Abstract
Neural crest (NC) precursors are stem cells that are capable of forming many cell types after migration to different locations in the embryo. NC and placodes form at the neural plate border (NPB). The Wnt pathway is essential for specifying NC versus placodal identity in this cell population. Here we describe the BTB domain-containing protein Potassium channel tetramerization domain containing 15 (Kctd15) as a factor expressed in the NPB that efficiently inhibits NC induction in zebrafish and frog embryos. Whereas overexpression of Kctd15 inhibited NC formation, knockdown of Kctd15 led to expansion of the NC domain. Likewise, NC induction by Wnt3a plus Chordin in Xenopus animal explants was suppressed by Kctd15, but constitutively active β-catenin reversed Kctd15-mediated suppression of NC induction. Suppression of NC induction by inhibition of Wnt8.1 was rescued by reduction of Kctd15 expression, linking Kctd15 action to the Wnt pathway. We propose that Kctd15 inhibits NC formation by attenuating the output of the canonical Wnt pathway, thereby restricting expansion of the NC domain beyond its normal range.
Keywords: Neural crest, Neural plate border, Wnt signaling, Pigmentation, BTB domain, Zebrafish, Xenopus
INTRODUCTION
Specification, maintenance and differentiation of neural crest (NC) cells depend on multiple signaling pathways. In Xenopus and zebrafish, low levels of Bmp and high levels of Wnt signaling cooperate in NC induction (Saint-Jeannet et al., 1997; Wilson et al., 1997; Dorsky et al., 1998; Lewis et al., 2004). As Wnt signaling regulates anterior-posterior (A-P) patterning of the neural plate (Kim et al., 2000; McGrew et al., 1997; McGrew et al., 1995), Wnt signals might induce NC through posteriorization. However, neural A-P patterning and NC induction are separable events (Wu et al., 2005). A recent study indicates that Wnt-mediated posteriorization of the neural plate border (NPB) rather than the neural plate is crucial in NC induction (Li et al., 2009). Signaling pathways active in NC specification are modulated by activators and inhibitors to regulate their strength and spatial distribution (Hong and Saint-Jeannet, 2007; Sauka-Spengler and Bronner-Fraser, 2008; Zhao et al., 2008). Here we introduce a factor, Potassium channel tetramerization domain containing 15 (Kctd15), that has a profound influence on NC formation in zebrafish and Xenopus embryos. KCTD15 was identified in humans (Hotta et al., 2009; Willer et al., 2009) as a BTB domain-containing protein of unknown function. We show that zebrafish kctd15a and kctd15b are expressed at the NPB at the end of gastrulation. Ectopic expression of Kctd15 inhibits NC specification, whereas knockdown leads to expansion of NC markers. Simultaneous attenuation of Wnt and Kctd15 expression rescues NC specification in zebrafish embryos. We suggest that Kctd15 restricts the NC domain by interfering with the functioning or output of the Wnt/β-catenin signaling pathway.
MATERIALS AND METHODS
Animal maintenance and embryonic staging
Zebrafish (Danio rerio) embryos from AB/TL and TOP-dGFP (Dorsky et al., 2002) lines were obtained (Westerfield, 2000), and stages are indicated as hours post fertilization (hpf) (Kimmel et al., 1995). Xenopus laevis were staged according to (Nieuwkoop and Faber, 1967).
Plasmids and DNA constructs
The open reading frames (ORFs) of zebrafish kctd15a (BC083478), zebrafish kctd15b (BC078294) and X. laevis kctd15 (BC077862) were subcloned into pCS2+. The zebrafish β-cat* construct has four point mutations; S33A, S37A, T41A and S45A.
MO and mRNA injection
Morpholino oligonucleotides (MO) were targeted to 5′UTR regions of zebrafish kctd15a (5′-TCCTTCCCTCCTTGGAAGACATAGC-3′) and zebrafish kctd15b (5′-AGCTCTCCTTCCCCCTCTTGATCTT-3′) (Gene Tools). Embryos at 1-2 cells were injected with 1.5 ng Kctd15a plus 0.5 ng Kctd15b MO; 2 ng Wnt8.1a MO (Lewis et al., 2004); or 2-4 ng standard control MO (5′-CCTCTTACCCTCAGTTACAATTTATA-3′) per embryo. 5′-capped mRNAs were prepared using the mMESSAGE mMACHINE Kit (Ambion). Each zebrafish embryo was injected with 50 pg kctd15a, kctd15b, ΔNkctd15a, ΔCkctd15a, kctd10, kctd6 or kctd13 mRNA, or 5 pg β-cat* mRNA. One blastomere of 2-cell Xenopus embryos was injected with 250 pg of Xkctd15 mRNA. For rescue experiments, 1-10 pg of mRNA was injected into fish embryos.
