Abstract
The inner side of the nuclear envelope (NE) is lined with lamins, a meshwork of intermediate filaments that provides structural support for the nucleus and plays roles in many nuclear processes. Lamins, classified as A- or B-types on the basis of biochemical properties, have a conserved globular head, central rod and C-terminal domain that includes an Ig-fold structural motif. In humans, mutations in A-type lamins give rise to diseases that exhibit tissue-specific defects, such as Emery-Dreifuss muscular dystrophy. Drosophila is being used as a model to determine tissue-specific functions of A-type lamins in development, with implications for understanding human disease mechanisms. The GAL4-UAS system was used to express wild-type and mutant forms of Lamin C (the presumed Drosophila A-type lamin), in an otherwise wild-type background. Larval muscle-specific expression of wild type Drosophila Lamin C caused no overt phenotype. By contrast, larval muscle-specific expression of a truncated form of Lamin C lacking the N-terminal head (Lamin C ΔN) caused muscle defects and semi-lethality, with adult ‘escapers’ possessing malformed legs. The leg defects were due to a lack of larval muscle function and alterations in hormone-regulated gene expression. The consequences of Lamin C association at a gene were tested directly by targeting a Lamin C DNA-binding domain fusion protein upstream of a reporter gene. Association of Lamin C correlated with localization of the reporter gene at the nuclear periphery and gene repression. These data demonstrate connections among the Drosophila A-type lamin, hormone-induced gene expression and muscle function.
Keywords: Drosophila, Gene expression, Lamins, Larval muscle, Nuclear envelope
INTRODUCTION
The nuclear lamina is a protein meshwork that lines the inner nuclear membrane. Type V intermediate filament proteins called lamins and membrane-associated proteins, many of which possess a LEM domain (named for founding members LAP2, Emerin and MAN1), make up the lamina (Cai et al., 2001; Herrmann and Aebi, 2004; Lin et al., 2000; McKeon et al., 1986; Pinto et al., 2008; Schulze et al., 2009; Stuurman et al., 1998; Wolff et al., 2001). Lamins have a conserved N-terminal globular head domain, a central coiled rod domain and a C-terminal tail that includes an Ig-fold structural motif (Fisher et al., 1986; McKeon et al., 1986). Lamins play roles in nuclear shape determination, nuclear positioning, chromosomal arrangement, heterochromatin organization, nuclear pore function, chromosome segregation, cytoskeletal organization and signal transduction (Capco and Penman, 1983; Furukawa and Hotta, 1993; Liu et al., 2000; Morris, 2003; Paddy et al., 1990; Schirmer et al., 2003; Starr and Fischer, 2005; Sullivan et al., 1999; Wilhelmsen et al., 2006).
Lamins are classified as A- and B-types (Burke and Gerace, 1986; Georgatos et al., 1994; Gerace and Blobel, 1980; Gerace and Burke, 1988; Riemer et al., 1995; Stuurman et al., 1998). A-type lamins are expressed in nearly all differentiated somatic cells and are not essential for cell viability (Fisher et al., 1986). By contrast, B-type lamins are ubiquitously expressed and are essential for cell viability (Biamonti et al., 1992; Harborth et al., 2001; Pollard et al., 1990). Mutations in LMNA, the gene encoding A-type lamins in humans, give rise to at least 12 disorders termed laminopathies, including autosomal dominant Emery-Dreifuss muscular dystrophy (AD-EDMD) (Bonne et al., 1999). Laminopathies have tissue-specific phenotypes, despite the global expression of LMNA. EDMD is also caused by mutations in the gene encoding emerin, a LEM domain protein (Bione et al., 1994; Manilal et al., 1996; Nagano et al., 1996). Emerin interacts with A-type lamins (Clements et al., 2000; Sakaki et al., 2001) and is mislocalized in many patients with LMNA mutations (Ben-Harush et al., 2009; Muchir et al., 2003; Sullivan et al., 1999).
Here, we focus on the role of the N-terminal head of A-type lamin with respect to muscle function and gene expression. Structural studies implicate the head domain in lamina assembly (Herrmann and Foisner, 2003; Isobe et al., 2007; Sasse et al., 1998; Shumaker et al., 2005; Stuurman et al., 1996). Lamins dimerize through the rod domain and form head-to-tail interactions to generate filaments (Sasse et al., 1998; Stuurman et al., 1996). In humans, a mutation in the LMNA gene, which is presumed to produce a truncated protein lacking the head domain, has been implicated in a form of EDMD possessing a neurological phenotype (Walter et al., 2005).
We have developed Drosophila melanogaster as a model to study tissue-specific functions of A-type lamins. Drosophila is the only invertebrate model organism known to possess the two types of lamins found in humans (Klapper et al., 1997; Riemer et al., 1995; Schulze et al., 2005). The Drosophila genome possesses two lamin genes: Lamin C, proposed to encode the A-type lamin, and Lamin Dm0, the B-type lamin (Riemer et al., 1995). Expression of mutant forms of Lamin C that alter the N- or C-terminal domain associate with the nuclear envelope (NE) and, in some cases, cause lethality when expressed in larval muscle but not in other tissues (Schulze et al., 2009). By contrast, alterations in the rod domain cause abnormal lamin aggregation within the nucleus but not lethality. Collectively, our studies support a role for the N-terminal head and C-terminal globular domain in larval muscle function.
To better understand the role of the N-terminal head domain, we generated transgenic flies expressing a mutant form of Lamin C lacking the first 42 amino acids (Lamin C ΔN) (Schulze et al., 2009). This mutant form localizes to the NE and causes semi-lethality when expressed in muscle but not in non-muscle tissues (Schulze et al., 2009). Here, we demonstrate that lethality is associated with larval body wall muscle defects that include nuclear and cytoplasmic abnormalities. ‘Adult escapers’ expressing Lamin C ΔN have leg defects that are consistent with a loss of muscle function and disruptions in ecdysone hormone signaling. Collectively, these studies demonstrate a role for the N-terminal head domain of Lamin C in muscle function and gene expression.
MATERIALS AND METHODS
Drosophila stocks and genetic analyses
Drosophila stocks were raised at room temperature on standard sucrose and cornmeal medium (Shaffer et al., 1994). Generation of transgenic stocks expressing wild type and Lamin C ΔN was previously reported (Schulze et al., 2005; Schulze et al., 2009). The GAL4-UAS system (Brand and Perrimon, 1993; Duffy, 2002) was used to drive expression of wild type Lamin C and Lamin C ΔN in specific tissues. The GAL4 driver stocks used in this study are described in Table 1.
Table 1.
Tissue-specific expression of wild-type and mutant forms of Lamin C
Immunostaining of muscles
Larval muscle preparations were performed according to published procedures (Budnik et al., 1990). After fixation, the dissected body wall preparations were blocked in PBS2+ (130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, 10 mM EGTA, 0.1% Triton-X 100) containing 0.1% BSA for 1 hour. Muscle preparations were stained with 66 nM fluorescent Phalloidin (Alexa Fluor 488 from Molecular Probes) in PBS2+ containing 0.1% BSA, rat anti-tyrosinated tubulin (1:50 dilution), rabbit anti-D-Titin (anti-52, 1:5000 dilution; a generous gift from Deborah J. Andrew) (Fabian et al., 2007) or rabbit anti-Mlp84b (B54, 1:100 dilution; a generous gift from K. Clark and M. Beckerle, University of Utah). Nuclear pore proteins were detected using MAb414 (anti-mouse, 1:3000 dilution; Covance; recognizes the FXFG repeat sequence in nucleoporins, a related family of NPC proteins including p62, p152 and p90). Otefin was detected using mouse anti-Otefin (1:100 dilution; a generous gift from Y. Gruenbaum, Hebrew University of Jerusalem) and Klaroid was detected using rat anti-Klaroid (1:20 dilution; anti-Koi, a generous gift for J. Fischer, University of Texas, Austin). Imaging was performed on a Bio-Rad MRC 1024 confocal microscope (Center for Microscopy, University of Iowa, IA, USA).
Phalloidin staining of leg imaginal discs
Six hours after pupal formation, pupae were collected off the walls of vials and dissected in Ringer's solution (pH 7.4) at room temperature. Leg discs were fixed in 4% formaldehyde solution for 30 minutes at room temperature. The discs were permeabilized for 1-2 hours with 0.5% Triton X-100 (w/v in PBS) at room temperature and stained with Phalloidin overnight at room temperature. Leg discs were rinsed three times with 0.5% Triton X-100 (w/v in PBS) for 15 minutes at room temperature and mounted in Vectashield (Vector Laboratories).
