Abstract
The cellular response to Nutlin-3, a small-molecule inhibitor of the p53 repressor MDM2, varies widely among human cancer-derived cell types. Whereas HCT116 colorectal carcinoma cells display sustained cell cycle arrest, BV173 leukemia cells undergo rapid apoptosis and other cell lines show an intermediate response. We found that the expression of the p53 target genes p21, 14-3-3σ and the microRNA miR-34a correlates tightly with the cell fate choice adopted. All three genes were strongly induced in arresting cells, but silenced in cells undergoing Nutlin-3-induced apoptosis. In contrast, key apoptotic p53 target genes were equally expressed in arresting and apoptotic cells. Interestingly, we establish that miR-34a cooperates with p21 and 14-3-3σ to override the apoptotic signals generated by p53 activation. Strikingly, p53 binding to chromatin and p53-mediated recruitment of certain coactivators to all three target loci does not vary among cell types. Instead, the cell type-specific silencing of these genes is due to enhanced p21 mRNA degradation, 14-3-3σ promoter DNA methylation and reduced processing of the miR-34a primary transcript. Thus, p53-independent events regulating expression of protein-coding genes and microRNAs within the network can define the cellular outcome of p53 activation.
Keywords: mRNA degradation, DNA methylation, microRNA processing, targeted therapies, treatment outcome
INTRODUCTION
Deciphering how complex gene networks determine cell behavior is of paramount importance for the biomedical sciences, as disruption of regulatory interactions within key networks is a hallmark of many disease states, including cancer. The p53 transcription factor enables cells to undergo cell cycle arrest, apoptosis or senescence in response to a variety of forms of cellular stress, which generates a selective pressure to inactivate this protein during tumor growth.1 In the absence of stress, cells express low levels of p53 protein due to rapid turnover mediated by the E3-ubiquitin ligase MDM2.2 Induction of cell cycle arrest by p53 is achieved mostly by transcriptional activation of the cell cycle inhibitors p21 and 14-3-3σ.3, 4 The molecular events that lead to p53-dependent apoptosis are more complex. p53 induces expression of numerous apoptotic genes involved in the mitochondrial apoptotic pathway and the death-receptor pathway, but there is also evidence for transcriptionally-independent functions of p53 in apoptosis.5
p53-inducible microRNAs constitute a newly recognized class of p53 target genes.6 miRNAs are small non-coding RNAs that post-transcriptionally regulate gene expression. miRNAs are transcribed as primary transcripts (pri-miRNAs) and then processed into mature miRNAs by the sequential action of the ribonucleases Drosha and Dicer. Mature miRNAs are incorporated into RISC (RNA-induced Silencing Complex) to inhibit expression of target mRNAs by varied mechanisms.7 Recently, miR-34a was identified as a transcriptional target of p53. The precise function of miR-34a in the p53 network is unclear at this point, as some reports implicate miR-34a in cell cycle arrest whereas others propose that miR-34a promotes apoptosis (6 and references therein).
What determines a particular cellular outcome after p53 activation? This question is of paramount importance in the clinical arena: understanding how a cell decides whether or not to die in response to p53 activation will enable us to envision therapeutic strategies to harness this pathway for selective elimination of cancer cells. The choice of cellular response to p53 activation seems to be influenced by a number of variables, including but not restricted to p53 post-translational modifications, p53 cofactors and the action of parallel signaling pathways.8 Recently, a small-molecule inhibitor of the p53-MDM2 interaction, known as Nutlin-3, has been developed.9 Nutlin-3 mimics three hydrophobic amino acids of p53 required for MDM2 binding, thus behaving as a competitive inhibitor that activates p53 in the absence of genotoxic stress. Intriguingly, Nutlin-3 treatment induces p53-dependent cell cycle arrest, rather than apoptosis, in most cell types tested.10 Here, we report that expression levels of p21, 14-3-3σ and miR-34a correlate tightly with the cellular response evoked by Nutlin-3 and we demonstrate that miR-34a plays an anti-apoptotic role in cooperation with p21 and 14-3-3σ. Most importantly, our mechanistic studies show that p53 target gene activity is ultimately defined by p53-independent regulatory events such as enhanced p21 mRNA turnover, 14-3-3σ promoter DNA methylation and pri-miR-34a processing.
