Abstract
Genetic studies in Drosophila melanogaster have revealed that IAP (Inhibitor of Apoptosis) proteins and IAP antagonists such as reaper play a pivotal role in controlling cell death in insects. Interestingly, while the sequences and structures of IAPs are highly conserved, the sequence of IAP antagonists diverged very rapidly during evolution, making their identification difficult. Using a customized bioinformatics approach, we identified an IAP antagonist, Ibm1, from the genome of the silkworm Bombyx mori. This is the first reaper/grim ortholog identified in a non-Dipteran insect. Previous analysis indicated that both Reaper and Grim induce cell death through their N-terminal IAP-binding motif (IBM) as well as the Grim_helix3 (GH3) domain. Functional studies indicated that Ibm1 binds to an IAP protein from Bombyx mori, BmIAP1, and induces apoptosis in insect cells via the IAP-binding motif, a 7 amino acid sequence that is highly conserved in all IAP antagonists. Interestingly, Ibm1 also contains a region that is a statistically significant match to the GH3 domain. Mutational analysis indicated that the GH3-like motif in Ibm1 has an important supportive role in IAP-antagonist function and can trigger cell death under certain conditions.
Introduction
Programmed cell death, often through the morphological ritual termed as apoptosis, is a fundamental biological process in all metazoans. In Drosophila melanogaster, the IAPs and IAP antagonists play a pivotal role in regulating apoptosis during normal development as well as in response to cytotoxic stimuli. IAPs were initially identified in insect viruses and later found in insects as well as mammals (Clem and Miller 1994; Hay et al. 1995; Verhagen et al. 2001). In Drosophila embryos mutated for diap1, the main caspase-inhibiting IAP in Drosophila, essentially all cells undergo apoptosis when the maternally deposited Diap1 is depleted (Wang et al. 1999; Goyal et al. 2000). In addition, silencing of diap1 expression in Drosophila cell lines results in rapid apoptosis (Igaki et al. 2002; Muro et al. 2002; Zimmermann et al. 2002). All IAPs contain one to three BIR (Baculovirus IAP Repeat) domains, which, in the case of certain IAPs, bind to and inhibit caspases. Some IAPs, such as Diap1, also contain a RING domain, which confers E3 ubiquitin ligase activity. The detailed functional mechanism of IAPs has been the subject of intense research effort in the last decade and was extensively reviewed recently (Ditzel and Meier 2005; Vaux and Silke 2005).
During development, diap1 is ubiquitously expressed in the embryo. The selection of specific cells to undergo apoptosis during development is achieved largely through cell-autonomous expression of IAP antagonists, namely reaper, hid, grim, and sickle. Interestingly, the four IAP antagonists in Drosophila melanogaster reside in the same chromosomal region spanning about 350 kb. A deletion that removes reaper, hid, and grim essentially blocks all developmental cell death and impedes irradiation induced cell death (White et al. 1994). Reaper, grim, and sickle are almost exclusively expressed in cells destined to die (White et al. 1994; Chen et al. 1996; Christich et al. 2002; Srinivasula et al. 2002; Wing et al. 2002). However, hid mRNA can be found in cells that do not undergo apoptosis. This may due to that fact that the pro-apoptotic function of Hid is subjected to phosphorylation and suppression by the MAP kinases (Bergmann et al. 1998). The only sequence motif shared by the four IAP antagonists is the 7-aa IAP-binding motif (IBM) at their N-termini. This heptapeptide motif specifically binds to a surface groove in the BIR domain of Diap1 and thus releases its inhibition of caspases (Chai et al. 2003).
Both caspases and IAPs are highly conserved at the sequence level. Typically when a genome is sequenced, caspases and IAPs can be predicted with considerable confidence using routine sequence analysis approaches (Waterhouse et al. 2007; Bryant et al. 2008). For instance, the sequence of the Anopheles gambiae genome predicted a significant increase in the number of caspases as well as IAPs as compared to the Drosophila melanogaster genome (Christophides et al. 2002). The specific expansion of Iap1 in A. gambiae indicated this pathway must be conserved, and likely extended for developmental processes as well as immunoresponse in the mosquitoes (Christophides et al. 2002). However, no IAP antagonist was identified in the Anopheles genome by the genome project, which reflects the fact that these genes have significantly diverged during evolution and often evade detection by routine sequence similarity searches. Using a customized search strategy and biochemical verification, we identified michelob_x (mx) as the missing IAP antagonist in several mosquito genomes (Zhou et al. 2005). The only significant sequence similarity between MX and Drosophila IAP antagonists is the conservation of the IAP-binding motif at their N-termini. Despite the very low level of overall sequence similarity, Mx can bind to Diap1 via its IAP-binding motif and induce cell death in Drosophila cells. Interestingly, similar to what was observed for reaper, expression of mx is also induced/up-regulated immediately following irradiation (Zhou et al. 2005).
One major difference between Mx and Reaper/Grim is the absence of a C-terminal pro-apoptotic domain. Both Reaper and Grim have remaining cell death inducing activity, albeit reduced, even when their N-terminal IAP-binding motifs are removed (Vucic et al. 1997; Wing et al. 1998). The pro-apoptotic activity of the IBM-less Reaper/Grim has been attributed to the GH3 (Grim Helix 3) domain that is shared between Reaper and Grim (Claveria et al. 2002). Expression of the GH3 domain induces cell death in mammalian cells through interacting with mitochondria and causing the release of cytochrome C (Claveria et al. 2002; Abdelwahid et al. 2007). Mx lacks a clearly discernable GH3 domain at the sequence level. Functional analysis also revealed that an Mx mutant without the IAP-binding motif has no detectable pro-apoptotic activity when expressed in Drosophila S2 or mosquito C6/36 cell lines (Zhou et al. 2005). This discrepancy between Mx and Reaper/Grim could, at a nominal level, be the result of a loss of the GH3 domain in mosquitoes, or alternatively, a joining of GH3 and IBM in Drosophilidae. Domain shuffling is fully possible when only mosquito and Drosophila are concerned, since mx in both Aedes and Anopheles have an intron while reaper and grim are single-exon genes. An ortholog from a non-Dipteran species, such as one from Bombyx mori, a Lepidopteran, would offer more insight into understanding the evolution of the IAP-antagonists and the functional role of the GH3 domain in IAP antagonist-induced cell death.