Wholemount in situ hybridization and Alcian Blue staining
Zebrafish embryos were hybridized with one or two probes (Hauptmann and Gerster, 1994) for kctd15a, kctd15b, pax3 (Seo et al., 1998), foxd3 (Kelsh et al., 2000), sox2 (Okuda et al., 2006), sox10 (Dutton et al., 2001), dlx2a (Akimenko et al., 1994), msxb (Phillips et al., 2006), dlx3b (Ekker et al., 1992), mitfa (Lister et al., 1999), gfp (Dorsky et al., 2002), foxi1 (Solomon et al., 2003), lim3 (Glasgow et al., 1997), prl (Herzog et al., 2003), cldna (Kollmar et al., 2001), eya1 (Sahly et al., 1999), six4.1 (Kobayashi et al., 2000) and gata1 (Detrich et al., 1995). INT-BCIP (red, Roche) and BM purple (blue, Roche) were used. Alcian Blue staining was as described (Barrallo-Gimeno et al., 2004).
Animal cap assay
Xenopus 2-cell embryos were injected with mRNA(s) for Xwnt3a (300 pg/embryo), Xchordin (300 pg/embryo), Xkctd15 (500 pg/embryo) or β-cat* (50 pg/embryo). Animal caps were dissected at stage 8-9, cultured to stage 18 and RNA was analyzed by RT-PCR (Zhao et al., 2008).
Luciferase assay
HEK293T cells in DMEM with 10% fetal calf serum were transfected using FuGENE (Roche). We used Xkctd15 and zβ-cat* in pCS2+ in addition to TOPflash luciferase and Renilla luciferase pRL-CMV (Promega) constructs. Assays were performed as described (Zhao et al., 2008).
RESULTS AND DISCUSSION
kctd15 expression in zebrafish embryos
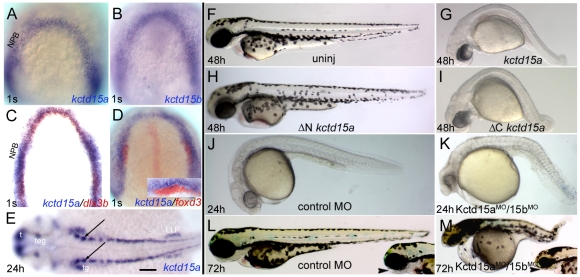
Cells in the NPB express kctd15a and kctd15b at the 1-somite (1s) stage (Fig. 1A,B). kctd15a colocalized with the preplacodal marker dlx3b (Toro and Varga, 2007) (Fig. 1C), but not with premigratory NC cells expressing foxd3 (Monsoro-Burq et al., 2005; Steventon et al., 2005) (Fig. 1D). Subsequently, pharyngeal arches express kctd15a; additional expression domains include pronephric ducts, cranial placode precursors and the brain (Fig. 1E; see Fig. S1C,D,G in the supplementary material). kctd15b transcripts occur maternally (see Fig. S1A,B in the supplementary material), and subsequently are seen in the olfactory placode, cranial NC, lateral line primordium, pharyngeal arches, fin buds and optic tectum (see Fig. S1E,F in the supplementary material).
Fig. 1.
Zebrafish kctd15 expression pattern, loss-of-function and gain-of-function phenotypes. (A-E) Wild-type embryos were hybridized in situ, with probes indicated at lower right, stages shown at lower left. Dorsal views, anterior towards the top (A-D) or left (E). (A,B) Cells in the neural plate border (NPB) express kctd15a and kctd15b. (C) dlx3b (red) and kctd15a (blue) are coexpressed at the NPB. (D,D inset) kctd15a (blue) is excluded from the premigratory neural crest (NC) expressing foxd3 (red). (E) kctd15a expression at 24 hpf. arrows, hindbrain neurons. (F-M) Embryos were injected with RNA (F-I) or MO (J-M). Lateral views, anterior to the left. Overexpression of kctd15a (G, 75/100) and ΔCkctd15a (I, 82/100) results in loss of pigmentation and a short tail, whereas ΔNkctd15a mRNA-injected embryos were normal (H, 60/60). Co-injection of Kctd15a/15b MO results in a small head and abnormal somites (K, 85/100) and subsequently increased dorsal pigmentation (M, 91/100) and loss of jaw elements (M inset, arrowhead). s, somite; t, telencephalon; teg, tegmentum; tg, trigeminal placode; LLP, lateral line primordia. Scale bars: 50 μm in A-D,L,M inset; 80 μm in E; 100 μm in F-M.
Kctd15 inhibits pigmentation in zebrafish
Embryos injected with kctd15a or b mRNA showed a loss of pigmentation, short tail, bent axis, and loss of yolk extension (Fig. 1F,G; see Fig. S2A-C in the supplementary material). The non-NC-derived retinal pigmented epithelium was unaffected, suggesting that Kctd15 blocks the development of NC-derived melanophores. The effect is specific as mRNA injection of the related kctd6, kctd10 and kctd13 genes had no effect (see Fig. S2 in the supplementary material).