RESULTS AND DISCUSSION
Distinct responses to p53 activation by Nutlin-3 correlate with differential expression of p21 and 14-3-3σ
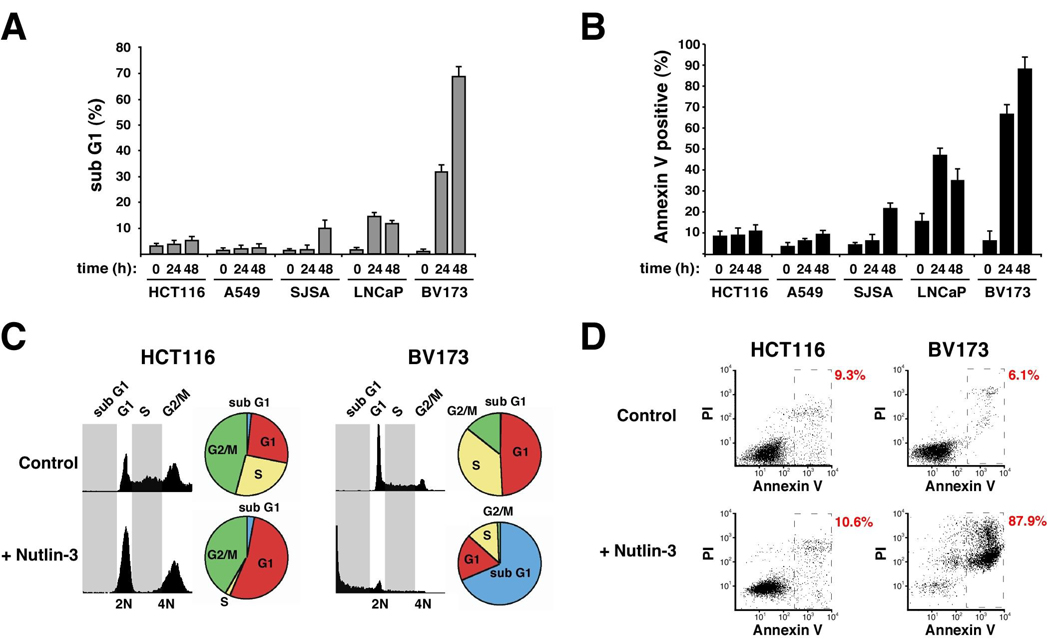
In order to study mechanisms of cell type-specific responses to p53 activation, we analyzed the cellular response to Nutlin-3 treatment in a panel of cell lines expressing wild type p53. Using FACS analysis of DNA content and phosphatidyl-serine exposure, we observed varied responses after identical treatment (Figure 1). At one end of the spectrum, HCT116 colorectal carcinoma cells show negligible signs of apoptosis even at 48 h post-treatment. DNA content profiles indicate that HCT116 cells undergo cell cycle arrest in G1 and G2/M with depletion of cells in S phase. In sharp contrast, chronic myelogenous leukemia (CML) BV173 cells undergo rapid apoptosis, as seen by increased sub G1 population and Annexin V staining. A549 lung cancer cells respond similarly to HCT116 cells, whereas SJSA osteosarcoma cells and LNCaP prostate cancer cells show an intermediate response. Expectedly, Nutlin-3 produced no effects on HCT116 p53 −/− and K562 cells (CML, p53 null) (data not shown).
Figure 1. Cell type-specific responses to Nutlin-3.
Cultures were treated with Nutlin-3 for the indicated time points before analysis of DNA content (A, C) and apoptotic index (B, D). Average levels of DNA degradation and phosphatidyl-serine exposure from three independent experiments are displayed for all five cell-lines (A, B). Raw data from one representative experiment is shown for HCT116 and BV173 cells (C-D, 48 h treatment).