It is also interesting to note that IAPs were originally identified in baculoviruses that infect Lepidoptera (Crook et al. 1993). It has been shown that the ability of these viruses to inhibit apoptosis is essential for their virulence (Clarke and Clem 2003). Conversely, this suggests that the IAP antagonist/IAP pathway is important for the organism to fend off virus infection. However, testing of such a hypothesis has been hindered by the lack of an identified IAP antagonist in a Lepidopteran species. Here, we have cloned and characterized the first Reaper ortholog, IAP-binding motif 1 (Ibm1), from the genome of the silkworm Bombyx mori. Structural and functional information obtained from this study indicated that the IAP-binding motif plays an essential role in the pro-apoptotic activity of Ibm1. The conservation of a sequence motif similar to the GH3 domain indicated that this motif also has an important role in IAP-antagonist function but has been subject to more evolutionary changes.
Results
Identification and cloning of an IAP antagonist in silkworm Bombyx mori
Using a bioinformatics strategy detailed previously (Zhou et al. 2005), we searched the silkworm genomic sequences for predicted open reading frames that contain either the IAP-binding motif or the GH3 motifs. The model for IBM was built using identified Drosophila and mosquito IAP antagonists. Since no GH3 domain has been verified in mosquito IAP antagonists, the model for GH3 was built with Reaper/Grim/Sickle orthologs from D. melanogaster, D. pseudoobscura, and D. virilis (Zhou 2005). From the search, no potential hit was found to have both IBM and GH3 motifs. The gene providing the best match for the IAP-binding motif was designated as IAP-binding motif 1 (Ibm1).
A cDNA encompassing the predicted ORF was first cloned via RT-PCR using mRNA extracted from silkworm pupae and larvae. Northern analysis indicated a transcript of about 1 kb in length (data not shown). The status of Ibm1 as an expressed gene was independently confirmed when two ESTs containing this sequence were released by the Silkworm Genome Research Program (BY922187, BY914780), both containing the complete ORF of 93 amino acids and a putative polyA tail. Remarkably, as with repear/grim/sickle in Drosophila, Ibm1 in Bombyx has only one exon. In contrast, michelob_x in both Aedes and Anopheles mosquitoes have an intron in the same position relative to their protein sequences (Zhou et al. 2005). The fact that Ibm1 is a single exon gene indicates that the intron observed in mosquito mx most likely evolved after the separation of the Lepidoptera and the Diptera, but before the divergence of the subfamilies Anophelinae and Culicinae (including Aedes). Global alignment of the identified Reaper/Grim–like IAP antagonists indicated that overall, the similarity beyond the IAP-binding motif region is low among these proteins. However, the C-terminal end of the Mx sequence after its intron has a clear resemblance to the C-terminal end of Ibm1 (Figure 1A), suggesting that the altered gene structure in mx is more likely due to an insertion of an intron rather than an addition of an exon.
Figure 1. Evolution of IAP antagonists.
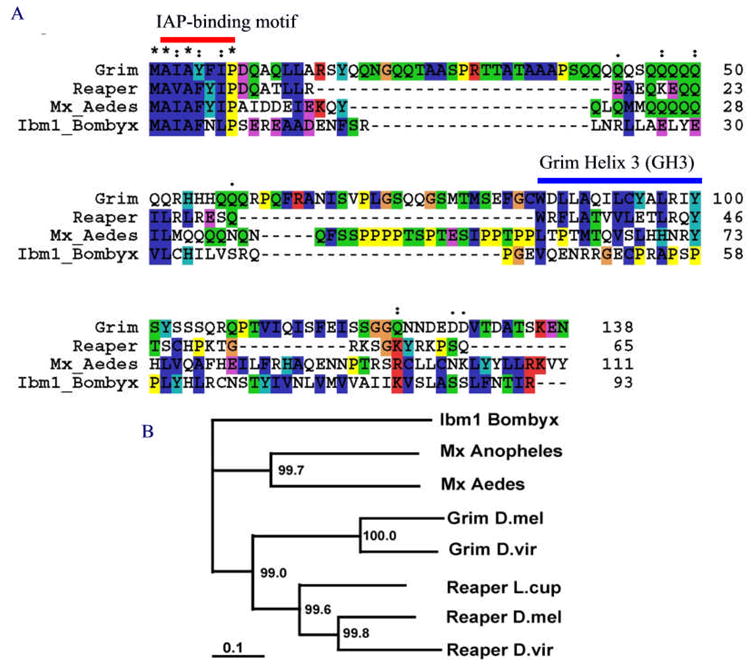
(A) Global alignment of Grim (D. melanogaster), Reaper (D. melanogaster), Mx (Aedes aegypti), and Ibm1 (B. mori). The IAP-binding motif is indicated by the red bar. The GH3 domain previously identified in Grim and Reaper is indicated by the blue bar on top of the Grim sequence. The GH3-like motifs in Mx and Ibm1 sequences are not aligned with the GH3 region in Reaper and Grim. Note that while there is a very strong conservation of the IAP-binding motif in all of the IAP-antagonists, the conservation of other regions, including the GH3 region, is minimal. (B) A distance tree of seven IAP-antagonists from D. melanogaster (D.mel), D. virilis (D. viri), Aedes aegypti (Aedes), Anopheles gambiae (Anopheles), and Bombyx mori (Bombyx). The numbers at each branch are the boot-strapping values (% confirmation). The scale bar represents 0.1 substitution per position.
When a neighbor-joining distance tree of the orthologs was constructed based on global alignment, it was obvious that it reflects the evolutionary history of the species (Figure 1B). It is rather interesting to note that the distance between Grim and Reaper is shorter than their relative distance to Mx or Ibm1. This raises the possibility that grim and reaper in Drosophila arose from the duplication of an ancestral gene that also gave rise to mx in mosquitoes and Ibm1 in moths. This duplication likely happened before the separation of Acalyptratae (including the fruit fly) and Calyptratae. A previously identified Reaper ortholog in Lucilia cuprina (Chen et al. 2004) is clearly in the same clade as Reapers from D. melanogester and D. virilis (Figure 1B).