Embryos co-injected with MOs against kctd15a and kctd15b (Kctd15aMO/15bMO) exhibited small heads, abnormal somites and increased cell death (Fig. 1J,K), an apparent increase in pigmentation in the dorsal hindbrain region (Fig. 1L,M) and abnormal pharyngeal arches at 72 hpf (Fig. 1M, inset; see Fig. S3 in the supplementary material). These phenotypes were rescued by injection of kctd15a mRNA (see Fig. S4 in the supplementary material). Thus, suppressing Kctd15 had a complementary effect on pigmentation as overexpression, whereas pharyngeal arch development was impaired in both cases (Fig. 1M; see Fig. S3 in the supplementary material), suggesting early and late Kctd15 functions.
The BTB region of Kctd15 is essential for its function
Kctd15 contains a BTB (POZ) domain (residues 32-119), a protein-protein interaction module (Stogios et al., 2005). Constructs encoding the N-terminal BTB domain (ΔCkctd15a) or the C-terminal region (ΔNkctd15a) were injected, showing that ΔCkctd15a caused a similar phenotype as full-length protein (Fig. 1G,I), whereas ΔNkctd15a had no effect (Fig. 1F,H). Thus, the BTB domain of Kctd15 is essential for its function in the embryo.
Kctd15 inhibits NC specification
The above experiments suggest a role for Kctd15 in melanophore and pharyngeal arch development, both involving NC cells. Therefore, we analyzed the NC-specifier pax3 and the pan-NC markers foxd3 and sox10 (Monsoro-Burq et al., 2005; Steventon et al., 2005) in Kctd15 morphants and overexpressing embryos. Expression of markers was reduced or lost in overexpressing embryos (Fig. 2A,C,D,F,G,I); similar results were obtained for msxb and sox9b (data not shown). Consistently, Kctd15aMO/15bMO-injected embryos showed upregulation and expansion of these markers (Fig. 2B,E,H). These effects on foxd3 expression are consistent at different stages (see Fig. S5 in the supplementary material). The pan-neural marker sox2 is expressed at normal levels in Kctd15-overexpressing or morphant embryos (Fig. 2J-L) but the sox2 domain is smaller in the morphants owing to a narrowing of the neural tube that might lead to the small head phenotype at later stages (Fig. 1K).
Fig. 2.
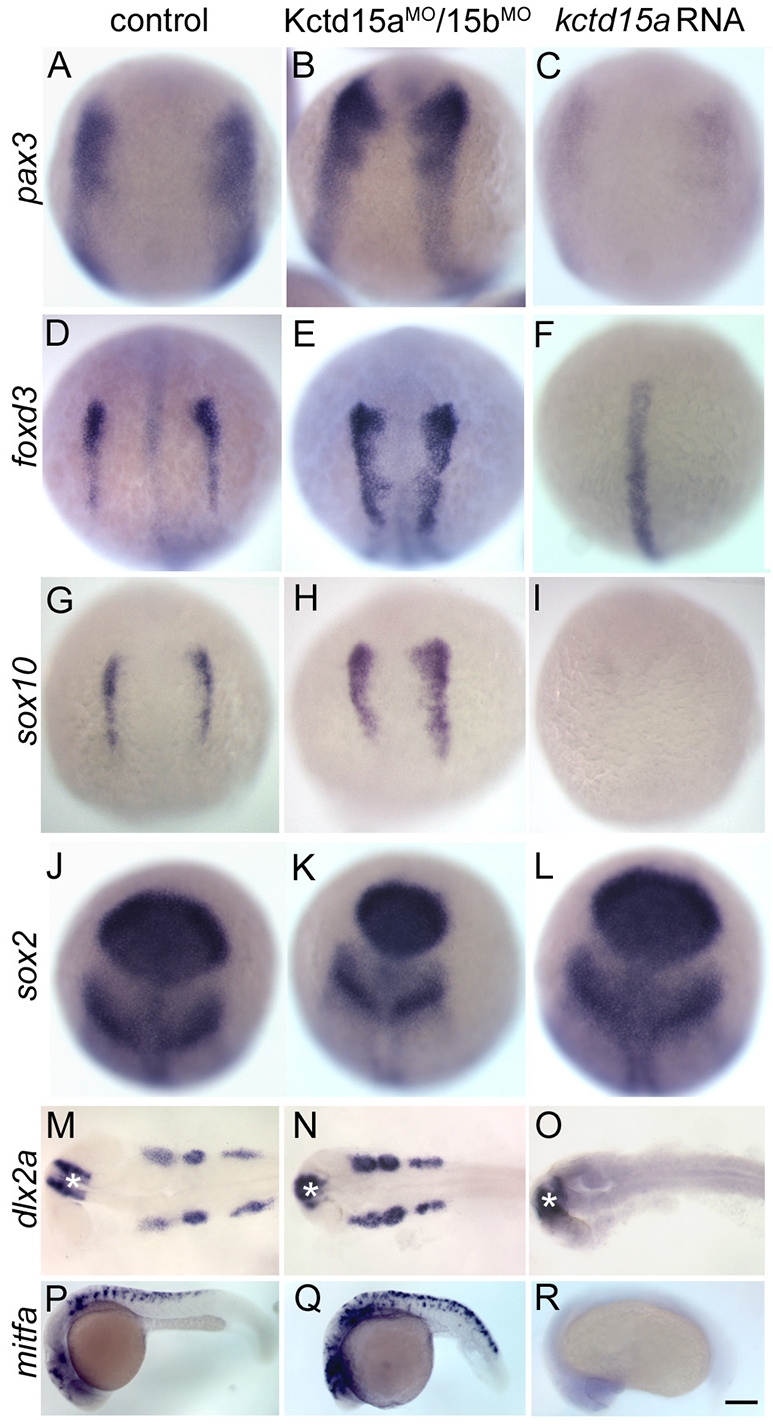