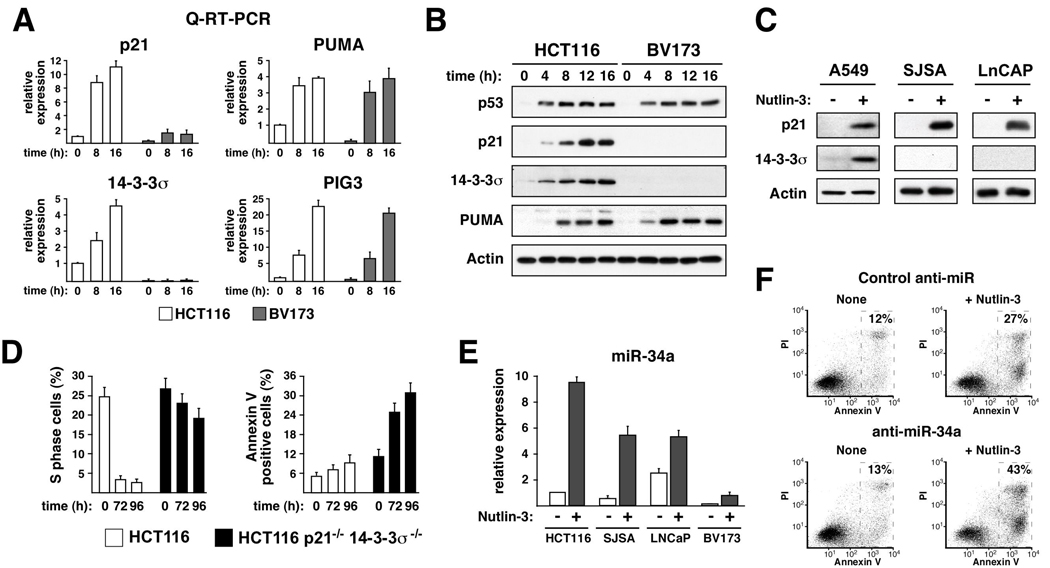
We hypothesized that differential expression of p53 target genes may drive the response adopted by different cell types. To test this, we analyzed expression of diverse p53 target genes at the mRNA and protein levels. We began with Q-RT-PCR analysis of HCT116 and BV173 cells, which revealed important differences between the two cell types (Figure 2A). Interestingly, BV173 cells display significantly lower levels of p21 and 14-3-3σ mRNAs both before and after Nutlin-3 treatment. These differences are clearly gene-specific, as the mRNAs for PUMA and PIG3 reach similar levels upon induction in both cell types. Several other p53 target genes, including MDM2, BTG2, GADD45a and FAS/APO1 also showed equivalent expression (Supp. Figure 1). Importantly, Western Blots revealed that Nutlin-3 treatment leads to equivalent p53 stabilization and PUMA induction in both cell types, yet accumulation of p21 and 14-3-36σ proteins is only observed in HCT116 cells (Figure 2B). Of note, concurrent activation of p21, 14-3-3σ and PUMA in HCT116 cells results in cell cycle arrest, suggesting that the apoptotic program is overridden in this scenario and that the dosage of cell cycle inhibitors could define the cellular response to Nutlin-3. This notion is further supported by analysis of other cell lines (Figure 2C). A549 cells, which respond similarly to HCT116 cells, express both p21 and 14-3-3σ. SJSA and LNCaP cells, which show an intermediate response, express p21 but not 14-3-3σ. Furthermore, a detailed kinetics analysis indicates that the delayed onset of apoptosis in SJSA cells correlates with decreased p21 expression over time (Supp. Figure 2). In summary, our analysis establishes a strong correlation between expression of p21 and 14-3-3σ and the cellular response adopted.
Figure 2. p21, 14-3-3σ and miR-34a protect cells from the apoptotic effects of Nutlin-3.
A. Q-RT-PCR analysis reveals differential expression of p21 and 14-3-3σ in HCT116 versus BV173 cells. Cultures were treated with Nutlin-3 for the indicated times before RNA extraction and RT-PCR. 18s rRNA was used for normalization. B. Western Blot analysis showing cell type-specific accumulation of p21 and 14-3-3σ proteins after Nutlin-3 treatment. C. Western Blot showing induction of p21 and/or 14-3-3σ expression in Nutlin-3-treated (24 h) A549, SJSA and LNCaP cells. D. Comparative analysis of parental versus double knock-out HCT116 cells demonstrates the anti-apoptotic effects of p21 and 14-3-3σ during Nutlin-3 treatment. E. Strong correlation between overall miR-34a expression and the cellular response to Nutlin-3 among cancer cell types. Cells were treated with Nutlin-3 for 16 h before extraction of small RNAs and Taqman assays. U6 snRNA was used as normalization control. F. Knock-down of miR-34a increases the apoptotic effects of Nutlin-3 in p21−/− 14-3-3−/− HCT116 cells. Control or anti-miR34a LNA-oligonucleotides were transfected 24 h prior to Nutlin-3 addition. Apoptotic indexes were performed at 96 h post-treatment. Error bars indicate standard deviations over three independent experiments.
