Lab, West Sussex, UK) antibodies overnight at 4°. Detection of primary antibodies was carried out using alkaline-phosphatase-conjugated secondary antibodies (Jackson Immunoresearch).
Cytology
Semi-squash preparations (Figure 1B) were performed as described by Fuchs and Loidl [32]. Meiotic chromosome spreads were carried out as described by Agarwal and Roeder [8]. FISH in conjunction with immunostaining was performed as described previously [9]. Additional information on cytology and imaging are provided in Supplemental Experimental Procedures.
Statistical Analysis
Fisher’s Exact Test was used for statistical analysis of polycomplex formation data in Figure 1 and homologous pairing data in Figure S2. The Mann-Whitney Test was used for statistical analysis of scatterplot data in Figures 5 and 6. Tests were performed using InStat3 and Prism software (www.Graphpad.com).
Supplementary Material
Acknowledgements
We thank Michael O’Dell for technical assistance, and B. Rockmill and T. Tsubouchi for thoughtful discussion.
Funding Supported by the Helen Hay Whitney Foundation (A.J.M), NIH grant K99 GM084293-01 (A.J.M.) and the Howard Hughes Medical Institute.
Footnotes
Supplementary Data Five figures, six movies and additional Experimental Procedures are provided in a supplemental file.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 2.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 3.Dresser ME, Ewing DJ, Harwell SN, Coody D, Conrad MN. Nonhomologous synapsis and reduced crossing over in a heterozygous paracentric inversion in Saccharomyces cerevisiae. Genetics. 1994;138:633–647. doi: 10.1093/genetics/138.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillies CB. The effect of Ph gene alleles on synaptonemal complex formation in Triticum aestivum × T. kotschyi hybrids. Theor. Appl. Gen. 1987;74:430–438. doi: 10.1007/BF00289817. [DOI] [PubMed] [Google Scholar]
- 5.Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 6.MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 9.Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- 10.Perry M, Kleckner N, Borner GV. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17594–17599. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinohara M, Oh SD, Hunter N, Shinohara A. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 2008;40:299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- 12.Sym M, Engebrecht J, Roeder GS. Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 13.Tsubouchi T, Zhao H, Roeder GS. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev. Cell. 2006;10:809–819. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 15.White EJ, Cowan C, Cande WZ, Kaback DB. In vivo analysis of synaptonemal complex formation during yeast meiosis. Genetics. 2004;167:51–63. doi: 10.1534/genetics.167.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochwagen A, Tham W-H, Brar GA, Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Benton BM, Zang JH, Thorner J. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae is a proline rotamase localized to the nucleolus. J. Cell Biol. 1994;127:623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng C-H, Lo Y-H, Liang S-S, Ti S-C, Lin F-M, Yeh C-H, Huang H-Y, Wang T-F. SUMO modification control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooker GW, Roeder GS. A role for SUMO in meiotic chromosome synapsis. Curr. Biol. 2006;16:1238–1243. doi: 10.1016/j.cub.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 20.Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- 21.Loidl J, Nairz K, Klein F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 1991;100:221–228. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- 22.Tsubouchi T, Macqueen AJ, Roeder GS. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 2008;22:3217–3226. doi: 10.1101/gad.1709408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 24.Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- 25.Jordens J, Janssens V, Longin S, Stevens I, Martens E, Bultynck G, Engelborghs Y, Lescrinier E, Waelkens E, Goris J, Van Hoof C. The protein phosphatase 2A phosphatase activator is a novel peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem. 2006;281:6349–6357. doi: 10.1074/jbc.M507760200. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda S, Koyasu S. Regulation of MAPK signaling pathways through immunophilin-ligand complex. Curr. Top. Med. Chem. 2003;3:1358–1367. doi: 10.2174/1568026033451916. [DOI] [PubMed] [Google Scholar]
- 27.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 29.Lambie EJ, Roeder GS. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs J, Loidl J. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 2004;12:427–438. doi: 10.1023/B:CHRO.0000034726.05374.db. [DOI] [PubMed] [Google Scholar]
- 33.Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rockmill B, Fung JC, Branda SS, Roeder GS. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 2003;13:1954–1962. doi: 10.1016/j.cub.2003.10.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.