RT-PCR analysis
RT-PCR was performed using the iCycler iQ Real-Time PCR System (BioRad). Total RNA was isolated using TRIzol reagent (Sigma), according to the manufacturer. Twenty micrograms of RNA were treated with two units of DNase I (Invitrogen) for 15 minutes at 37°C to eliminate genomic DNA. The reaction was quenched with 2.5 mM EDTA and incubation at 65°C for 10 minutes. cDNA synthesis was performed using 6 μg of RNA and reverse transcriptase in 20 μl with a SuperScript First-Strand Synthesis System for RT-PCR Kit (Invitrogen). Aliquots (3.2 μl) of cDNA were added to a 78 μl reaction mixture containing 39 μl of 2× SYBR Green PCR Master Mix (Applied Biosystems) and 1 mM of each primer for triplicate reactions in a 25 μl volume per well. Triplicate reactions were performed and averaged. The reaction was first incubated at 95°C for 3 minutes, followed by 40 cycles at 95°C for 30 seconds and 60°C for 1 minute. The sequences for the RT-PCR are shown in Table S1 in the supplementary material. At least three independent experiments were performed for each primer set on three independent RNA samples. Relative levels of mRNA expression were calculated using the cycle threshold (CT) method (Valasek and Repa, 2005). Individual expression values were normalized by comparison with rp49 mRNA.
Immunohistochemical staining of salivary glands and imaginal disc nuclei
Larvae were raised at room temperature in vials, administered a 45 minute heat-shock at 37°C and allowed to recover for 2 hours. Salivary glands and imaginal disc tissues were dissected (the total dissection time for an experiment was less than 1.5 hours), placed in welled slides and fixed with 2% formaldehyde for 15-20 minutes, followed by three washes of 5 minutes each in PBS2+. Tissues were blocked in PBS2+ containing 0.1% BSA for 1 hour and incubated overnight with primary antibodies at 4°C. After three 15 minute washes with PBS2+, tissues were incubated for 2 hours with the relevant secondary antibodies in the dark at room temperature. Tissues were washed three times for 10 minutes in PBS2+ and mounted in Vectashield H-1000 or Vectashield with DAPI (Vector Laboratories, Burlingame, CA, USA). The primary antibodies used were: mouse anti-Lamin C (LC28.26, 1:500 dilution; University of Iowa Hybridoma Core Facility, IA, USA), mouse anti-LacI (clone 9A5, 1:200 dilution; Upstate Biotechnology), rabbit anti-LacI (1:200 dilution; Stratagene).
Fluorescent in situ hybridization
Salivary glands and diploid tissues were dissected in 1× PBS and fixed in 4% formaldehyde in 1× PBS. The tissue was washed with 2× SSCT (0.3 M NaCl, 0.03 M sodium citrate and 0.1% Tween-20) and treated with RNase A in 2× SSCT for 30 minutes. The tissues were washed with gradually increasing (20%, 40% and 50%) formamide concentration in 2× SSCT at 37°C. Then the tissues were incubated with 100 ng of a DNA fragment representing the lac operator repeats (gift of Y. Rong, NIH) in hybridization solution (10% dextran sulfate, 3× SSC, 50% formamide) at 92°C for 2 minutes, then at 37°C overnight. The tissues were washed with gradually decreasing (50%, 40%, 20%) formamide concentration, counterstained with anti-LacI antibodies and DAPI, and mounted on to glass coverslips.
RESULTS
Expression of Lamin C ΔN causes nuclear organization and muscle defects
In previous studies, we demonstrated muscle sensitivity to mutant forms of Drosophila A-type lamin (Schulze et al., 2009). Here, we extend these studies by testing the effects of expressing Lamin C ΔN in additional tissues and stages of development, with emphasis on muscle. Expression of wild-type Lamin C caused no aberrant morphological phenotype or loss of viability for all drivers tested (Table 1). By contrast, expression of Lamin C ΔN caused semi- or complete lethality when expressed by the larval muscle drivers tested but not by adult muscle or non-muscle drivers (Table 1). Lethality was attributed to the loss of the head domain and not overexpression; western analysis showed that wild-type Lamin C and Lamin C ΔN were expressed at nearly identical levels (see Fig. S1 in the supplementary material). These data demonstrate the importance of the head domain in larval muscle.
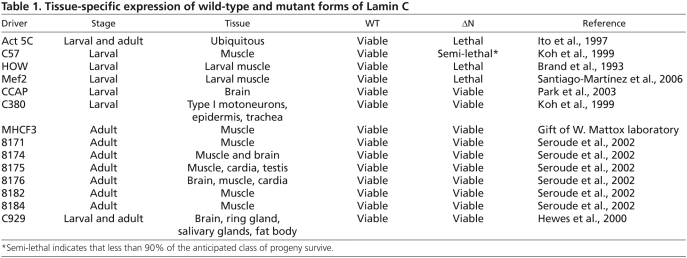
To investigate the mechanism by which Lamin C ΔN caused lethality, third instar larval body wall muscles were fixed and stained with antibodies to Lamin C. Expression of wild-type Lamin C by the C57 larval muscle-specific driver resulted in proper localization of Lamin C (both endogenous and exogenously expressed) at the nuclear periphery of normal shaped nuclei (Fig. 1A). By contrast, up to 13% of the nuclei (n=300) in larvae expressing Lamin C ΔN by the C57 driver exhibited Lamin C aggregation, misshapen and elongated nuclei in which the chromatin appeared to be highly condensed by DAPI staining (Fig. 1A). Given the disruption in A-type lamin localization, we examined the distribution of the nuclear pore complexes (NPCs; Fig. 1B). Larvae expressing wild-type Lamin C showed the anticipated rim pattern of staining around the nucleus (Fig. 1B). By contrast, nuclei of larvae expressing Lamin C ΔN showed NPC aggregation that was enriched at one side of the nucleus in 9% of the nuclei (n=300; Fig. 1B). Lamins interact with LEM domain proteins; therefore, we investigated the localization of Otefin, a potential homolog of mammalian emerin. Larval muscle expressing wild-type Lamin C stained with anti-Otefin antibodies showed peripheral staining around the nucleus. By contrast, the muscle cells of larvae expressing Lamin C ΔN showed aggregation of Otefin in 11% of the nuclei (n=300; Fig. 1C). Lamin proteins interact with the nuclear envelope proteins containing SUN domains (Haque et al., 2006; Hasan et al., 2006; Mejat et al., 2009); therefore, we examined the localization of Klaroid, a Drosophila SUN domain protein (Kracklauer et al., 2007). Klaroid localized to the nuclear periphery in larval muscle nuclei expressing wild-type Lamin C (Fig. 1D). In larval muscle expressing Lamin C ΔN, nuclear aggregates of Klaroid were apparent in 12% of the nuclei (n=300). In addition, atypical cytoplasmic localization was observed (Fig. 1D). Thus, the localization pattern of nuclear proteins belonging to different classes was disrupted upon expression of the headless A-type lamin.
Fig. 1.
Larval muscle expression of Lamin C ΔN causes alterations in nuclear organization and muscle defects. (A-D) Staining of larval body wall muscle with anti-Lamin C (A, green), anti-NPC (B, green), anti-Otefin (C, green) or anti-Klaroid (D, green) antibodies, Phalloidin (purple) and DAPI (blue) from larvae expressing wild-type Lamin C (wt) or Lamin C ΔN (ΔN) transgenes by the muscle-specific driver C57.