Kctd15 inhibits NC induction and differentiation. (A-R) Control embryos (control MO-injected and uninjected embryos were indistinguishable; A,D,G,J,M,P), Kctd15aMO/15bMO-injected (B,E,H,K,N,Q) and kctd15a mRNA-injected (C,F,I,L,O,R) embryos were fixed at the 1-somite stage (A-L) or at 24 hpf (M-R). Expression of pax3 (A-C), foxd3 (D-F), sox10 (G-I), sox2 (J-L), dlx2a (M-O) and mitfa (P-R) was analyzed by in situ hybridization. Morphants showed expansion of pax3 (B, 42/50), foxd3 (E, 28/35) and sox10 (H, 25/35). kctd15a mRNA-injected embryos showed inhibition of pax3 (C, 38/45), foxd3 (F, 45/50) and sox10 (I, 32/37) in the NC domain, whereas sox2 expression was preserved (K, 25/25; L, 20/20). Expression of dlx2a in the branchial arches was expanded in morphants (N, 32/44) and lost in overexpressing embryos (O, 35/40); mitfa was expanded in morphants (Q, 30/35) and lost in overexpressing embryos (R, 40/40). (A-L) Dorsal views, anterior towards the top; (M-O) dorsal views; (P-R) lateral views, anterior towards the left. Asterisk in M-O indicates dlx2a expression in the forebrain. Scale bar: 100 μm in A-L; 200 μm in M-R.
To study NC precursor differentiation, we tested dlx2a and mitfa. At 24 hpf, we observed a loss of dlx2a in pharyngeal arches and of mitfa in melanophore precursors in kctd15a mRNA-injected embryos (Fig. 2O,R). foxd3 in the notochord and dlx2a in the telencephalon were unaltered (Fig. 2F,O), illustrating the specificity of NC inhibition. In Kctd15aMO/15bMO-injected embryos, the expression of dlx2a in pharyngeal arches was maintained (Fig. 2M,N) and expression of mitfa was increased (Fig. 2P,Q). Whereas the increased mitfa expression correlates with the intense pigmentation of MO-injected embryos (Fig. 1L,M), the inhibition of pharyngeal arch formation in these embryos (see Fig. S3B in the supplementary material) points to a late function of Kctd15 in this lineage.
At early stages, kctd15 is expressed in the preplacodal domain but excluded from NC precursors; thus, it might have a role in delineating preplacodal versus NC identity. We find that kctd15a RNA-injected embryos show enhanced expression of the preplacodal markers eya1 and six4.1 in the anterior domain (see Fig. S6A-D in the supplementary material). Similarly, pan-pituitary markers lim3 and prl were expanded (see Fig. S6E-H in the supplementary material). Expression of foxi1, a marker for otic progenitors, was unaffected and the later otic marker cldna was reduced (see Fig. S6I-L in the supplementary material). These results suggest that loss of NC caused by Kctd15 overexpression was compensated by expansion of the anterior preplacodal domain, whereas both NC and placode development were inhibited by Kctd15 in posterior regions.