Lack of p21 and 14-3-3σ sensitizes colorectal carcinoma cells to Nutlin-3-induced apoptosis
The anti-apoptotic effects of p21 and 14-3-3σ have been previously established when analyzing the cellular response to genotoxic p53-activating agents.11 To define the role of these two cell cycle inhibitors in cell fate choice upon non-genotoxic activation of p53, we employed HCT116 cells where both genes have been knocked-out.11 After prolonged Nutlin-3 treatment, parental HCT116 cells remain arrested and show low levels of cell death (Figure 2D). In contrast, p21−/− 14-3-3σ−/− cells show reduced arrest and increased apoptosis, thus confirming the notion that p21 and 14-3-3σ play a protective role against Nutlin-3-induced apoptosis. However, the level of apoptosis in the double knock-out cells did not reach that observed for BV173 cells, which lead us to hypothesize the existence of additional anti-apoptotic factors.
miR-34a cooperates with p21 and 14-3-3σ in cell fate choice to Nutlin-3 treatment
Recently, miR-34a was identified as a p53-inducible miRNA, but its role in cell fate choice has not been defined. Using Taqman assays, we measured miR-34a levels in several cell types (Figure 2E). As expected, miR-34a was induced by Nutlin-3 in all p53-expressing cell lines. However, important differences in overall miR-34a levels were evident, with HCT116 cells showing the highest expression and BV173 cells the lowest. Clearly, miR-34a levels correlate negatively with the apoptotic efficacy of Nutlin-3, which led us to hypothesize that miR-34a may also play anti-apoptotic roles. To test this, we utilized Locked-Nucleic Acid (LNA) technology to decrease endogenous miR-34a activity in HCT116 p21−/− 14-3-3σ−/− cells. Transfection of anti-miR-34a, but not of control oligonucleotides, enhanced Nutlin-3-induced apoptosis as seen by increased numbers of Annexin V-positive cells (27% to 43%, Figure 2F). Overall, we conclude that p21, 14-3-3σ and miR-34a collaborate to suppress the p53-dependent apoptotic program in Nutlin-3-treated cells.
These results indicate that determining the expression levels of p21, 14-3-3σ and miR-34a in a tumor sample could have prognostic value in terms of Nutlin-3 response. However, it is likely that additional factors also contribute to the heterogeneous responses to the drug. Recent reports suggest that cell lines expressing higher levels of MDM2 seem to be more susceptible to Nutlin-3-induced apoptosis10. Other reports postulate that the levels of MDM4, a p53 repressor acting independently of MDM2, can also influence the cellular response to the drug.12–14 Furthermore, cancer cells employ different strategies to block the apoptotic machinery15, which could determine the efficacy of apoptotic genes induced by Nutlin-3-activated p53. Overall, we can hypothesize that a ‘molecular signature’ capable of predicting the response to Nutlin-3 would include information on the status of p21, 14-3-3σ, miR-34a, MDM2, MDM4, as well as several components of the apoptotic machinery.