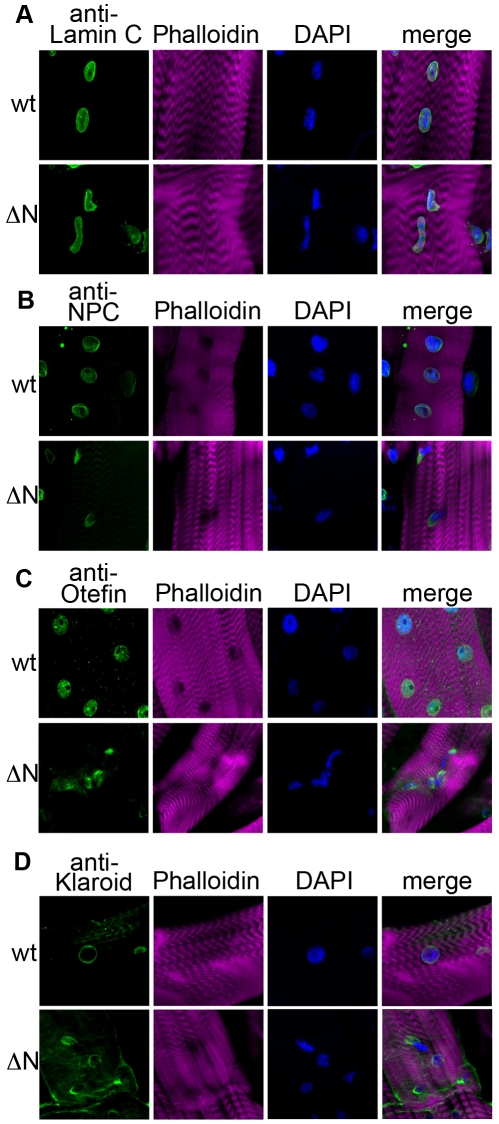
In mammals, SUN domain proteins interact with nesprin proteins within the perinuclear space (Padmakumar et al., 2005). Nesprins extend into the cytoplasm and associate with cytoskeletal components such as actin (Crisp et al., 2006). The mislocalization of Klaroid upon expression of Lamin C ΔN suggested the possibility that cytoplasmic connections might be disrupted. Therefore, body wall muscle from larvae expressing wild-type Lamin C and Lamin C ΔN were stained with antibodies to tyrosinated tubulin, which recognizes newly incorporated tubulin. In the case of wild-type Lamin C, a dense meshwork of microtubules was detected around the nucleus (Fig. 2). This localization pattern is similar to that observed in non-transgenic larvae (data not shown). By contrast, muscle from larvae expressing Lamin C ΔN showed an absence of tyrosinated tubulin meshwork around the misshapen nuclei (n=144; Fig. 2). Given these alterations in tubulin localization, we assayed the organization of actin using Phalloidin, which recognizes filamentous actin. Larvae expressing wild-type Lamin C showed a typical striated muscle fiber pattern expected of Phalloidin staining, whereas ~21% of the muscles in larvae expressing Lamin C ΔN showed disruptions in the striated pattern (n=180; Fig. 2). In addition, Phalloidin staining fibers were observed in ~27% of the nuclei of the Lamin C ΔN expressing larvae (n=1157; Fig. 2), suggesting the presence of actin filament formation in the nucleus, similar to those observed in Lamin C null mutants (Schulze et al., 2009). Such fibers were not apparent in nuclei expressing wild-type Lamin C. These data are consistent with Lamin C regulating actin polymerization in the nucleus, a hypothesis supported showing that lamin bundles actin in vitro (Simon et al., 2010).
Fig. 2.
Larval muscle expression of Lamin C ΔN causes alterations in cytoskeletal organization and muscle defects. Larval body wall muscle stained with an anti-tyrosinated tubulin antibody (green, left), anti-Lamin C antibody (purple, left) and Phalloidin (right) in transgenic larvae expressing wild-type Lamin C (wt) and Lamin C ΔN (ΔN) by the C57 driver. Arrows indicate the presence (wt) or absence (ΔN) of the microtubular network surrounding the nucleus. Expression of Lamin C ΔN causes nuclear actin fibers (small arrowhead) and disrupted muscle striation (large arrowhead).
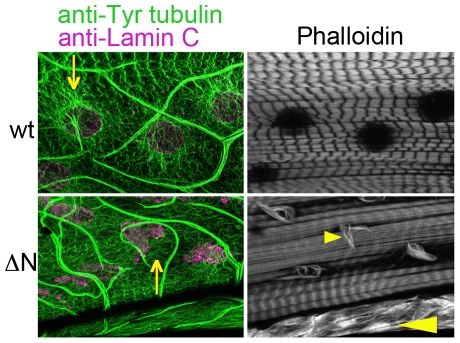
The presence of intranuclear actin filaments in larvae expressing Lamin C ΔN prompted us to examine the localization of Mlp84b (Muscle LIM protein 84b), which has the ability to bundle actin and contribute to muscle cell differentiation (Louis et al., 1997). Mlp84b is a member of the cysteine-rich protein (CRP) family, a group of cytoskeletal proteins that have conserved amino acid sequences that include one or more LIM domains, glycine-rich modules (Liebhaber et al., 1990; Louis et al., 1997; Weiskirchen and Gunther, 2003). Mlp84b is found in the nuclear and cytoplasmic compartments. Drosophila Mlp84b shares features with vertebrate MLPs including muscle-specific expression (Stronach et al., 1999). Third instar larvae expressing wild-type Lamin C in muscle stained with antibodies against Mlp84b showed proper localization resembling the sarcomeric striation pattern observed with Phalloidin (Fig. 3A). By contrast, larvae expressing Lamin C ΔN in muscle showed colocalization of Mlp84b and Phalloidin-positive actin fibers in muscle nuclei (Fig. 3A). These data suggest that, in the presence of mutant forms of Lamin C, Mlp84b might play a role in nuclear actin bundling.
Fig. 3.
Cytoskeletal elements organization is perturbed in the muscles of larvae expressing Lamin C ΔN. Staining of larval body wall muscle nuclei with (A) anti-Mlp84b or (B) anti-D-Titin antibodies (green) and Phalloidin (purple) in larvae expressing wild-type (wt) or Lamin C ΔN (ΔN) by the muscle-specific driver C57. Expression of Lamin C ΔN causes accumulation of nuclear Mlp84b that colocalizes with nuclear actin fibers (yellow arrow, A) and a network of D-Titin around the nucleus (yellow arrows, B).
These findings prompted us to examine the localization of Titin, a cytoskeletal protein that cooperates with Mlp84b to maintain muscle structural integrity (Clark et al., 2007). Titin binds lamins and has been proposed to contribute to nuclear organization during interphase (Zastrow et al., 2006). Drosophila Titin (D-Titin) is a large spring-like protein that shares sequence identity with vertebrate titins, spans half the length of the sarcomere (Clark et al., 2007) and localizes to both the cytoplasm and nucleus in early Drosophila embryos (Machado and Andrew, 2000). Staining of third instar larvae expressing wild-type Lamin C in muscle with antibodies against D-Titin showed a typical striation pattern within the cytoplasm with no nuclear defects (Fig. 3B). Larvae expressing Lamin C ΔN in muscle showed filamentous structures surrounding the muscle nuclei without any detectable Titin staining of the nuclear Phalloidin rods (Fig. 3B). Thus, D-Titin localization is disrupted by the expression of Lamin C ΔN, but does not appear to play a direct role in nuclear actin filament formation.
Larval muscle-specific expression of Lamin C ΔN causes defects in leg morphogenesis
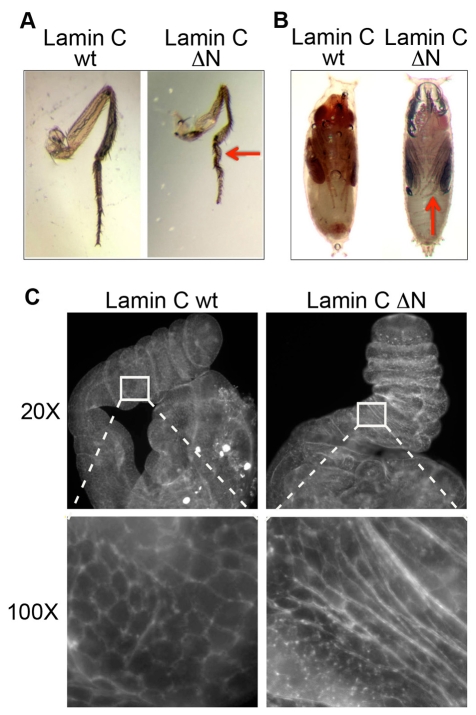
Expression of Lamin C ΔN using the GAL4 larval muscle-specific drivers frequently resulted in complete or semi-lethality, with at most 10% of the anticipated progeny surviving to adulthood (Table 1). The adult ‘escapers’ appeared normal upon expression of Lamin C ΔN with most drivers. However, in the case of the C57 driver, more than 50% of the escapers (n=326) showed defects in the third leg. These flies had a correct number of legs, all of which were properly segmented; however, the femur and tibia of the third leg were shorter and thinner in flies expressing Lamin C ΔN compared with flies expressing wild-type Lamin C. The basitarsus and the last tarsal segments were twisted and shorter in flies expressing Lamin C ΔN (Fig. 4A). These defects were apparent at the pupal stage, where the legs were observed to curl around the wing, rather than point to the distal end of the pupal case, as in wild type (Fig. 4B). Thus, Lamin C ΔN expression in the larval muscle hindered third leg morphogenesis.
Fig. 4.