Kctd15 antagonizes Wnt signaling during NC formation
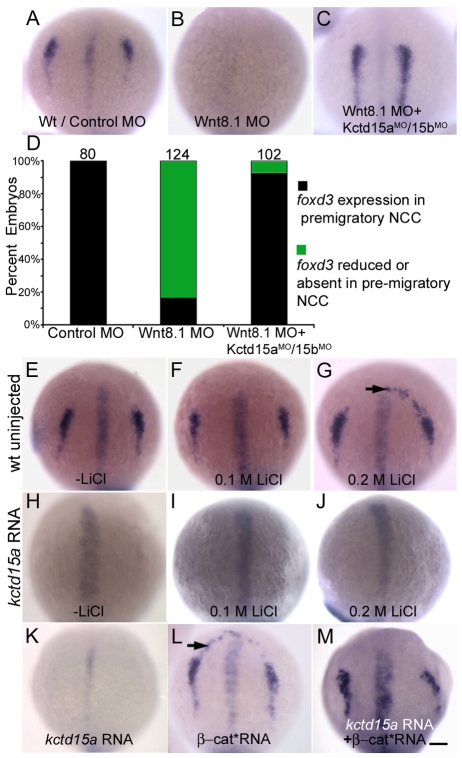
Wnt signaling is necessary for NC specification and later differentiation (Dorsky et al., 1998; Garcia-Castro et al., 2002; LaBonne and Bronner-Fraser, 1998; Saint-Jeannet et al., 1997). We injected embryos with Wnt8.1 MO or with MOs for Wnt8.1, Kctd15a and Kctd15b and assayed NC specification. As reported (Lewis et al., 2004), foxd3 expression was lost in Wnt8.1 morphants (Fig. 3A,B). Importantly, we observed a dramatic rescue of foxd3 expression in embryos injected with a combination of Wnt8.1 and Kctd15a/b MOs (Fig. 3C,D). Similar results were obtained using sox10 (data not shown). kctd15a expression in Wnt8.1 morphants was unaffected (see Fig. S7A,B in the supplementary material). These observations indicate that Kctd15 interferes with Wnt signal transduction or output, such that the reduction in ligand availability in the Wnt8.1 morphants is compensated for by alleviating the inhibition of pathway output by Kctd15.
Fig. 3.
Kctd15 inhibits the output of canonical Wnt signaling. (A) foxd3 expression in control embryos. (B) Loss of foxd3 expression in wnt8.1 morphants. (C) Injection of Wnt8.1 MO together with Kctd15a/15b MOs rescued foxd3 expression. (D) Quantification of A-C; number of embryos (n) is given above bars. (E-J) foxd3 expression in embryos treated with LiCl (E-G). Arrow in G indicates expanded expression in 26/30 embryos. (H-J) LiCl did not rescue foxd3 expression in kctd15a mRNA-injected embryos (I, 40/40; J, 50/50; numbers indicate embryos with appearance as shown in the figure). (K-M) Inhibition of foxd3 by kctd15a (K) was rescued by β-cat* mRNA (M, 41/50 showing foxd3 expression in NC domain). β-cat* mRNA alone induced ectopic expression of foxd3 (L, arrow, 25/31). (A-C,E-M) 1-somite stage; dorsal views, anterior towards top. Scale bar: 100 μm.
To probe the relationship between Kctd15 and Wnt signaling, we treated embryos with LiCl, which inhibits Gsk3β activity leading to β-catenin stabilization (Klein and Melton, 1996; van de Water et al., 2001). Embryos were treated with 0.1 M LiCl (shield to bud) or 0.2 M LiCl (shield to 80% epiboly) and assayed for foxd3 expression. Although 0.2 M LiCl led to ectopic foxd3 expression, Kctd15 overexpression abolished all foxd3 expression (Fig. 3E-J), indicating that Kctd15 acts downstream of Gsk3β in NC induction. By contrast, constitutively active β-catenin (β-cat*) rescued foxd3 expression in kctd15 RNA-injected embryos, although ectopic foxd3 expression was abolished (Fig. 3K-M). These results indicate that Kctd15 acts upstream of or in concert with β-catenin in NC induction.
Kctd15 function is conserved and inhibits NC induction in explants
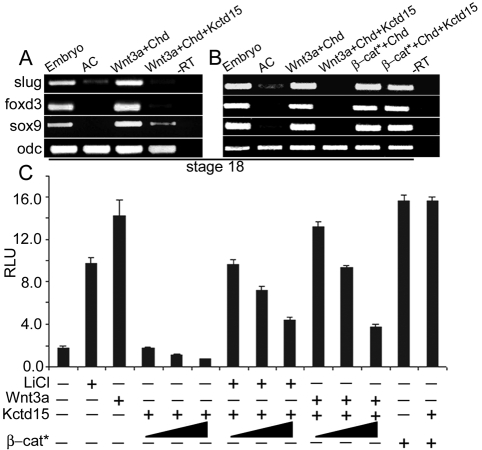
Explants from Xenopus embryos (animal caps) are induced to the NC by expression of Wnt and a Bmp antagonist (Saint-Jeannet et al., 1997). We used this system to further explore the function of Kctd15. Kctd15 function is conserved in frogs, as injection of Xenopus kctd15 mRNA into one blastomere of 2-cell embryos suppressed the NC marker slug (snail2) (see Fig. S8 in the supplementary material). For explant experiments, embryos were injected with wnt3a plus chordin (chrd) mRNAs, with or without kctd15 mRNA, and animal caps were assayed for NC markers. Induced slug, foxd3 and sox9 expression was strongly inhibited by the co-injection of kctd15 mRNA (Fig. 4A). In zebrafish embryos, β-cat*-mediated induction of NC proved largely resistant to Kctd15 inhibition. This is also true in Xenopus animal caps (Fig. 4B). Thus, the explant and whole embryo experiments are consistent, indicating that Kctd15 inhibits NC induction at or above the level of β-catenin.