Despite overall low similarity, Reaper and Grim share two motifs, the IAP-binding motif and the GH3. The GH3 regions of Reaper and Grim are quite similar to each other in structure and have been shown to possess similar function (Figure 1A) (Claveria et al. 2002; Olson et al. 2003; Claveria et al. 2004). Another conserved feature among the orthologs from different insect orders is the enrichment of basic residues, especially lysine, toward the very C-terminal end of the protein. This is consistent with ubiquitination of Reaper/Grim-like IAP-antagonists being a fundamental regulatory mechanism for the interaction between IAP and IAP antagonists (Olson et al. 2003).
Conservation of protein motifs
While global alignment of protein sequences helps to measure the relative distance among orthologs, the drawback of such an alignment is that it can overlook the conservation of functional motifs if their relative positions in the sequences have shifted during evolution. Since protein functions are mainly carried out by individual domain(s), it is possible that some proteins can tolerate many insertion/deletion events between (among) domains but still remain functional. To analyze the conservation of functional domains regardless of their relative positions in the protein, we searched for shared protein motifs among the identified IAP-antagonists using the MEME (Multiple EM for Motif Elicitation) methodology (Bailey and Elkan 1994).
A total of seven sequences, including Reaper, Hid, Grim, and Sickle from D. melanogaster, Mx from Aedes aegypti (Mx_Aedes) and Anopheles gambiae (MX_Anopheles), and Ibm1 from Bombyx mori (Ibm1_Bombyx) were fed into the program for identification of shared sequence motifs. The program was instructed that a shared motif may or may not be present in all of the sequences, thus whether a motif is reported for a particular sequence is based on unbiased statistical analysis (Bailey and Gribskov 1998). As shown in Figure 2, two motifs were identified as being shared by more than three of the proteins. Not surprisingly, all of the seven proteins were found to share a motif at their N-termini, which encompasses the IAP-binding motif (Figure 2A). The IAP-binding motif from Mx and Ibm1 is almost indistinguishable from that of Reaper or Grim. However, it was somewhat unexpected to see that both Mx and Ibm1 contain a region that is significantly similar to the identified GH3 region in Reaper and Grim (Figure 2B).
Figure 2. Motif analysis of IAP antagonists.
Seven IAP antagonist sequences were subjected to detection of shared sequence motifs using MEME. The program was run in the Zero or One Occurrence Per Sequence (zoops) mode. (A) All of the seven sequences were detected as having a shared motif at their N-terminal end, which corresponds to the IAP-binding motif. (B) Six of the seven sequences, but not Hid, were detected as having a second shared motif, which corresponds to the GH3 domain identified in Grim and Reaper. For both A & B, the number in front of each sequence indicates the starting amino acid position of the motif in the corresponding protein. The number after the sequence indicates the p value reported by the program as the possibility to identify such a match in random sequences. (C) A helix wheel drawing of the second shared motif detected in Ibm1 (aa 21–35) indicates that it has the potential to form an amphipathic helix, similar to the GH3 motifs in Grim and Reaper.
The second motif identified by the program encompassed 15 amino acids. The identified region matched exactly with the GH3 domains of Reaper and Grim, which have been functionally characterized as having independent pro-apoptotic function (Claveria et al. 2002). This region is also part of the R3 region in Reaper that has been shown by Chen et al as having pro-apoptotic activity when fused with GFP (Chen et al. 2004). Both Mx and Ibm1 have a statistically significant match of this motif (Figure 2B). In contrast, no significant match was found in the Hid sequence that was evaluated at the same time. GH3 regions in both Reaper and Grim were predicted to form an amphipathic helix structure. The corresponding region in Ibm1, amino acids 21–35, also was predicted to form an amphipathic helix (Figure 2C), further indicating that Ibm1 is an ortholog of Reaper/Grim.
However, as was stated above, when we searched sequences with a GH3 model built with Reaper/Grim/Sickle sequences from 3 Drosophila species we were not able to identify Ibm1 as having a GH3 domain. This is probably because we have over-trained the model so that it became too restrictive, i.e. it has very high specificity for GH3 domains in Drosophila Reaper and Grim but consequentially has low sensitivity for identifying distant matches such as the one in Ibm1. In all of the Drosophila sequences that we used to build the GH3 model, the first amino acid of the domain was invariably Tryptophan (W) (Zhou 2005) (supplementary information), however, that was not the case for the motif identified in Mx or Ibm1. In addition, several other positions in the GH3 domain of Reaper/Grim, such as Alanine at position 5, are also highly conserved among the Drosophila orthologs but were not conserved in the corresponding region in Mx and Ibm1 (Figure 2B).
The main feature of the second motif is the conservation of hydrophobic amino acids in positions 4, 8, 12, and 15 of the motif (Figure 2B). Position 4 corresponds to Grim L89 and Reaper L35, which have been shown to be the most important position for GH3 pro-apoptotic function. Mutation of Grim L89 or Reaper L35 to a hydrophilic amino acid abolished most of the pro-apoptotic activity of GH3 (Claveria et al. 2002; Olson et al. 2003; Claveria et al. 2004). Notably, leucine is conserved at this position in all of the proteins identified as having a potential GH3 domain, including Ibm1 (Figure 2B).
The IAP-binding motif is essential for the pro-apoptotic activity of Ibm1
Both Grim and Reaper have been shown to be able to induce cell death even when their IAP-binding motifs were removed (Wing et al. 1998). However, we have previously shown that this is not the case for their mosquito ortholog Mx. In both Drosophila S2 and Aedes albopictus C6/36 cell lines, the pro-apoptotic activity of Mx is totally abolished when its IAP-binding motif is removed (Zhou et al. 2005). Correspondingly, co-transfection of MX and Diap1 at a ratio of 1:2 can totally block the pro-apoptotic activity of Mx in C6/36 cells. In contrast, similar co-transfection, even with a higher ratio of Diap1, fails to completely block the pro-apoptotic activity of Grim or Reaper in S2 or C6/36 cells (Zhou et al. 2005). All of these data indicated that, unlike Reaper and Grim, Mx does not have a C-terminal pro-apoptotic domain that can function independently of its IAP-binding motif.