Larval muscle-specific expression of Lamin C ΔN causes leg defects in adult legs and leg imaginal discs. (A,B) Third leg from adult flies (A) and pupae (B) expressing wild-type Lamin C (wt) or Lamin C ΔN (ΔN) by the C57 driver. Note the twisting of the leg in pupae and adult flies (red arrows) expressing Lamin C ΔN. (C) Leg discs from 6-hour pupae expressing wild-type (Lamin C wt) or N terminal deletion of Lamin C (Lamin C ΔN) by the C57 driver were dissected, fixed and stained with Phalloidin, which outlines the cell borders. Note the shortened leg length and unusual cell shape upon expression of Lamin C ΔN.
The malformed adult leg phenotype suggested defects during leg disc elongation (Kiss et al., 1988; von Kalm et al., 1995). To determine when during morphogenesis this defect became apparent, we examined leg imaginal discs from wild-type and mutant prepupae. The shape of the leg imaginal disc cells can be examined by Phalloidin staining (Fortier et al., 2003). Discs from carefully staged pupae expressing wild-type Lamin C and Lamin C ΔN via the C57 driver were dissected and stained with Phalloidin to visualize filamentous actin. Discs were dissected six hours after puparium formation (APF). Pupae that developed from larvae expressing Lamin C ΔN via the C57 driver possessed leg discs that were misshaped and stunted in elongation compared with pupae that developed from larvae expressing wild-type Lamin C (Fig. 4C). In high magnification images of the control and Lamin C ΔN mutant leg discs, differences in cell shape were observed (Fig. 4C).
Leg defects are consistent with alterations in ecdysone signaling
Leg imaginal discs are determined in embryogenesis and proliferate during larval development (Cohen et al., 1993). Leg elongation, morphogenesis and terminal differentiation depend on the steroid hormone ecdysone (20-hydroxyecdysone) (Fristrom et al., 1993; von Kalm et al., 1995). Ecdysone induces early response genes, which in turn regulate late genes directing downstream biological responses. The leg phenotype observed in pupae and adult escapers expressing Lamin C ΔN by the C57 driver was remarkably similar to the phenotype observed in βFtz-F1 mutant flies (Broadus et al., 1999). βFTZ-F1 is an orphan nuclear receptor expressed throughout the organism during larval molts and briefly during the mid-prepupal period (Lavorgna et al., 1993; Woodard et al., 1994; Yamada et al., 2000). Unlike other hormone-induced genes that are expressed upon maximum hormone levels, βFtz-F1 is induced after ecdysone levels begin to decline. This temporal expression is necessary for the progression of metamorphosis (Broadus et al., 1999; Woodard et al., 1994; Yamada et al., 2000).
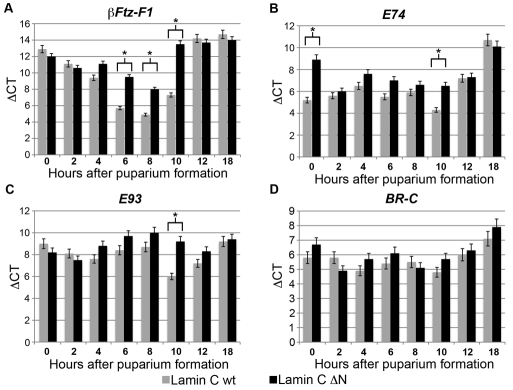
Given the striking similarity in phenotype of the Lamin C ΔN-expressing flies and the βFtz-F1 mutant, we examined the βFtz-F1 mRNA levels in flies expressing wild-type Lamin C and Lamin C ΔN by the C57 driver. RT-PCR analyses were performed on total RNA isolated from staged pupae staged at 2-hour intervals post-puparium formation. Quantitation of steady-state levels of mRNA were represented as ΔCT between the βFtz-F1 mRNA and mRNA from rp49 used for normalization. ΔCT inversely correlates with copy number of an mRNA species; therefore, more abundant mRNAs have lower ΔCT values than less abundant mRNAs. When wild-type Lamin C was expressed by the C57 driver, βFtz-F1 is expressed during the mid-prepupal period of development (6-8 hours APF; Fig. 5). This temporal pattern of regulation is consistent with that published for wild-type flies (D'Avino and Thummel, 1999). In flies expressing Lamin C ΔN via the C57 driver, βFtz-F1 mRNA levels remained low at mid-prepupal phase (6-8 hours APF), suggesting a failure of induction (Fig. 5). βFTZ-F1 acts as a competence factor for ecdysone induction of early genes E74, E93 and BR-C (Woodard et al., 1994). Loss-of-function alleles of βFtz-F1 result in pupal lethality, with reduced expression of E74, E93 and BR-C in the late prepupal stage (Broadus et al., 1999). Given the lack of induction of βFtz-F1 mRNA upon expression of Lamin C ΔN, we analyzed the expression levels of these downstream genes using RT-PCR. In flies expressing wild-type Lamin C, both E74 and E93 genes are maximally transcribed at mid-to-late prepupal stage (10 hours APF). At this stage, flies expressing Lamin C ΔN showed suboptimal levels of induction of E74 and E93 mRNA (reduced by ~8-fold). By contrast, BR-C mRNA was present at similar levels in wild-type Lamin C- and Lamin ΔN-expressing flies (data not shown). A possible explanation for the lack of change in BR-C expression is that activation can occur via βFTZ-F1-dependent and -independent pathways (Broadus et al., 1999; Karim and Thummel, 1992). Collectively, these findings show that larval muscle-specific expression of mutant Lamin C ΔN results in lack of induction of βFtz-F1 at the prepupal stage of development, which in turn fails to induce E74 and E93 early response genes.
Fig. 5.
Expression of ecdysone-responsive genes in staged pupae. Quantitative RT-PCR analyses of mRNA from ecdysone-responsive genes. The y-axis indicates steady-state levels of (A) βFtz-F1,(B) E74, (C) E93 and (D) BR-C mRNA as ΔCT normalized to rp49 CT. The x-axis shows time relative to puparium formation (set at zero). Asterisk indicates P≤0.001 as calculated by an unpaired Students' t-test.
A role for Lamin C in gene regulation
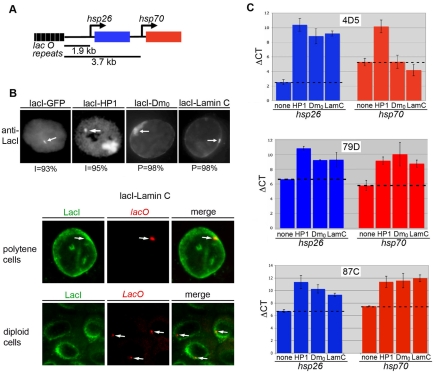
Given that expression of Lamin C ΔN caused alterations in gene expression, we were interested in determining whether Lamin C could directly regulate gene expression. Lamins participate in genome organization and gene regulation. Lamina-genome interactions control gene expression during lineage commitment and terminal differentiation (Peric-Hupkes et al., 2010; Schneider and Grosschedl, 2007). The nuclear periphery is frequently lined with heterochromatin and is generally considered as a transcriptionally repressive environment. However, recent studies have suggested that in some cases active genes reside near the NE, perhaps in microdomains (Luo et al., 2009; Shimi et al., 2008). In the absence of known direct target genes for Lamin C, we developed a system that allowed us to determine the consequences of association of Lamin C with a reporter gene. This system involved the generation of ‘expressor’ stocks in which the hsp70 promoter was used to drive expression of a fusion protein comprising the DNA binding domain of the E. coli LacI repressor and full-length Lamin C. Using this system, we targeted Lamin C to a site upstream of two reporter genes (Fig. 6A). To determine whether effects were specific for A-type lamins, we generated a stock expressing the DNA-binding domain of the LacI repressor fused to Lamin Dm0. A stock expressing green fluorescent protein (GFP) fused to the DNA binding domain of the LacI repressor served as a negative control (Danzer and Wallrath, 2004). A stock expressing a fusion of LacI and Heterochromatin protein 1 (HP1) served as a positive control for gene silencing (Danzer and Wallrath, 2004). Each reporter stock possessed a P-element transposon containing a tagged version of hsp26 (hsp26-tag) and an hsp70-white reporter gene (Fig. 6A). Two heat-shock reporter genes were present at 1.9 and 3.7 kb distances downstream of 256 copies of lac operator repeats (Danzer and Wallrath, 2004; Robinett et al., 1996) (Fig. 6A). Reporter stocks differed only by the site of insertion of the lac-hsp26-hsp70 transposon as described in Table S2 in the supplementary material. Flies from the expressor stocks were crossed to flies from three separate reporter stocks. Progeny that resulted from crossing the expressor flies to the reporter flies were examined for nuclear position of the reporter genes and gene expression. Immunofluorescence staining using LacI antibodies was performed on polytene third instar salivary gland nuclei and diploid imaginal disc nuclei. The nuclear position of the reporter gene was scored as peripheral when intense staining at the reporter gene colocalized with anti-LacI staining at the nuclear periphery. Antibodies against LacI showed a focus of staining that localized within the nuclear interior in 93% of the cells expressing GFP-LacI (n=150; Fig. 6B). Similarly, a focus of staining was observed within the nuclear interior of 95% of cells expressing LacI-HP1 (n=150; Fig. 6B). In addition, these nuclei also showed staining at the chromocenter (the site of fusion of the centromeres), the major site of HP1 localization, which is located at the nuclear periphery (Fig. 6B). The anti-LacI focus localized at the nuclear periphery in the remaining small percentage of the cells expressing GFP-Lac and LacI-HP1. In contrast to these findings, staining with LacI antibodies showed a single focus at the periphery in 96% and 98% of the cells expressing LacI-Lamin C and LacI-Lamin Dm0, respectively (n=150; Fig. 6B). This single focus at the nuclear periphery was observed in both diploid and polytene nuclei. The remaining small percentage of the cells showed localization within the nuclear interior. Fluorescent in situ hybridization using a labeled oligonucleotide recognizing the lac repeats demonstrated colocalization of the anti-LacI signal and the reporter gene at the nuclear periphery (Fig. 6B). Thus, association of the reporter gene cassette with lamins, but not with GFP or HP1, caused the reporter genes to localize at the nuclear periphery.