p53-dependent induction of p21 is suppressed at post-transcriptional steps in BV173 cells
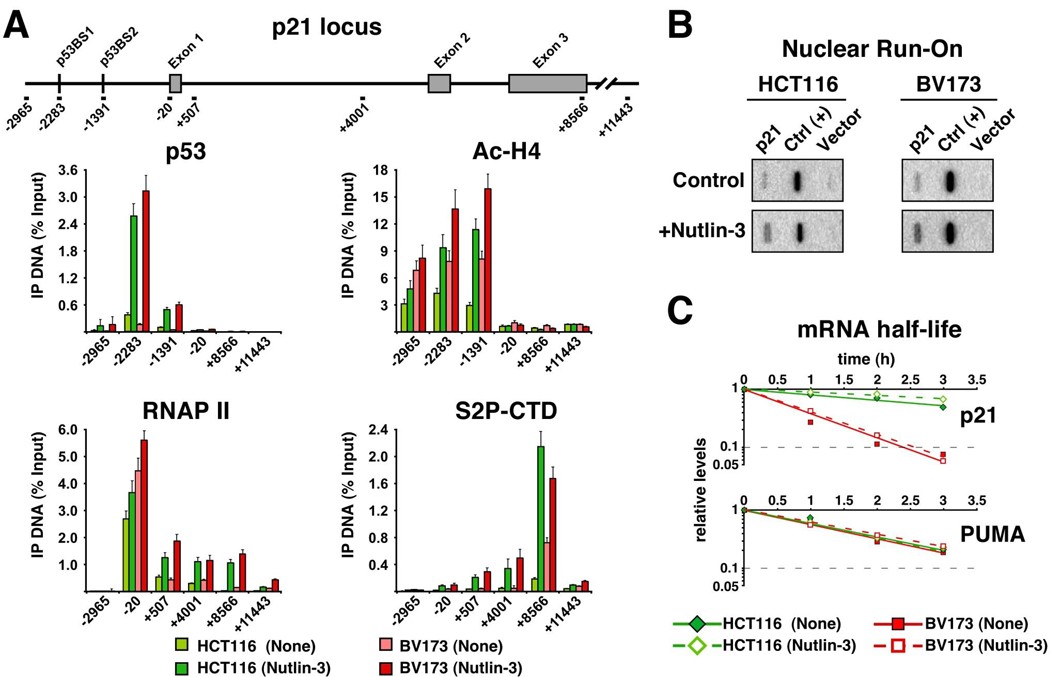
Having defined a role for p21, 14-3-3σ and miR-34a in cell fate choice upon Nutlin-3 treatment, we investigated the mechanisms driving cell type-specific expression of these genes. Influenced by our past work on stimulus-specific transcriptional regulation of p53 target genes16–18, we hypothesized that differential accumulation of p21 mRNA in HCT116 versus BV173 cells could be due to transcriptional events. To test this, we performed ChIP analysis of the p21 locus (Figure 3A). We found that induced levels of chromatin-bound p53 were equivalent in both cell types. Transcriptional activation by p53 involves recruitment of histone acetyl-transferases (HATs) to the promoter region of target genes.19 Importantly, ChIP assays detected high levels of acetylated histone H4 (Ac-H4) upstream of the p21 promoter in both cell types, suggesting that p53-mediated recruitment of HATs is not impaired in BV173 cells. Transcriptional activation of p21 occurs at post-initiation steps and involves conversion of paused RNAP II into the elongating form of the enzyme.16 Accordingly, significant amounts of pre-loaded RNAP II were detected at the p21 proximal promoter in both HCT116 and BV173 cells. Upon Nutlin-3 treatment, a significant increase in RNAP II occupancy was observed at the intragenic region in both cell types (amplicons +507, +4001 and +8566), indicative of RNAP II activation. RNAP II elongation is more easily visualized when using antibodies against phosphorylated Serine 2 within the C-terminal domain repeats of its largest subunit (S2P-CTD), a modification that accumulates toward the 3’ end of genes. Our ChIP analysis showed that S2P-CTD signal is effectively induced in both HCT116 and BV173 cells, suggesting that the p21 gene is transcriptionally activated to similar extent in both cell types.
Figure 3. p21 induction is suppressed at post-transcriptional steps in BV173 cells.
A. ChIP analysis of the p21 locus indicates that p53 binding, histone acetylation and RNAP II activation is equivalent in HCT116 and BV173 cells. Cells were treated with Nutlin-3 for 8 h before protein extract preparation. After immunoprecipitation with the indicated antibodies, ChIP-enriched DNA was analyzed by Q-PCR using the amplicons shown on top. Amplicon number indicates relative position to the transcription start site (base pairs). Error bars indicate standard deviations over three independent experiments. B. Nuclear run-on assays demonstrate that p21 mRNA is synthesized at similar rates in both HCT116 and BV173 cells. C. mRNA half-life assays indicate that the p21 mRNA half-life is much longer in HCT116 cells (~195 min) than in BV173 cells (~43 min).