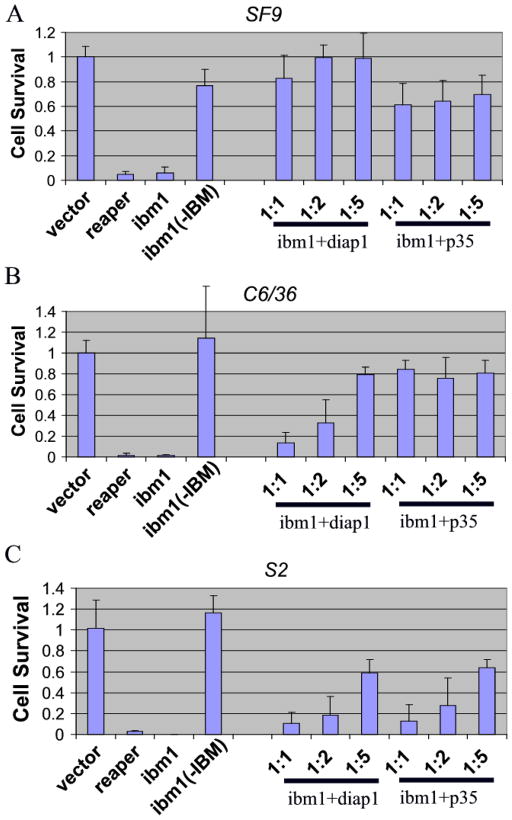
Expression of Ibm1 in the insect cell lines SF9 (Spodoptera), C6/36 (Aedes), and S2 (Drosophila) induced cell death in a fashion very similar to that observed for Reaper and Grim (Figure 3). Since the first Alanine of the IAP-binding motif is absolutely required for interacting with the BIR domain of IAP, we generated a mutated form of Ibm1 that removes amino acids 2–4 (“Ala-Iso-Ala”), which we designated as Ibm1[ΔIBM]. No detectable pro-apoptotic activity was observed when Ibm1[ΔIBM] was expressed in the C6/36 or the S2 cell lines (Figure 3). There seemed to be a difference between Ibm1[ΔIBM] and the vector control in the SF9 cell line, which was marginally significant (p=0.02538, t- test).
Figure 3. Under normal conditions, the pro-apoptotic activity of Ibm1 requires the IAP-binding motif.
Plasmids expressing Ibm1 or Ibm1[Δ2–4] (−IBM) were transfected into Lepidopteran SF9 (A), mosquito C6/36 (B), and Drosophila S2 (C) cell lines. Ibm1 was also cotransfected with increasing ratios of Diap1 or P35-expressing plasmids. Cell survival of vector (pie3)-transfected samples was set as 100% and the relative cell survival rate was calculated for each gene or combination of genes. The results are the average of 3–4 experiments and the error bars reflect standard deviation. pie-reaper was included as a positive control.
The cellular sensitivity to Ibm1-induced cell death varied slightly among the three tested cell lines. The same amount of DNAs were transfected into each cell line. However, while almost all S2 cells were killed by Ibm1, about 10% of transfected SF9 cells survived. The difference in sensitivity was also reflected in the effectiveness of co-tranfected Diap1 and P35 in blocking Ibm1-induced cell death. While 1:1 co-transfection with diap1 essentially blocked all Ibm1-induced cell death in SF9 cells, co-transfection of Ibm1/diap1 at a 1:5 ratio in S2 cells failed to restore cell survival to the control (vector-tranfected) levels (Figure 3).
These results indicated that the IAP-binding motif is essential for the pro-apoptotic activity of Ibm1. Since this motif mediates IAP binding, we examined whether Ibm1 could bind to an IAP protein encoded by Bombyx mori, BmIAP1 (GenBank accession AF281073). FLAG-tagged Ibm1 or Ibm1[ΔIBM] were co-transfected with HA-tagged BmIAP1 into SF21 cells, which were used since we were not able to achieve the necessary transfection efficiency in the BmN cell line from Bombyx mori. Immunoprecipitation using anti-FLAG revealed that Ibm1 bound to BmIAP1, and this interaction depended on the IBM motif (Fig. 4). BmIAP1 was also heavily ubiquitinated when co-expressed with Ibm1, while co-expression with Ibm1[ΔIBM] did not result in ubiquitination, presumably because the proteins could not interact (Fig 4). It should be noted that it was necessary to treat the transfected cells with proteosome and caspase inhibitors in order to detect BmIAP1 protein; the presence of these inhibitors were presumably responsible for the apparent increase in BmIAP1 protein when co-transfected with Ibm1.
Figure 4. Ibm1 interacts with BmIAP1 through its IAP-binding motif (IBM).
(A) Expression of FLAG-tagged Ibm1 and Ibm1[ΔIBM] and HA-tagged BmIAP1 was assessed (either singly or in combination) by western blotting. Note that BmIAP1 is heavily ubiquitinated in the presence of Ibm1 (as indicated by the bracket). (B) Interaction between Ibm1 and BmIAP1 was detected by immunoprecipitation with α-FLAG followed by western blotting with α-HA. A non-specific background band is designated by an asterisk. An arrow indicates band of interest and an asterisk designates a non-specific background band. The upper blot contains immunoprecipitated material, while the lower blot contains the unbound material that remained in the supernatant. Note that BmIAP1 levels are increased when co-expressed with Ibm1.
The C-terminal region of Ibm1 can induce cell death under sensitized conditions
To further explore whether the IBM-less Ibm1 can have pro-apoptotic activity, we tested its killing ability in SF21 cells following heat shock induction of the transgene, which has been shown to sensitize these cells to apoptosis (Hozak et al. 2000). In this system, the Ibm1[ΔIBM] was capable of inducing cell death, albeit with less efficiency (Figure 5A). This IBM-independent cell killing activity was mediated by caspases, as shown by the ability of zVADfmk, a pan-caspase inhibitor, to suppress death, and by an increase in caspase 3-like activity following heat shock induction of Ibm1 or Ibm1[ΔIBM]. This observation indicated that under sensitizing conditions, IBM-less Ibm1 can induce cell death. However, its pro-apoptotic activity is severely compromised without the IBM.
Figure 5. An Ibm1 mutant lacking the IAP-binding motif has a mild pro-apoptotic phenotype under sensitized conditions.
A) SF21 cell viability following transfection of Ibm1 or Ibm1[ΔIBM]. The cells were heat shocked prior to measuring viability, which sensitizes them to apoptosis. B) Caspase activity in SF21 cells transfected with Ibm1 or Ibm1[ΔIBM] was determined by incubation of cell lysate with DEVDafc (Human caspase 3 substrate). Transfections were done in the presence or absence of the pan-caspase inhibitor zVADfmk.