Fig. 6.
Effects of tethering Lamin C upstream of reporter genes. (A) Diagram of the reporter gene cassette comprising two heat-shock transgenes at 1.9 and 3.7 kb downstream of 256 copies of the lac operator. (B) Top: Salivary gland (polytene) and imaginal disc (diploid) nuclei of transgenic larvae expressing LacI-GFP, LacI-Lamin Dm0 and LacI-Lamin C tethered at genomic location 4D5 were stained with anti-LacI antibody. White arrows show tethered locus. Bottom: Fluorescent in situ hybridization using a lacO probe and staining with the LacI antibody was performed on polytene and diploid cells. White arrows indicate lacO foci. (C) Expression of heat-shock reporter genes upon tethering lamins at genomic positions 4D5 (upper), 87C (middle) and 79D (lower). RT-PCR was performed on RNA isolated from larvae possessing the reporter gene cassette alone (none) or with expression of LacI-HP1, LacI-Lamin Dm0 and LacI-Lamin C. mRNA levels of the hsp26-tag transgene (blue bars) and hsp70-white transgene (red bars) are shown as a ΔCT normalized to rp49 CT.
To determine whether peripheral localization altered gene expression, transcript levels from the reporter genes were measured using RT-PCR analyses following a brief heat-shock treatment. RT-PCR analysis was performed using primers that uniquely recognize the message from the hsp26 and hsp70 reporters (see Table S1 in the supplementary material). The results show that expression of LacI-HP1 caused silencing at hsp26 and hsp70 at the three insertion sites tested relative to the GFP-LacI control stock (Fig. 6C). These data were consistent with published findings (Danzer and Wallrath, 2004). Similar to HP1, silencing was observed upon targeting either Lamin Dm0 or Lamin C upstream of hsp26 at all genomic sites tested (Fig. 6C). Both lamins caused silencing of hsp70 at positions 79D and 87C but not at position 4D5. Taken together, these data demonstrate that lamin association caused the reporter gene to mislocalize to the nuclear periphery and be silenced in a genomic context dependent manner.
A possible explanation for the different outcomes is that the occupancy of the LacI-lamin proteins varies depending on the site of insertion. We tested for this by quantitating the fluorescent intensity of LacI-Lamin C bound at position 4D5, 79D and 87C. These sites were found to have remarkably similar levels of association (see Fig. S2 in the supplementary material), suggesting that other features within the genomic region, and not LacI-Lamin C occupancy, was the explanation for the differential effects on gene expression.
DISCUSSION
The function of the N-terminal domain of A-type lamins
The N-terminal head domain of A-type lamins in humans comprises ~33 amino acids that are not predicted to fold into a distinct structure (Herrmann and Aebi, 2004). Several studies have suggested that the head and tail domains are involved in lamin assembly (Herrmann and Foisner, 2003; Isobe et al., 2007; Sasse et al., 1998; Shumaker et al., 2005; Stuurman et al., 1998).
We have determined the consequences of expressing the Drosophila A-type lamins lacking a head domain (Lamin C ΔN) in specific tissues. Most tissues were unaffected, whereas larval muscle was sensitive (Schulze et al., 2009). Similar to the cell culture studies (Izumi et al., 2000; Spann et al., 2002; Spann et al., 1997), the muscle cell nuclei were misshapen and exhibited aggregation of both A- and B-type lamins (Fig. 1) (Schulze et al., 2009). Expression of the headless A-type lamin caused disorganization of the nuclear pore complexes, Otefin, a SUN domain protein and the actin-tubulin cytoskeletal network, suggesting physical connections exist between the nucleus and cytoplasm. The fact that Lamin C ΔN caused these phenotypic consequences in the presence of endogenous wild-type Lamin C is consistent with a dominant-negative function, as has been suggested for several laminopathies (Burke and Stewart, 2006; Gotzmann and Foisner, 2006; Muchir et al., 2004; Walter et al., 2005).
Actin in the nucleus
Expression of Lamin C ΔN caused numerous defects in larval body wall muscles. One of the most striking defects was the presence of intranuclear Phalloidin staining fibers (Fig. 2). The formation of similar nuclear rods has been observed in stressed cells (Domazetovska et al., 2007; Fukui and Katsumaru, 1979; Ohta et al., 1989; Pendleton et al., 2003; Stuven et al., 2003; Welch and Suhan, 1985), in which Cofilin serves as a nuclear import factor (Ohta et al., 1989; Pendleton et al., 2003). In the Drosophila larval body wall muscle, we observed nuclear actin rod formation in the absence of Lamin C (Schulze et al., 2009) and upon expression of Lamin C ΔN. In Lamin C nulls, actin rods were apparent in muscle, but not diploid imaginal disc tissue, suggesting that formation is possibly linked with mechanical stress (Schulze et al., 2009). These findings are consistent with Lamin C playing a role in regulating the polymerization state of nuclear actin. Actin binds two regions in the Lamin A/C tail: residues 461-536 of the Ig-fold domain (55% amino acid identity with Drosophila Lamin C) and residues 563-646 (no significant identity with Drosophila Lamin C) (Zastrow et al., 2004). A growing body of evidence proposes that NE lamina-spanning complexes are structures capable of transmitting outside mechanical signals to the genome (Razafsky and Hodzic, 2009) through interactions made between nuclear actin and lamins, Emerin or nuclear myosin 1 (Gieni and Hendzel, 2009). The possibility that actin polymers are architectural partners of lamin meshwork is gaining support (Simon et al., 2010; Zastrow et al., 2004). In connection with human disease, actin fibers in both the cytoplasm and nucleus have been observed in several myopathies (Domazetovska et al., 2007; Hutchinson et al., 2006; Sparrow et al., 2003).
Connecting Lamin C to ecdysone signaling
Overt phenotypes caused by expression of Lamin C ΔN by the C57 larval muscle-specific driver, were death and the twisted leg phenotype of adult escapers (Table 1; Fig. 3). The remarkable similarity of the leg defect to that of ecdysone mutants (Broadus et al., 1999; Ward et al., 2003) suggested a connection between Lamin C and hormone-induced gene expression. In Drosophila melanogaster, fluctuations in ecdysone concentration coordinate major developmental processes such as morphogenesis during metamorphosis, gene expression and cell death. A high ecdysone concentration at the end of third instar larva initiates leg imaginal disc elongation and eversion. During leg elongation, ecdysone induces cell shape changes, which are thought to account for the elongaton of basitarsal and tibial leg segments between 0 and 6 hours APF (Condic et al., 1991). Additionally, the contraction of larval body wall muscles is essential for leg elongation (Fortier et al., 2003). In directing leg formation and other developmental processes that occur during Drosophila metamorphosis, ecdysone acts to regulate a number of genes, including the orphan nuclear receptor βFtz-F1 (Broadus et al., 1999; Woodard et al., 1994). βFtz-F1 is expressed throughout the animal during the mid-prepupal period, reaching a peak at 8-12 hours APF (Lavorgna et al., 1993; Woodard et al., 1994; Yamada et al., 2000). βFtz-F1 induction occurs following a decline of ecdysone concentration at 3 hours APF (Broadus et al., 1999; Woodard et al., 1994; Yamada et al., 2000). βFTZ-F1, a nuclear hormone receptor, carries out many functions that include directing a muscular response to the prepupal ecdysone pulses (Broadus et al., 1999; Fortier et al., 2003) and providing the E74 and E93 early genes with the competence to respond to the high ecdysone concentration in the late prepupal stage, which direct head eversion, final leg and wing extension and shortening of the body during the prepupal-to-pupal transition. Consistent with this established transcriptional cascade, both E74 and E93 showed suboptimal induction in the mid-to-late prepupal stage (Fig. 5B,C), most likely owing to the failure of induction of βFtz-F1 at mid-prepupal stage upon expression of Lamin C ΔN (Fig. 5A). Lack of βFtz-F1 expression at this stage of development is known to limit muscle contractions necessary for leg extension (Fortier et al., 2003). Therefore, the leg phenotype resulting from reduced muscle function is probably due to two defects, disruption in the ecdysone signaling pathway and physical abnormalities in the muscle contractile apparatus.