Fig. 4.
Kctd15 inhibits NC induction by antagonizing the Wnt signal. (A,B) Kctd15 suppressed NC-specific marker gene induction by Chrd + Wnt3a, but not by Chrd + β-cat* in Xenopus animal caps. Injected RNAs are indicated at the top; expression of slug, foxd3, sox9 and of odc as control was assayed by RT-PCR. (C) Wnt signaling in HEK293T cells was measured by TOPflash luciferase. Kctd15 moderately inhibited Wnt signaling stimulated by Wnt3a or LiCl but not by β-cat*. AC, uninjected animal caps; −RT, without reverse transcriptase; RLU, relative luciferase units.
To determine whether Kctd15 is a general Wnt inhibitor, we tested Kctd15 in the Xenopus axis duplication assay (Smith and Harland, 1991; Sokol et al., 1991). Co-injection of Xkctd15 did not reduce the frequency or completeness of Wnt8-induced axis duplications (data not shown). A recent study showed that an activator of Wnt signaling, Skip (1-341), was inefficient in inducing axis duplications (Wang et al., 2010), suggesting that certain modulators of Wnt signaling are inefficient in early frog embryos. In zebrafish, cells responsive to β-catenin can be monitored in the transgenic TOP-dGFP reporter line (Dorsky et al., 2002). GFP expression was reduced in kctd15a mRNA-injected transgenic embryos compared with controls (see Fig. S9A,B in the supplementary material). More detailed inspection of the NPB region revealed that kctd15 mRNA-injected transgenic embryos showed a loss of GFP expression in the NC domain (see Fig. S9C,D in the supplementary material). Inhibition of the Wnt pathway could also be observed using TOPflash luciferase in HEK293T cells. Kctd15 reduced basal, Wnt3a or LiCl-stimulated TOPflash activity by about 3-fold, but not β-cat*-stimulated activity (Fig. 4C).
Kctd15 is a novel regulatory factor in NC formation
In this report, we introduce Kctd15 as a novel regulator of NC formation in zebrafish and Xenopus. We provide evidence that Kctd15 acts as an inhibitor of NC specification by interfering with the output of Wnt signaling. We show that: (1) ectopic expression of Kctd15 leads to a dramatic reduction in the induction of premigratory NC cells in whole embryos and of early NC marker genes in induced animal caps; (2) conversely, the expression domain of NC-specific genes is expanded in Kctd15 morphants; (3) loss of NC induction in zebrafish Wnt8.1 morphants was rescued by attenuating Kctd15 expression; (4) constitutively active β-catenin rescued the suppression of NC induction by Kctd15. These results show that Kctd15 is a potent regulator of NC formation.
Precursors of the NC and the preplacodal region arise from an initially mixed population of cells at the NPB (Streit, 2002) and are separated as a result of Wnt signaling and the action of Gbx2 (Li et al., 2009; Litsiou et al., 2005). Although the induction of Gbx2 in the NC domain is a key feature in its formation (Li et al., 2009), placode-specific inhibitors of NC induction might be required to assist in the separation of the two domains. We suggest that Kctd15 represents such a factor with a role in defining the NC versus the anterior preplacodal domain at the NPB. This suggestion is supported by the enhanced expression of several anterior preplacodal markers in kctd15-overexpressing embryos (see Fig. S6 in the supplementary material).
The BTB domain is found in many functional classes of proteins (Stogios et al., 2005) and various properties and associations have been reported for Kctd proteins (Bayon et al., 2008; Ding et al., 2009; Ding et al., 2008). Here, we have linked the inhibitory role of Kctd15 to Wnt pathway signal transduction or output. Our results show that Kctd15 is a novel, highly effective and specific regulatory component in NC induction with an apparent role in restricting the NC domain in the developing embryo.