The GH3-like motif has auxiliary function
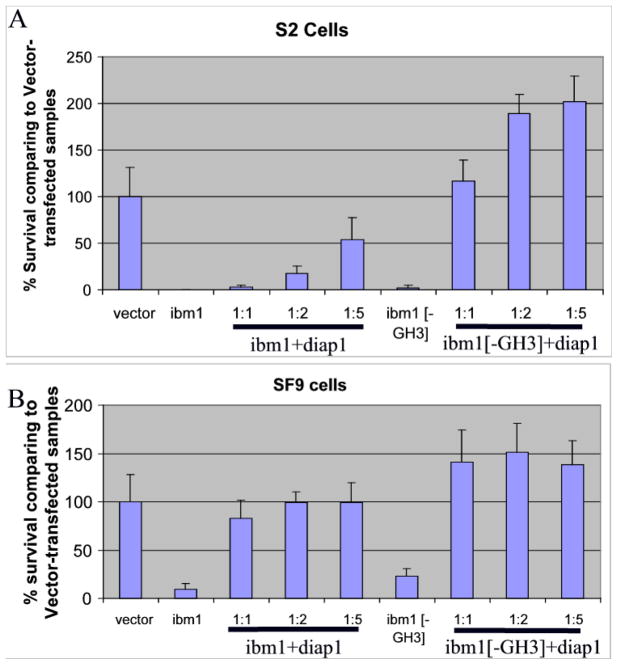
The observation that Ibm1[ΔIBM] has little pro-apoptotic function except under sensitizing conditions seemingly contradicts the motif analysis result showing that the GH3 domain was conserved during evolution (Figure 2B). To further test the functional significance of the GH3-like motif in Ibm1, we removed the region of aa 21–35 of Ibm1 (Ibm1[ΔGH3]) and tested its pro-apoptotic function in insect cell lines. We found that Ibm1[ΔGH3] induced cell death in both SF9 and S2 cells with similar efficiency as the wild type Ibm1 (Figure 6A & B). When transfected alone, there was no statistical difference between Ibm1 or Ibm1[ΔGH3]-induced cell death in either cell line.
Figure 6. Auxiliary role of the GH3-like motif for Ibm1 pro-apoptotic activity in insect cells.
Ibm1[ΔGH3], when transfected alone, induced cell death in S2 (A) and SF9 (B) cells similar to that induced by Ibm1. However, Ibm1[ΔGH3] was much more sensitive to the inhibitory effect of Diap1 in co-transfections.
However, without the GH3-like motif, Ibm1 pro-apoptotic activity was much more sensitive to inhibition by Diap1. In S2 cells, co-transfection of Ibm1 and Diap1 at the ratio of 1:5 only restored cell survival to about 50% of that of vector-transfected cells. However, co-transfection of Ibm1[ΔGH3] and Diap1 at the ratio of 1:1 restored survival to over 100% as compared to the vector-transfected samples. Co-transfection of Ibm1[ΔGH3] and Diap1 at the ratio of 1:2 or 1:5 resulted in a survival rate comparable to that of transfecting Diap1 alone, indicating that the pro-apoptotic activity of Ibm1[ΔGH3] is completely inhibited by the presence of Diap1 (Figure 6A & B).
Taken together, our results with Ibm1[ΔIBM] and Ibm1[ΔGH3] indicate that while the GH3 domain has little independent pro-apoptotic activity, it has an auxiliary function in the pro-apoptotic activity of Ibm1. It has been shown by Olson et al that the GH3 domain in Reaper most likely augments the function of the IAP-binding motif by transporting the protein to mitochondria (Olson et al. 2003).
Expression of Ibm1 is developmentally regulated
All four IAP antagonists in Drosophila are implicated in the regulation of developmental cell death. The expression of reaper and hid is significantly induced by ecdysone during metamorphosis (Jiang et al. 1997), and the increased level of Reaper and Hid is responsible for the massive destruction of larval tissues (Yin and Thummel 2004). To see if Ibm1 is also regulated during metamorphosis, we examined the level of Ibm1 mRNA in larval as well as pupal stages (Figure 7).
Figure 7. Transcriptional regulation of Ibm1 during metamorphosis.
The level of Ibm1 mRNA relative to that of gapdh (A) or actin (B) was determined by quantitative RT-PCR (represent as percent to the control). The day larvae hatched or cocoons appeared was counted as day 1 of larval and pupal stages, respectively. The shortening of larvae was observed on day 27 or 28 of the larva stage. Data are represented as Mean±SE of 3 – 4 QPCR measurements.
The level of Ibm1 expression remained low during all larval stages until just before pupal formation, when the larvae shorten in body length and become less agile in movement. After pupa formation, the level of Ibm1 mRNA continued to rise to very high levels and reached a peak at about 4–8 days post pupal formation. The relative ratio of Ibm1 mRNA to GAPDH mRNA showed a greater than 50-fold increase between the larval stages and day 8 pupae. The change in the relative ratio of Ibm1 to actin was even greater, probably reflecting a decrease in actin expression accompanying the increase of Ibm1 expression in degenerating tissues. This surge in relative concentration of Ibm1 mRNA during pupal stage suggests that, like reaper in Drosophila, Ibm1 is involved in tissue degeneration during metamorphosis.
Discussion
Divergence and conservation of IAP antagonists as pro-apoptotic initiators
The rapid divergence of IAP antagonists in insects is both fascinating and puzzling. The availability of many sequenced animal genomes has revealed that while downstream apoptosis regulators, such as caspases and IAPs, are very well conserved during evolution, the upstream initiators, such as IAP antagonists and BH3-only proteins, diverged amazingly fast. The strong conservation of downstream regulators is not at all surprising given the fundamental importance of apoptosis in animal development and the shared morphological/biochemical changes associated with the dying cell. It is surprising, however, that the most important upstream regulators, such as IAP antagonists in insects and BH3-only proteins in mammals, which control the initiation of cell death during development, actually diverged very quickly. Comparison of Reaper/Grim-like IAP antagonists in insects and BH3-only proteins in mammals reveals several shared properties of these two classes of upstream pro-apoptotic regulators. These include 1) the pro-apoptotic function is carried out by a small peptide motif; 2) sequences other than the pro-apoptotic motif(s) are almost dispensable for pro-apoptotic function and are not very well conserved; and 3) expression is often regulated at the transcription level.