The role of lamins in gene regulation
Lamins play a major role in genome organization and gene regulation (Schneider and Grosschedl, 2007). The nuclear periphery, which is generally considered a transcriptionally repressive environment, in some cases accommodates active genes (Luo et al., 2009; Shimi et al., 2008). Several studies have examined the interaction of lamins with DNA in living cells (Guelen et al., 2008; Pickersgill et al., 2006; Shevelyov et al., 2009). These studies used DamID, a process in which the E. coli DNA adenine methyl transferase (Dam) was fused to a B-type lamin and expressed in cultured cells. Methylated genomic DNA fragments are then presumed to be in contact with the NE. Results from such studies showed that lamin-associated domains (LADs) include large intergenic regions with a low gene density (Guelen et al., 2008).
The direct position of reporter loci near the NE in mammalian cells has been shown in three independent studies using the lacO-LacI system. Expression of LacI fused to either Lamin B1 (Kumaran and Spector, 2008), Emerin (Reddy et al., 2008) or the lamina-associated polypeptide 2β (LAP2β) (Finlan et al., 2008) were used to test the effects of tethering at the nuclear periphery on gene expression. Expression of LacI-Lamin B1 brought an inducible transgene array into juxtaposition with the nuclear lamina, but did not inhibit inducible gene activation (Kumaran and Spector, 2008). Repositioning of loci to the nuclear periphery using a LacI-Emerin fusion protein resulted in transcriptional repression of most, but not all, associated genes on the chromosome (Reddy et al., 2008). Similarly, peripheral tethering via a LAP2β-LacI fusion protein influenced the expression of some, but not all, endogenous genes near the lac operator repeats (Finlan et al., 2008). Collectively these data suggest that the repressive effects of the NE are gene-dependent. Perhaps weak promoters are silenced, whereas strong promoters retain the ability to recruit transcriptional machinery.
Our studies complement and expand upon these findings. For the first time, both A- and B-type lamins were used to tether reporter genes at the nuclear periphery. We used two reporter genes with similar promoters inserted at three different genomic sites. Targeting lamins caused the reporter gene to localize to the nuclear periphery (Fig. 6). The hsp26 promoter positioned 1.9 kb downstream of the lacO repeats was silenced by lamins at all genomic sites tested. This was not the case for the hsp70 promoter positioned 3.7 kb downstream of the repeats, which was silenced at 79D and 87C but not 4D5 (Fig. 6), despite that fact that nearly identical levels of LacI-Lamin C were associated at each genomic site (see Fig. S2 in the supplementary material). A plausible explanation for these results is that the transcriptional activity within the region 4D5 is relatively low during late third instar (the time at which RNA was collected) compared with the 79D and 87C genomic regions. The chromatin within of the region 4D5 is likely to be in a more closed configuration, as suggested by the low transcription levels (http://Flybase.org) and low abundance epigenetic modifications found in transcriptionally active chromatin (http://modENCODE.org). By contrast, region 79D shows higher levels of transcriptional activity and higher abundance of H3K9 mono- and trimethylation. Similarly, region 87C contains heat-shock-induced transcripts, which are robustly transcribed upon the heat-shock induction required to produce the LacI-Lamin proteins. Our studies demonstrate that, in addition to promoter strength, genomic context dictates the ability of a gene to be transcribed at the nuclear periphery.
Supplementary Material
Acknowledgements
We thank members of the Wallrath laboratory for comments regarding the manuscript and D. Cryderman for technical assistance. We are grateful to R. Ward (University of Kansas) and L. von Kalm (University of Central Florida) for insightful discussions on adult leg development. We are thankful for the kind gifts of reagents from K. Clark and M. Beckerle (University of Utah), D. Andrew (Johns Hopkins University School of Medicine), Y. Gruenbaum (The Hebrew University of Jerusalem), Y. Rong (NIH) and J. Fischer (The University of Texas at Austin). We are grateful for research support from the National Institutes of Health (GM61513) and the Muscular Dystrophy Association Grant (MDA3605) to L.L.W., the American Heart Association Postdoctoral Fellowship (0825794G) to G.D. and the National Institutes of Health grant (NS063228) to V.B. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.048231/-/DC1
References
- Ben-Harush K., Wiesel N., Frenkiel-Krispin D., Moeller D., Soreq E., Aebi U., Herrmann H., Gruenbaum Y., Medalia O. (2009). The supramolecular organization of the C. elegans nuclear lamin filament. J. Mol. Biol. 386, 1392-1402 [DOI] [PubMed] [Google Scholar]
- Biamonti G., Giacca M., Perini G., Contreas G., Zentilin L., Weighardt F., Guerra M., Della Valle G., Saccone S., Riva S., et al. (1992). The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell. Biol. 12, 3499-3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S., Maestrini E., Rivella S., Mancini M., Regis S., Romeo G., Toniolo D. (1994). Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 8, 323-327 [DOI] [PubMed] [Google Scholar]
- Bonne G., Di Barletta M. R., Varnous S., Becane H. M., Hammouda E. H., Merlini L., Muntoni F., Greenberg C. R., Gary F., Urtizberea J. A., et al. (1999). Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21, 285-288 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 [DOI] [PubMed] [Google Scholar]
- Broadus J., McCabe J. R., Endrizzi B., Thummel C. S., Woodard C. T. (1999). The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell 3, 143-149 [DOI] [PubMed] [Google Scholar]
- Budnik V., Zhong Y., Wu C. F. (1990). Morphological plasticity of motor axons in Drosophila mutants with altered excitability. J. Neurosci. 10, 3754-3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Gerace L. (1986). A cell free system to study reassembly of the nuclear envelope at the end of mitosis. Cell 44, 639-652 [DOI] [PubMed] [Google Scholar]
- Burke B., Stewart C. L. (2006). The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu. Rev. Genomics Hum. Genet. 7, 369-405 [DOI] [PubMed] [Google Scholar]
- Cai M., Huang Y., Ghirlando R., Wilson K. L., Craigie R., Clore G. M. (2001). Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 20, 4399-4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capco D. G., Penman S. (1983). Mitotic architecture of the cell: the filament networks of the nucleus and cytoplasm. J. Cell Biol. 96, 896-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. A., Bland J. M., Beckerle M. C. (2007). The Drosophila muscle LIM protein, Mlp84B, cooperates with D-titin to maintain muscle structural integrity. J. Cell Sci. 120, 2066-2077 [DOI] [PubMed] [Google Scholar]
- Clements L., Manilal S., Love D. R., Morris G. E. (2000). Direct interaction between emerin and lamin A. Biochem. Biophys. Res. Commun. 267, 709-714 [DOI] [PubMed] [Google Scholar]
- Cohen B., Simcox A. A., Cohen S. M. (1993). Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development 117, 597-608 [DOI] [PubMed] [Google Scholar]
- Condic M. L., Fristrom D., Fristrom J. W. (1991). Apical cell shape changes during Drosophila imaginal leg disc elongation: a novel morphogenetic mechanism. Development 111, 23-33 [DOI] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J. B., Shanahan C., Burke B., Stahl P. D., Hodzic D. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172, 41-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Avino P. P., Thummel C. S. (1999). Ectopic expression systems in Drosophila. Methods Enzymol. 306, 129-142 [DOI] [PubMed] [Google Scholar]
- Danzer J. R., Wallrath L. L. (2004). Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131, 3571-3580 [DOI] [PubMed] [Google Scholar]
- Domazetovska A., Ilkovski B., Kumar V., Valova V. A., Vandebrouck A., Hutchinson D. O., Robinson P. J., Cooper S. T., Sparrow J. C., Peckham M., et al. (2007). Intranuclear rod myopathy: molecular pathogenesis and mechanisms of weakness. Ann. Neurol. 62, 597-608 [DOI] [PubMed] [Google Scholar]
- Duffy J. B. (2002). GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34, 1-15 [DOI] [PubMed] [Google Scholar]
- Fabian L., Xia X., Venkitaramani D. V., Johansen K. M., Johansen J., Andrew D. J., Forer A. (2007). Titin in insect spermatocyte spindle fibers associates with microtubules, actin, myosin and the matrix proteins skeletor, megator and chromator. J. Cell Sci. 120, 2190-2204 [DOI] [PubMed] [Google Scholar]