Supplementary Material
Acknowledgements
We thank Hyunju Ro for the β-catenin mutant and other reagents, Martha Rebbert for technical support, Robert Cornell (University of Iowa) for plasmids, Brian Brooks for support and members of the Dawid laboratory for discussion. This research was supported by the Intramural Research Program of the NICHD, NIH. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.047548/-/DC1
References
- Akimenko M. A., Ekker M., Wegner J., Lin W., Westerfield M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475-3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Holzschuh J., Driever W., Knapik E. W. (2004). Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development 131, 1463-1477 [DOI] [PubMed] [Google Scholar]
- Bayon Y., Trinidad A. G., de la Puerta M. L., Del Carmen Rodriguez M., Bogetz J., Rojas A., De Pereda J. M., Rahmouni S., Williams S., Matsuzawa S., et al. (2008). KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 275, 3900-3910 [DOI] [PubMed] [Google Scholar]
- Detrich H. W., 3rd, Kieran M. W., Chan F. Y., Barone L. M., Yee K., Rundstadler J. A., Pratt S., Ransom D., Zon L. I. (1995). Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA 92, 10713-10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X. F., Luo C., Ren K. Q., Zhang J., Zhou J. L., Hu X., Liu R. S., Wang Y., Gao X., Zhang J. (2008). Characterization and expression of a human KCTD1 gene containing the BTB domain, which mediates transcriptional repression and homomeric interactions. DNA Cell Biol. 27, 257-265 [DOI] [PubMed] [Google Scholar]
- Ding X., Luo C., Zhou J., Zhong Y., Hu X., Zhou F., Ren K., Gan L., He A., Zhu J., et al. (2009). The interaction of KCTD1 with transcription factor AP-2alpha inhibits its transactivation. J. Cell. Biochem. 106, 285-295 [DOI] [PubMed] [Google Scholar]
- Dorsky R. I., Moon R. T., Raible D. W. (1998). Control of neural crest cell fate by the Wnt signalling pathway. Nature 396, 370-373 [DOI] [PubMed] [Google Scholar]
- Dorsky R. I., Sheldahl L. C., Moon R. T. (2002). A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev. Biol. 241, 229-237 [DOI] [PubMed] [Google Scholar]
- Dutton K. A., Pauliny A., Lopes S. S., Elworthy S., Carney T. J., Rauch J., Geisler R., Haffter P., Kelsh R. N. (2001). Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128, 4113-4125 [DOI] [PubMed] [Google Scholar]
- Ekker M., Akimenko M. A., Bremiller R., Westerfield M. (1992). Regional expression of three homeobox transcripts in the inner ear of zebrafish embryos. Neuron 9, 27-35 [DOI] [PubMed] [Google Scholar]
- Garcia-Castro M. I., Marcelle C., Bronner-Fraser M. (2002). Ectodermal Wnt function as a neural crest inducer. Science 297, 848-851 [DOI] [PubMed] [Google Scholar]
- Glasgow E., Karavanov A. A., Dawid I. B. (1997). Neuronal and neuroendocrine expression of lim3, a LIM class homeobox gene, is altered in mutant zebrafish with axial signaling defects. Dev. Biol. 192, 405-419 [DOI] [PubMed] [Google Scholar]
- Hauptmann G., Gerster T. (1994). Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10, 266 [DOI] [PubMed] [Google Scholar]
- Herzog W., Zeng X., Lele Z., Sonntag C., Ting J. W., Chang C. Y., Hammerschmidt M. (2003). Adenohypophysis formation in the zebrafish and its dependence on sonic hedgehog. Dev. Biol. 254, 36-49 [DOI] [PubMed] [Google Scholar]
- Hong C. S., Saint-Jeannet J. P. (2007). The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell 18, 2192-2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K., Nakamura M., Nakamura T., Matsuo T., Nakata Y., Kamohara S., Miyatake N., Kotani K., Komatsu R., Itoh N., et al. (2009). Association between obesity and polymorphisms in SEC16B, TMEM18, GNPDA2, BDNF, FAIM2 and MC4R in a Japanese population. J. Hum. Genet. 54, 727-731 [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., Dutton K., Medlin J., Eisen J. S. (2000). Expression of zebrafish fkd6 in neural crest-derived glia. Mech. Dev. 93, 161-164 [DOI] [PubMed] [Google Scholar]
- Kim C. H., Oda T., Itoh M., Jiang D., Artinger K. B., Chandrasekharappa S. C., Driever W., Chitnis A. B. (2000). Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 407, 913-916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310 [DOI] [PubMed] [Google Scholar]
- Klein P. S., Melton D. A. (1996). A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 93, 8455-8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Osanai H., Kawakami K., Yamamoto M. (2000). Expression of three zebrafish Six4 genes in the cranial sensory placodes and the developing somites. Mech. Dev. 98, 151-155 [DOI] [PubMed] [Google Scholar]
- Kollmar R., Nakamura S. K., Kappler J. A., Hudspeth A. J. (2001). Expression and phylogeny of claudins in vertebrate primordia. Proc. Natl. Acad. Sci. USA 98, 10196-10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C., Bronner-Fraser M. (1998). Neural crest induction in Xenopus: evidence for a two-signal model. Development 125, 2403-2414 [DOI] [PubMed] [Google Scholar]