One simple, but possibly superficial, explanation for the fast divergence of pro-apoptotic initiators is that there are multiple paralogs in each genome and thus each one of them is under less evolutionary pressure to maintain its structure. However, the fact that there are also multiple paralogs in the caspase and IAP families argues against this reasoning. Another explanation is that the function of the pro-apoptotic initiators is analogous to a key that ignites the apoptotic engine. While the IAP-binding motif (or the BH3 domain) is like the teeth of the key, the rest of the protein is just the handle or the key chain and can be changed without significant functional consequence in pro-apoptotic activity. Needless to say, it is also fully possible that these regions have regulatory functions that have yet to be understood. This key and engine hypothesis is supported by the fact that, even though IAP antagonists in different insect orders have diverged almost beyond recognition at the whole protein level, their short IAP-binding motifs are highly conserved. Consequently, the pro-apoptotic function is highly conserved. As we showed in this study, Ibm1 is equally potent in inducing cell death in Dipteran or Lepidopteran cell lines. Also supporting this hypothesis is the finding that the uniqueness of the IAP-binding motifs is highly conserved in orthologs of Reaper, Hid, Grim, and sickle in divergent Drosophila species (Zhou 2005). The functions of the paralogs are not simply redundant but each seems to have distinct properties in inducing cell death (Wing et al. 1998; Wing et al. 2001; Zachariou et al. 2003). Thus it seems that paralogs of IAP antagonists serve as unique, rather than general, keys for the fine tuning of cell death regulation.
In sharp contrast with the rapid divergence of the coding sequences of IAP-antagonists, many aspects of their transcriptional regulation are conserved over very long evolutionary distances. For instance, similar to that of reaper, transcriptional regulation of mx in mosquitoes is responsive to radiation (Zhou et al. 2005). In this study, we have shown that, similar to reaper, Ibm1 in Bombyx is significantly up-regulated during metamorphosis. Programmed cell death has been described for several physiological systems in Bombyx mori (Kakei et al. 2005; Mpakou et al. 2006). The role of Ibm1 and other IAP antagonists in regulating apoptosis in these systems remains to be addressed.
Pro-apoptotic mechanism of Reaper/Grim–like IAP antagonists
The functional mechanism of the IAP-binding motif has been very well studied and there is little controversy. The main controversy regarding Reaper/Grim–like IAP antagonists concerns the role of the GH3 motifs. In this study, we showed that a distant ortholog of Reaper/Grim, Ibm1, has a conserved motif similar to the GH3 motif characterized in Reaper and Grim. The C-terminus of Ibm1, in the absence of the N-terminal IAP-binding motif, has very weak pro-apoptotic activity (in SF9 cells) and could only induce significant cell death when the cells are sensitized. Under non-heat shock conditions, the ability of Ibm1 to induce cell death was not changed when the GH3-like motif was deleted. However, Ibm1 without the GH3-like domain was much more sensitive to Diap1 levels, indicating that the GH3 domain has an auxiliary role for the pro-apoptotic function of Ibm1. Our data support the model proposed by Olson et al (Olson et al. 2003) that the GH3 domain in Reaper is required for mitochondrial localization and facilitates the degradation of the IAP. Indeed, the GH3-like motif in Ibm1 is required for mitochondrial localization (Figure 7). Without the GH3-like motif, Ibm1 is much more sensitive to the inhibitory effect of Diap1. Although we can not exclude the possibility, it is unlikely due to protein stability of Ibm1[ΔGH3], since GH3-less Reaper and Grim are stable (Claveria et al. 2002; Olson et al. 2003). Detailed analysis of the potential role of GH3 in stabilizing Reaper demands more thorough biochemical analysis.
Although the auxiliary role of the GH3-like motif did not significantly alter the potency of Ibm1 in the absence of the co-transfected Diap1, it is conceivable that it is very important under in vivo circumstances. In Drosophila, Diap1 is ubiquitously expressed. Thus the potency of an IAP antagonist against Diap1 should be very significant in shifting the balance and mediating selective cell death. This is probably why the GH3 domains in Reaper and Grim are conserved among distantly related Drosophila species and in Ibm1. However, it is also apparent that the GH3 domain (or GH3-like motif) in IAP antagonists outside of the Drosophila order has diverged much faster than the IAP-binding motif. The GH3-like region identified in Mx_Anopheles has so many alterations that it is not clear whether it still possesses similar functions. In fact, before the addition of the Ibm1 sequence, no shared region corresponding to the GH3 domain had been identified by the same analysis described here (Zhou et al. 2005). The identification of Ibm1 thus served as a bridge for the identification of the GH3-like motifs in these proteins. It is very possible that in some orders/species, such as Anopheles, the relative importance of GH3-like motifs could be rather diminished compared to their role in Drosophila species.
In summary, identification of distant orthologs of Reaper has revealed interesting insights into the functional conservation and evolution of this class of important apoptosis initiators. It is expected that the identification of additional IAP antagonists in these species will allow systematic comparison of cell death regulation and reveal important aspects of the organization and evolution of the cell death machinery.
Materials and methods
Bioinformatics
Genomic sequences of Bombyx mori were downloaded from NCBI. The protein motif search strategy has been described in detail in our previous publications (Zhou et al. 2005). Protein sequence alignment and calculation of distance tree were performed using Clustal X (Chenna et al. 2003). Identification of shared motifs was performed either using the MEME web server or a copy of the program installed in a Linux workstation.
Gene Cloning and mutagenesis
A cDNA encoding Ibm1 was first cloned via RT-PCR using mRNA extracted from silk worm pupae and larvae. The ORF was cloned into the Pie3 vector for transfection assays in insect cell lines. For subcellular localization assays, Ibm1 ORF was cloned into a modified pRK5 vector so that it was in frame with a C-terminal fusion of GFP+FLAG tag. Ibm1[Δ2–4] and Ibm1[Δ21–35] were generated by PCR mutagenesis and verified by sequencing. The Bombyx mori IAP1 cDNA was obtained as a generous gift from Qihong Huang (Wistar Institute). The cDNA was cloned into pHSEpiOpIAPVI+ (Vucic et al. 1997) after removing Op-iap from the vector using XmaI at 5′ terminus and EcoRI at 3′ terminus, and the construct was named pHSBmIAP1VI+. This vector contains a Drosophila hsp70 promoter and an N-terminal HA epitope tag.