- Finlan L. E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J. R., Bickmore W. A. (2008). Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4, e1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D. Z., Chaudhary N., Blobel G. (1986). cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. USA 83, 6450-6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier T. M., Vasa P. P., Woodard C. T. (2003). Orphan nuclear receptor betaFTZ-F1 is required for muscle-driven morphogenetic events at the prepupal-pupal transition in Drosophila melanogaster. Dev. Biol. 257, 153-165 [DOI] [PubMed] [Google Scholar]
- Fristrom D., Wilcox M., Fristrom J. (1993). The distribution of PS integrins, laminin A and F-actin during key stages in Drosophila wing development. Development 117, 509-523 [DOI] [PubMed] [Google Scholar]
- Fukui Y., Katsumaru H. (1979). Nuclear actin bundles in Amoeba, Dictyostelium and human HeLa cells induced by dimethyl sulfoxide. Exp. Cell Res. 120, 451-455 [DOI] [PubMed] [Google Scholar]
- Furukawa K., Hotta Y. (1993). cDNA cloning of a germ cell specific lamin B3 from mouse spermatocytes and analysis of its function by ectopic expression in somatic cells. EMBO J. 12, 97-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgatos S. D., Meier J., Simos G. (1994). Lamins and lamin-associated proteins. Curr. Opin. Cell Biol. 6, 347-353 [DOI] [PubMed] [Google Scholar]
- Gerace L., Blobel G. (1980). The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell 19, 277-287 [DOI] [PubMed] [Google Scholar]
- Gerace L., Burke B. (1988). Functional organization of the nuclear envelope. Annu. Rev. Cell Biol. 4, 335-374 [DOI] [PubMed] [Google Scholar]
- Gieni R. S., Hendzel M. J. (2009). Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem. Cell Biol. 87, 283-306 [DOI] [PubMed] [Google Scholar]
- Gotzmann J., Foisner R. (2006). A-type lamin complexes and regenerative potential: a step towards understanding laminopathic diseases? Histochem. Cell Biol. 125, 33-41 [DOI] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., et al. (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948-951 [DOI] [PubMed] [Google Scholar]
- Haque F., Lloyd D. J., Smallwood D. T., Dent C. L., Shanahan C. M., Fry A. M., Trembath R. C., Shackleton S. (2006). SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 26, 3738-3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Elbashir S. M., Bechert K., Tuschl T., Weber K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557-4565 [DOI] [PubMed] [Google Scholar]
- Hasan S., Guttinger S., Muhlhausser P., Anderegg F., Burgler S., Kutay U. (2006). Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 580, 1263-1268 [DOI] [PubMed] [Google Scholar]
- Herrmann H., Foisner R. (2003). Intermediate filaments: novel assembly models and exciting new functions for nuclear lamins. Cell. Mol. Life Sci. 60, 1607-1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H., Aebi U. (2004). Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu. Rev. Biochem. 73, 749-789 [DOI] [PubMed] [Google Scholar]
- Hewes R. S., Schaefer A. M., Taghert P. H. (2000). The cryptocephal gene (ATF4) encodes multiple basic-leucine zipper proteins controlling molting and metamorphosis in Drosophila. Genetics 155, 1711-1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson D. O., Charlton A., Laing N. G., Ilkovski B., North K. N. (2006). Autosomal dominant nemaline myopathy with intranuclear rods due to mutation of the skeletal muscle ACTA1 gene: clinical and pathological variability within a kindred. Neuromuscul. Disord. 16, 113-121 [DOI] [PubMed] [Google Scholar]
- Isobe K., Gohara R., Ueda T., Takasaki Y., Ando S. (2007). The last twenty residues in the head domain of mouse lamin A contain important structural elements for formation of head-to-tail polymers in vitro. Biosci. Biotechnol. Biochem. 71, 1252-1259 [DOI] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D. (1997). The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761-771 [DOI] [PubMed] [Google Scholar]
- Izumi M., Vaughan O. A., Hutchison C. J., Gilbert D. M. (2000). Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol. Biol. Cell 11, 4323-4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F. D., Thummel C. S. (1992). Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J. 11, 4083-4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss I., Beaton A. H., Tardiff J., Fristrom D., Fristrom J. W. (1988). Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics 118, 247-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper M., Exner K., Kempf A., Gehrig C., Stuurman N., Fisher P. A., Krohne G. (1997). Assembly of A- and B-type lamins studied in vivo with the baculovirus system. J. Cell Sci. 110, 2519-2532 [DOI] [PubMed] [Google Scholar]
- Koh Y. H., Popova E., Thomas U., Griffith L. C., Budnik V. (1999). Regulation of DLG localization at synapses by CaMKII-dependent phosphorylation. Cell 98, 353-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracklauer M. P., Banks S. M., Xie X., Wu Y., Fischer J. A. (2007). Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly 1, 75-85 [DOI] [PubMed] [Google Scholar]
- Kumaran R. I., Spector D. L. (2008). A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180, 51-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G., Karim F. D., Thummel C. S., Wu C. (1993). Potential role for a FTZ-F1 steroid receptor superfamily member in the control of Drosophila metamorphosis. Proc. Natl. Acad. Sci. USA 90, 3004-3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhaber S. A., Emery J. G., Urbanek M., Wang X. K., Cooke N. E. (1990). Characterization of a human cDNA encoding a widely expressed and highly conserved cysteine-rich protein with an unusual zinc-finger motif. Nucleic Acids Res. 18, 3871-3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Blake D. L., Callebaut I., Skerjanc I. S., Holmer L., McBurney M. W., Paulin-Levasseur M., Worman H. J. (2000). MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 275, 4840-4847 [DOI] [PubMed] [Google Scholar]
- Liu J., Rolef Ben-Shahar T., Riemer D., Treinin M., Spann P., Weber K., Fire A., Gruenbaum Y. (2000). Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell 11, 3937-3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis H. A., Pino J. D., Schmeichel K. L., Pomies P., Beckerle M. C. (1997). Comparison of three members of the cysteine-rich protein family reveals functional conservation and divergent patterns of gene expression. J. Biol. Chem. 272, 27484-27491 [DOI] [PubMed] [Google Scholar]
- Luo L., Gassman K. L., Petell L. M., Wilson C. L., Bewersdorf J., Shopland L. S. (2009). The nuclear periphery of embryonic stem cells is a transcriptionally permissive and repressive compartment. J. Cell Sci. 122, 3729-3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C., Andrew D. J. (2000). D-Titin: a giant protein with dual roles in chromosomes and muscles. J. Cell Biol. 151, 639-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manilal S., Nguyen T. M., Sewry C. A., Morris G. E. (1996). The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 5, 801-808 [DOI] [PubMed] [Google Scholar]
- McKeon F. D., Kirschner M. W., Caput D. (1986). Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature 319, 463-468 [DOI] [PubMed] [Google Scholar]
- Mejat A., Decostre V., Li J., Renou L., Kesari A., Hantai D., Stewart C. L., Xiao X., Hoffman E., Bonne G., et al. (2009). Lamin A/C-mediated neuromuscular junction defects in Emery-Dreifuss muscular dystrophy. J. Cell Biol. 184, 31-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris N. R. (2003). Nuclear positioning: the means is at the ends. Curr. Opin. Cell Biol. 15, 54-59 [DOI] [PubMed] [Google Scholar]
- Muchir A., van Engelen B. G., Lammens M., Mislow J. M., McNally E., Schwartz K., Bonne G. (2003). Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp. Cell Res. 291, 352-362 [DOI] [PubMed] [Google Scholar]
- Muchir A., Medioni J., Laluc M., Massart C., Arimura T., van der Kooi A. J., Desguerre I., Mayer M., Ferrer X., Briault S., et al. (2004). Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve 30, 444-450 [DOI] [PubMed] [Google Scholar]
- Nagano A., Koga R., Ogawa M., Kurano Y., Kawada J., Okada R., Hayashi Y. K., Tsukahara T., Arahata K. (1996). Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat. Genet. 12, 254-259 [DOI] [PubMed] [Google Scholar]
- Ohta Y., Nishida E., Sakai H., Miyamoto E. (1989). Dephosphorylation of cofilin accompanies heat-shock-induced nuclear accumulation of cofilin. J. Biol. Chem. 264, 16143-16148 [PubMed] [Google Scholar]
- Paddy M. R., Belmont A. S., Saumweber H., Agard D. A., Sedat J. W. (1990). Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell 62, 89-106 [DOI] [PubMed] [Google Scholar]
- Padmakumar V. C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A. A., Gotzmann J., Foisner R., Karakesisoglou I. (2005). The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118, 3419-3430 [DOI] [PubMed] [Google Scholar]