- Lewis J. L., Bonner J., Modrell M., Ragland J. W., Moon R. T., Dorsky R. I., Raible D. W. (2004). Reiterated Wnt signaling during zebrafish neural crest development. Development 131, 1299-1308 [DOI] [PubMed] [Google Scholar]
- Li B., Kuriyama S., Moreno M., Mayor R. (2009). The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development 136, 3267-3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister J. A., Robertson C. P., Lepage T., Johnson S. L., Raible D. W. (1999). nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126, 3757-3767 [DOI] [PubMed] [Google Scholar]
- Litsiou A., Hanson S., Streit A. (2005). A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development 132, 4051-4062 [DOI] [PubMed] [Google Scholar]
- McGrew L. L., Lai C. J., Moon R. T. (1995). Specification of the anteroposterior neural axis through synergistic interaction of the Wnt signaling cascade with noggin and follistatin. Dev. Biol. 172, 337-342 [DOI] [PubMed] [Google Scholar]
- McGrew L. L., Hoppler S., Moon R. T. (1997). Wnt and FGF pathways cooperatively pattern anteroposterior neural ectoderm in Xenopus. Mech. Dev. 69, 105-114 [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq A. H., Wang E., Harland R. (2005). Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell 8, 167-178 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. (1967). Normal Table of Xenopus laevis (Daudin). Amsterdam: North-Holland Publishing Company; [Google Scholar]
- Okuda Y., Yoda H., Uchikawa M., Furutani-Seiki M., Takeda H., Kondoh H., Kamachi Y. (2006). Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 235, 811-825 [DOI] [PubMed] [Google Scholar]
- Phillips B. T., Kwon H. J., Melton C., Houghtaling P., Fritz A., Riley B. B. (2006). Zebrafish msxB, msxC and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev. Biol. 294, 376-390 [DOI] [PubMed] [Google Scholar]
- Sahly I., Andermann P., Petit C. (1999). The zebrafish eya1 gene and its expression pattern during embryogenesis. Dev. Genes Evol. 209, 399-410 [DOI] [PubMed] [Google Scholar]
- Saint-Jeannet J. P., He X., Varmus H. E., Dawid I. B. (1997). Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc. Natl. Acad. Sci. USA 94, 13713-13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T., Bronner-Fraser M. (2008). A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557-568 [DOI] [PubMed] [Google Scholar]
- Seo H. C., Saetre B. O., Havik B., Ellingsen S., Fjose A. (1998). The zebrafish Pax3 and Pax7 homologues are highly conserved, encode multiple isoforms and show dynamic segment-like expression in the developing brain. Mech. Dev. 70, 49-63 [DOI] [PubMed] [Google Scholar]
- Smith W. C., Harland R. M. (1991). Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell 67, 753-765 [DOI] [PubMed] [Google Scholar]
- Sokol S., Christian J. L., Moon R. T., Melton D. A. (1991). Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell 67, 741-752 [DOI] [PubMed] [Google Scholar]
- Solomon K. S., Kudoh T., Dawid I. B., Fritz A. (2003). Zebrafish foxi1 mediates otic placode formation and jaw development. Development 130, 929-940 [DOI] [PubMed] [Google Scholar]
- Steventon B., Carmona-Fontaine C., Mayor R. (2005). Genetic network during neural crest induction: from cell specification to cell survival. Semin. Cell Dev. Biol. 16, 647-54 [DOI] [PubMed] [Google Scholar]
- Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Prive G. G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A. (2002). Extensive cell movements accompany formation of the otic placode. Dev. Biol. 249, 237-254 [DOI] [PubMed] [Google Scholar]
- Toro S., Varga Z. M. (2007). Equivalent progenitor cells in the zebrafish anterior preplacodal field give rise to adenohypophysis, lens, and olfactory placodes. Semin. Cell Dev. Biol. 18, 534-542 [DOI] [PubMed] [Google Scholar]
- van de Water S., van de Wetering M., Joore J., Esseling J., Bink R., Clevers H., Zivkovic D. (2001). Ectopic Wnt signal determines the eyeless phenotype of zebrafish masterblind mutant. Development 128, 3877-3888 [DOI] [PubMed] [Google Scholar]
- Wang Y., Fu Y., Gao L., Zhu G., Liang J., Gao C., Huang B., Fenger U., Niehrs C., Chen Y. G., et al. (2010). Xenopus skip modulates Wnt/beta-catenin signaling and functions in neural crest induction. J. Biol. Chem. 285, 10890-10901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book: a Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press; [Google Scholar]
- Willer C. J., Speliotes E. K., Loos R. J., Li S., Lindgren C. M., Heid I. M., Berndt S. I., Elliott A. L., Jackson A. U., Lamina C., et al. (2009). Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P. A., Lagna G., Suzuki A., Hemmati-Brivanlou A. (1997). Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development 124, 3177-3184 [DOI] [PubMed] [Google Scholar]
- Wu J., Yang J., Klein P. S. (2005). Neural crest induction by the canonical Wnt pathway can be dissociated from anterior-posterior neural patterning in Xenopus. Dev. Biol. 279, 220-232 [DOI] [PubMed] [Google Scholar]
- Zhao H., Tanegashima K., Ro H., Dawid I. B. (2008). Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development 135, 1283-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.