Cell culture and cell death assays
Lepidopteran (Spodoptera frugiperda) SF9 cells were maintained with SF900 media plus 10% FBS, while the mosquito C6/36 (Aedes albopictus) and the Drosophila S2 cell lines were maintained as previously described (Zhou et al. 2005). Transfection assays of pro-apoptotic activity were carried as previous described (Zhou et al. 2005). Briefly, a total of 0.9 ug testing DNA(s) and 0.1 ug of pie-lacZ was tranfected to cells seeded in two wells of a 24-well plate. About 20 hours after transfection the cells were fixed and stained for β-Gal. Blue cells were counted for each well and survival rate plotted using the vector (pie3) transfected samples as control. Statistical analysis was done with the open source R package (http://www.r-project.org/).
Spodoptera frugiperda SF21 cells were maintained in TC100 (Invitrogen) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals). Transfections were carried out as follows. Briefly, 6 × 105 cells were plated in one well of a six-well plate. Transfections were done using a total of 8 μg DNA (4 μg of each plasmid) and 6 μl lipofectin, in the presence of 100 μM caspase inhibitor zVAD-fmk (MP-Biomedicals) and 50 μg/ml of proteosome inhibitor MG-132 (Sigma). MG-132 was included because BmIAP1 was highly unstable due to ubiquitination, while zVADfmk was included because expression of BmIAP1 caused apoptosis in SF21 cells. Apoptosis has been previously observed when certain IAPs are expressed at high levels, presumably due to interference with endogenous IAP function (Hozak et al. 2000). For efficient expression, cells were heat-shocked at 42°C for 30 minutes at 16 hrs post-transfection. Four hours post heat-shock, cells were lysed in 150 μl IP buffer (contents described below) or counted (viability assay). After lysis, samples were centrifuged at 13,000 rpm at 4 °C for 15 minutes in a microcentrifuge and supernatant was used for analysis.
Immunoprecipitation
For immunoprecipitation preparation, protein G beads (Sigma) were prepared as follows. Per sample used, ~40 μl of beads were washed twice with 1 ml of immunoprecipitation (IP) buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1 % NP-40) by centrifugation at 5,000 rpm in a microcentrifuge at 4 °C. The supernatant was aspirated and 300 μl IP buffer and 10 μg of α-FLAG antibody (Sigma) were added. This mixture was then rocked at 4°C for 1 hr to pre-bind antibody to the beads. The beads were then washed with IP buffer again, as described above, and 70 μl lysate was added to the beads and incubated overnight by rocking at 4 °C. The beads were then centrifuged at 5,000 rpm for 5 min in a microcentrifuge at room temperature. The supernatant was collected and 25 μl was reserved to analyze unbound protein. The pellet containing the immunoprecipitation reaction was washed twice with 500 μl IP buffer at 15,000 rpm for 5 min in a microcentrifuge. The beads were then resuspended in 30 μl SDS PAGE sample buffer containing 0.93 % DTT and heated at 100 °C for 10 min to elute the immunoprecipitated proteins from the beads. The samples were then centrifuged for 15,000 rpm for 5 min in a microcentrifuge and the supernatant was collected and analyzed by SDS-PAGE. For whole lysate detection, 25 μl lysate was analyzed by SDS-PAGE. After SDS-PAGE, the proteins were transferred to PVDF membrane. To determine expression of FLAG-tagged proteins, membranes were probed with primary antibody (α-FLAG, Sigma) at 1/1,000 followed by secondary antibody (α-mouse-HRP, Pierce) at 1/5,000. For HA-tagged proteins, membranes were probed with primary antibody (α-HA, Covance) at 1/1,000 followed by secondary antibody (α-mouse-HRP, Pierce) at 1/10,000.
Caspase activity and viability assays
Caspase activity was determined by incubating lysate with 40 μM N-acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethyl coumarin (Ac-DEVD-AFC, MP-Biomedicals) for 15 min at 37 °C as previously described (Muro et al. 2004). Fluorescence was measured at 15-min intervals to determine enzyme kinetics. Activity assays were performed in triplicate and the results are presented as mean ± SE. To measure cell viability, cells were transfected as described above with a plasmid expressing eGFP and either Ibm1 or Ibm1[ΔIBM]. Transfections were done in the presence or absence of 100 μM zVAD-FMK. Viability was determined at 4 hrs post heat shock by the proportion of GFP-positive cells in high power fields of view from three wells. A total of 150–300 cells were counted per treatment.
Animal rearing and RNA extraction
Fertilized eggs of Bombyx mori were purchased from the Carolina Biological Supply Company. Hatched larvae were maintained on fresh mulberry leaves in ambient temperature. Under our culture conditions, the larval stage was about 27–29 days and the pupal stage about 15–16 days. The day that a visible cocoon appeared was set as day 1 of the pupal stage. For RNA extraction, larvae or pupae extracted from the cocoon were snap frozen in a dry ice/ethanol bath and stored in a −70°C freezer or liquid nitrogen cryogenic tank. To reduce variance, frozen samples of different stages were processed in batch. After homogenizing with a 15 or 30 ml Wheaton Potter-Elvehjem tissue grinder, PolyA mRNA was extracted using the MicroPoly (A) Purist kit (Ambion Inc, Austin, TX) according to the instructions provided by the manufacturer.
Quantitative PCR
For quantitative PCR, 5 ug of each RNA sample was reverse transcribed using the High Capacity cDNA Archive kit (Applied Biosystem). Relative expression of the Ibm1 gene, normalized to either actin or GAPDH, was determined in triplicate assays with SYBR Green nucleic acid dye. Primer pairs were selected using the Primer 3 program. For Ibm1, CCCACGTTTCCGTAGAAAAA (forward) and AGGATGTGGCACAGAACCTC (reverse); for actin, TGCGTGACATCAAGGAGAAG (forward) and ATCTTTCGTTTCCGATGGTG (reverse); for GAPDH, TTCCTGCCTCTACTGGTGCT (forward) and AGCTTGCAGGTTTTCCAAGA (reverse).
Acknowledgments
This work was supported by NIAID R21 AI067555 and R56 AI079074 (LZ) and R21 AI067642 (RJC). We thank Dr. Al Handler for comments on an earlier draft of this manuscript.