- Park J. H., Schroeder A. J., Helfrich-Forster C., Jackson F. R., Ewer J. (2003). Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development 130, 2645-2656 [DOI] [PubMed] [Google Scholar]
- Pendleton A., Pope B., Weeds A., Koffer A. (2003). Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J. Biol. Chem. 278, 14394-14400 [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S. W., Solovei I., Brugman W., Graf S., Flicek P., Kerkhoven R. M., van Lohuizen M., et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603-613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H., Kalverda B., de Wit E., Talhout W., Fornerod M., van Steensel B. (2006). Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005-1014 [DOI] [PubMed] [Google Scholar]
- Pinto B. S., Wilmington S. R., Hornick E. E., Wallrath L. L., Geyer P. K. (2008). Tissue-specific defects are caused by loss of the Drosophila MAN1 LEM domain protein. Genetics 180, 133-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K. M., Chan E. K., Grant B. J., Sullivan K. F., Tan E. M., Glass C. A. (1990). In vitro posttranslational modification of lamin B cloned from a human T-cell line. Mol. Cell. Biol. 10, 2164-2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D., Hodzic D. (2009). Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 186, 461-472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K. L., Zullo J. M., Bertolino E., Singh H. (2008). Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243-247 [DOI] [PubMed] [Google Scholar]
- Riemer D., Stuurman N., Berrios M., Hunter C., Fisher P. A., Weber K. (1995). Expression of Drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. J. Cell Sci. 108, 3189-3198 [DOI] [PubMed] [Google Scholar]
- Robinett C. C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A. S. (1996). In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135, 1685-1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki M., Koike H., Takahashi N., Sasagawa N., Tomioka S., Arahata K., Ishiura S. (2001). Interaction between emerin and nuclear lamins. J. Biochem. 129, 321-327 [DOI] [PubMed] [Google Scholar]
- Santiago-Martinez E., Soplop N. H., Kramer S. G. (2006). Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proc. Natl. Acad. Sci. USA 103, 12441-12446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse B., Aebi U., Stuurman N. (1998). A tailless Drosophila lamin Dm0 fragment reveals lateral associations of dimers. J. Struct. Biol. 123, 56-66 [DOI] [PubMed] [Google Scholar]
- Schirmer E. C., Florens L., Guan T., Yates J. R., 3rd, Gerace L. (2003). Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380-1382 [DOI] [PubMed] [Google Scholar]
- Schneider R., Grosschedl R. (2007). Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 21, 3027-3043 [DOI] [PubMed] [Google Scholar]
- Schulze S. R., Curio-Penny B., Li Y., Imani R. A., Rydberg L., Geyer P. K., Wallrath L. L. (2005). Molecular genetic analysis of the nested Drosophila melanogaster lamin C gene. Genetics 171, 185-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S. R., Curio-Penny B., Speese S., Dialynas G., Cryderman D. E., McDonough C. W., Nalbant D., Petersen M., Budnik V., Geyer P. K., et al. (2009). A comparative study of Drosophila and human A-type lamins. PLoS ONE 4, e7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroude L. (2002). GAL4 drivers expression in the whole adult fly. Genesis 34, 34-38 [DOI] [PubMed] [Google Scholar]
- Seroude L., Brummel T., Kapahi P., Benzer S. (2002). Spatio-temporal analysis of gene expression during aging in Drosophila melanogaster. Aging Cell 1, 47-56 [DOI] [PubMed] [Google Scholar]
- Shaffer C. D., Wuller J. M., Elgin S. C. (1994). Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol. 44, 99-108 [DOI] [PubMed] [Google Scholar]
- Shevelyov Y. Y., Lavrov S. A., Mikhaylova L. M., Nurminsky I. D., Kulathinal R. J., Egorova K. S., Rozovsky Y. M., Nurminsky D. I. (2009). The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl. Acad. Sci. USA 106, 3282-3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Pfleghaar K., Kojima S., Pack C. G., Solovei I., Goldman A. E., Adam S. A., Shumaker D. K., Kinjo M., Cremer T., et al. (2008). The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22, 3409-3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D. K., Lopez-Soler R. I., Adam S. A., Herrmann H., Moir R. D., Spann T. P., Goldman R. D. (2005). Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc. Natl. Acad. Sci. USA 102, 15494-15499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D. N., Zastrow M. S., Wilson K. L. (2010). Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus 1, 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann T. P., Moir R. D., Goldman A. E., Stick R., Goldman R. D. (1997). Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 136, 1201-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann T. P., Goldman A. E., Wang C., Huang S., Goldman R. D. (2002). Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 156, 603-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow J. C., Nowak K. J., Durling H. J., Beggs A. H., Wallgren-Pettersson C., Romero N., Nonaka I., Laing N. G. (2003). Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1). Neuromuscul. Disord. 13, 519-531 [DOI] [PubMed] [Google Scholar]
- Starr D. A., Fischer J. A. (2005). KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. BioEssays 27, 1136-1146 [DOI] [PubMed] [Google Scholar]
- Stronach B. E., Renfranz P. J., Lilly B., Beckerle M. C. (1999). Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis. Mol. Biol. Cell 10, 2329-2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N., Sasse B., Fisher P. A. (1996). Intermediate filament protein polymerization: molecular analysis of Drosophila nuclear lamin head-to-tail binding. J. Struct. Biol. 117, 1-15 [DOI] [PubMed] [Google Scholar]
- Stuurman N., Heins S., Aebi U. (1998). Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122, 42-66 [DOI] [PubMed] [Google Scholar]
- Stuven T., Hartmann E., Gorlich D. (2003). Exportin 6, a novel nuclear export receptor that is specific for profilin actin complexes. EMBO J. 22, 5928-5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C. L., Burke B. (1999). Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913-920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek M. A., Repa J. J. (2005). The power of real-time PCR. Adv. Physiol. Educ. 29, 151-159 [DOI] [PubMed] [Google Scholar]
- von Kalm L., Fristrom D., Fristrom J. (1995). The making of a fly leg: a model for epithelial morphogenesis. BioEssays 17, 693-702 [DOI] [PubMed] [Google Scholar]
- Walter M. C., Witt T. N., Weigel B. S., Reilich P., Richard P., Pongratz D., Bonne G., Wehnert M. S., Lochmuller H. (2005). Deletion of the LMNA initiator codon leading to a neurogenic variant of autosomal dominant Emery-Dreifuss muscular dystrophy. Neuromuscul. Disord. 15, 40-44 [DOI] [PubMed] [Google Scholar]
- Ward R. E., Evans J., Thummel C. S. (2003). Genetic modifier screens in Drosophila demonstrate a role for Rho1 signaling in ecdysone-triggered imaginal disc morphogenesis. Genetics 165, 1397-1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskirchen R., Gunther K. (2003). The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. BioEssays 25, 152-162 [DOI] [PubMed] [Google Scholar]
- Welch W. J., Suhan J. P. (1985). Morphological study of the mammalian stress response: characterization of changes in cytoplasmic organelles, cytoskeleton, and nucleoli, and appearance of intranuclear actin filaments in rat fibroblasts after heat-shock treatment. J. Cell Biol. 101, 1198-1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K., Ketema M., Truong H., Sonnenberg A. (2006). KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 119, 5021-5029 [DOI] [PubMed] [Google Scholar]
- Wolff N., Gilquin B., Courchay K., Callebaut I., Worman H. J., Zinn-Justin S. (2001). Structural analysis of emerin, an inner nuclear membrane protein mutated in X-linked Emery-Dreifuss muscular dystrophy. FEBS Lett. 501, 171-176 [DOI] [PubMed] [Google Scholar]
- Woodard C. T., Baehrecke E. H., Thummel C. S. (1994). A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell 79, 607-615 [DOI] [PubMed] [Google Scholar]
- Yamada M., Murata T., Hirose S., Lavorgna G., Suzuki E., Ueda H. (2000). Temporally restricted expression of transcription factor betaFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development 127, 5083-5092 [DOI] [PubMed] [Google Scholar]
- Zastrow M. S., Vlcek S., Wilson K. L. (2004). Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 117, 979-987 [DOI] [PubMed] [Google Scholar]
- Zastrow M. S., Flaherty D. B., Benian G. M., Wilson K. L. (2006). Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J. Cell Sci. 119, 239-249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.