To confirm that the p21 locus was transcriptionally active in BV173 cells, we performed nuclear run-on assays. These experiments detected de novo synthesis of p21 mRNA in both HCT116 and BV173 cells, with the transcription rate increasing about 3-fold upon Nutlin-3 treatment (Figure 3B). Based on these results, we hypothesized that differences in steady-state p21 mRNA levels could be due to effects on mRNA turnover. To test this, we performed mRNA half-life assays, which demonstrated that p21 mRNA half-life is drastically shorter in BV173 cells (~43 minutes) than in HCT116 cells (~195 minutes) (Figure 3C). In contrast, PUMA mRNA half-life is identical in both cell types, thus revealing the action of gene-specific mechanisms. Importantly, mRNA half-life for p21 and PUMA is similar before and after Nutlin-3 treatment, indicating that the differences in mRNA stability are not dependent on p53 activity. In summary, these results indicate that repression of p21 expression in BV173 cells occurs at post-transcriptional steps, in a p53-independent process likely involving enhanced mRNA degradation. Additional post-transcriptional mechanisms silencing p21 expression, such as translation inhibition or enhanced protein degradation, can not be discarded at this point.
Despite significant advances in the elucidation of post-transcriptional regulatory mechanisms, not much is known about their role in cell fate choice upon p53 activation. Several studies have identified cis-regulatory elements in the 3’ UTR of the p21 mRNA, as well as RNA binding proteins recognizing these sequences. For example, the RNA binding protein HuR binds to and stabilizes the p21 mRNA during myogenic differentiation.20 In contrast, binding of the AUF1 protein to the p21 3’ UTR leads to its destabilization.21 Future work will be required to identify cis-elements and trans-acting factors affecting p21 mRNA stability in different cancer cell types and, consequently, their impact on the efficacy of p53-based therapies.
14-3-3σ is repressed via promoter DNA methylation
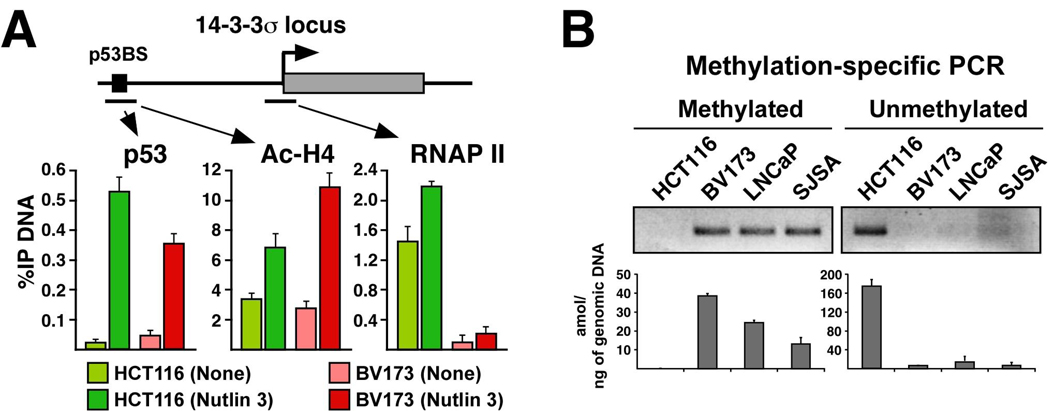
Next, we investigated the mechanism mediating differential expression of 14-3-3σ. 14-3-3σ is an intronless gene harboring a p53 response element ~1.8 kb upstream of the transcription start site (Figure 4A). As for p21, ChIP assays detected equivalent levels of p53 and increased histone acetylation at the 14-3-3σ regulatory region in HCT116 and BV173 cells (Figure 4A). However, RNAP II occupancy at the proximal promoter is greatly reduced in BV173 cells, suggesting that a repressive event downstream of p53 binding and histone acetylation is taking place at the 14-3-3σ locus in these cells. It has been reported that 14-3-3σ transcription is silenced in some cancers via aberrant DNA methylation of its promoter region.22, 23 To investigate this possibility, we performed methylation-specific PCR assays. Results in Figure 4B indicate that the 14-3-3σ promoter is indeed methylated in BV173, LNCaP and SJSA cells (none of which expresses 14-3-3σ), but not in HCT116 cells.