References
- Abdelwahid E, Yokokura T, et al. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12(5):793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Methods and statistics for combining motif match scores. J Comput Biol. 1998;5(2):211–21. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, et al. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95(3):331–41. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Bryant B, Blair CD, et al. Annotation and expression profiling of apoptosis-related genes in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2008;38(3):331–45. doi: 10.1016/j.ibmb.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Yan N, et al. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10(11):892–8. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chen P, Ho SI, et al. Bifunctional killing activity encoded by conserved reaper proteins. Cell Death Differ. 2004;11(7):704–13. doi: 10.1038/sj.cdd.4401406. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, et al. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christich A, Kauppila S, et al. The Damage-Responsive Drosophila Gene sickle Encodes a Novel IAP Binding Protein Similar to but Distinct from reaper, grim, and hid. Curr Biol. 2002;12(2):137–40. doi: 10.1016/s0960-9822(01)00658-3. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298(5591):159–65. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Clarke TE, Clem RJ. Insect defenses against virus infection: the role of apoptosis. Int Rev Immunol. 2003;22(5–6):401–24. doi: 10.1080/08830180305215. [DOI] [PubMed] [Google Scholar]
- Claveria C, Caminero E, et al. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. Embo J. 2002;21(13):3327–36. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claveria C, Martinez AC, et al. A Bax/Bak-independent mitochondrial death pathway triggered by Drosophila Grim GH3 domain in mammalian cells. J Biol Chem. 2004;279(2):1368–75. doi: 10.1074/jbc.M309819200. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Miller LK. Control of Programmed Cell Death by the Baculovirus Genes p35 and iap. Mol and Cell Biol. 1994;14(8):5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, et al. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67(4):2168–74. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M, Meier P. Ubiquitylation in apoptosis: DIAP1’s (N-)en(d)igma. Cell Death Differ. 2005;12(9):1208–12. doi: 10.1038/sj.cdd.4401711. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, et al. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. Embo J. 2000;19(4):589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, et al. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83(7):1253–62. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hozak RR, Manji GA, et al. The BIR motifs mediate dominant interference and oligomerization of inhibitor of apoptosis Op-IAP. Mol Cell Biol. 2000;20(5):1877–85. doi: 10.1128/mcb.20.5.1877-1885.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Kanda H, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. Embo J. 2002;21(12):3009–18. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Baehrecke EH, et al. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 1997;124(22):4673–83. doi: 10.1242/dev.124.22.4673. [DOI] [PubMed] [Google Scholar]
- Kakei M, Iwami M, et al. Death commitment in the anterior silk gland of the silkworm, Bombyx mori. J Insect Physiol. 2005;51(1):17–25. doi: 10.1016/j.jinsphys.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Mpakou VE, I, Nezis P, et al. Programmed cell death of the ovarian nurse cells during oogenesis of the silkmoth Bombyx mori. Dev Growth Differ. 2006;48(7):419–28. doi: 10.1111/j.1440-169X.2006.00878.x. [DOI] [PubMed] [Google Scholar]
- Muro I, Hay BA, et al. The Drosophila DIAP1 Protein Is Required to Prevent Accumulation of a Continuously Generated, Processed Form of the Apical Caspase DRONC. J Biol Chem. 2002;277(51):49644–50. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- Muro I, Monser K, et al. Mechanism of Dronc activation in Drosophila cells. J Cell Sci. 2004;117(Pt 21):5035–41. doi: 10.1242/jcs.01376. [DOI] [PubMed] [Google Scholar]
- Olson MR, Holley CL, et al. A GH3-like domain in reaper required for mitochondrial localization and induction of IAP degradation. J Biol Chem. 2003;13:13. doi: 10.1074/jbc.M308055200. [DOI] [PubMed] [Google Scholar]
- Olson MR, Holley CL, et al. Reaper is regulated by IAP-mediated ubiquitination. J Biol Chem. 2003;278(6):4028–34. doi: 10.1074/jbc.M209734200. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Datta P, et al. sickle, a Novel Drosophila Death Gene in the reaper/hid/grim Region, Encodes an IAP-Inhibitory Protein. Curr Biol. 2002;12(2):125–30. doi: 10.1016/s0960-9822(01)00657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6(4):287–97. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Coulson EJ, et al. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2001;2(7):REVIEWS3009. doi: 10.1186/gb-2001-2-7-reviews3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Seshagiri S, et al. Characterization of reaper- and FADD-induced apoptosis in a lepidopteran cell line. Mol Cell Biol. 1997;17(2):667–76. doi: 10.1128/mcb.17.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, et al. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98(4):453–63. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316(5832):1738–43. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, et al. Genetic control of programmed cell death in Drosophila. Science. 1994;264(5159):677–83. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Wing JP, Karres JS, et al. Drosophila sickle Is a Novel grim-reaper Cell Death Activator. Curr Biol. 2002;12(2):131–5. doi: 10.1016/s0960-9822(01)00664-9. [DOI] [PubMed] [Google Scholar]
- Wing JP, Schwartz LM, et al. The RHG motifs of Drosophila Reaper and Grim are important for their distinct cell death-inducing abilities. Mech Dev. 2001;102(1–2):193–203. doi: 10.1016/s0925-4773(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Wing JP, Zhou L, et al. Distinct cell killing properties of the Drosophila reaper, head involution defective, and grim genes. Cell Death Differ. 1998;5(11):930–9. doi: 10.1038/sj.cdd.4400423. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thummel CS. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc Natl Acad Sci U S A. 2004;101(21):8022–7. doi: 10.1073/pnas.0402647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariou A, Tenev T, et al. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. Embo J. 2003;22(24):6642–52. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. The Unique Key Feature of The Iap-binding Motifs in RHG Proteins. Cell Death and Differentiation. 2005 May 20;12(8):1148–51. doi: 10.1038/sj.cdd.4401637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jiang G, et al. Michelob_x is the missing inhibitor of apoptosis protein antagonist in mosquito genomes. EMBO Rep. 2005;6(8):769–74. doi: 10.1038/sj.embor.7400473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann KC, Ricci JE, et al. The role of ARK in stress-induced apoptosis in Drosophila cells. J Cell Biol. 2002;156(6):1077–87. doi: 10.1083/jcb.20112068. [DOI] [PMC free article] [PubMed] [Google Scholar]