Figure 4. The 14-3-3σ promoter DNA is methylated in several cancer cell types.
A. ChIP analysis of the 14-3-3σ locus reveals equal binding of p53 and histone acetylation in HCT116 and BV173 cells, but absence of RNAP II in the latter. B. Methylation-specific PCR demonstrates that the 14-3-3σ promoter DNA is hypermethylated in BV173, LNCaP and SJSA cells. Genomic DNA was analyzed by PCR with primer pairs recognizing the alternative methylation states. Reactions were analyzed by agarose electrophoresis (top) and Q-PCR (bottom).
Silencing of tumor suppressor genes via promoter DNA methylation is a frequent event during cancer development. The mechanism driving 14-3-3σ promoter methylation remains to be elucidated. A recent report demonstrated that the protein kinase IKKα shields 14-3-3σ from DNA methylation, as IKKα−/− keratinocytes display repressive histone modifications and recruitment of DNA methyltransferases to this locus.24 It will be interesting to determine if differential IKKα activity contributes to cell type-specific silencing of 14-3-3σ and, consequently, to the apoptotic efficacy of Nutlin-3.
Differential expression of miR-34a is due to differences in pri-miRNA processing
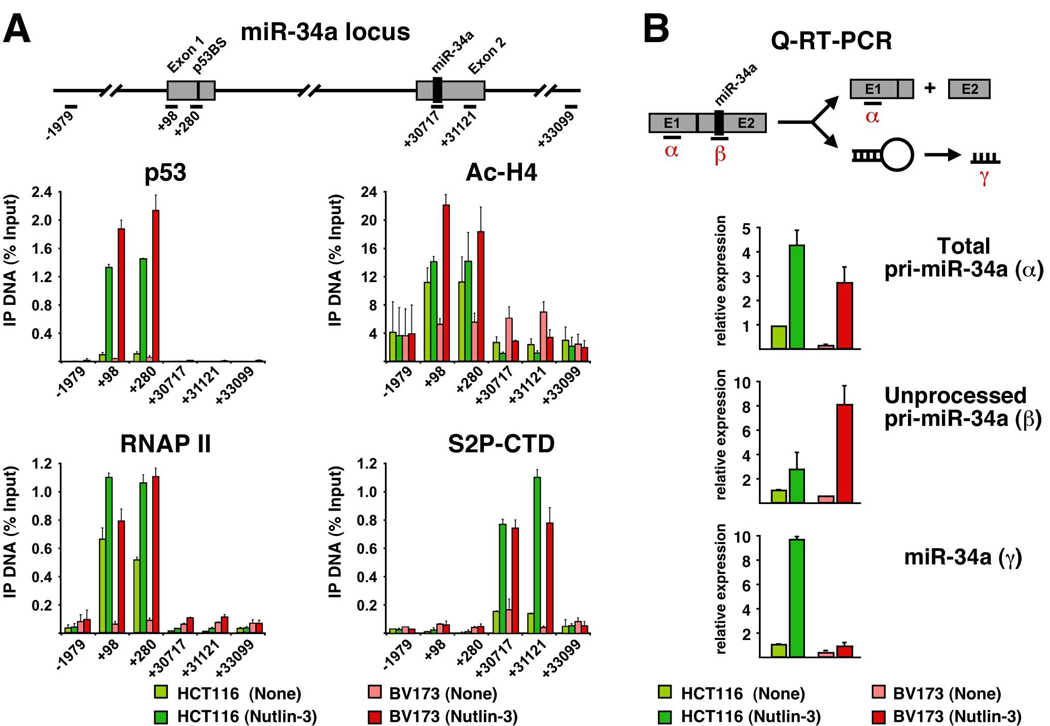
Lastly, we investigated the mechanism driving differential expression of miR-34a in HCT116 versus BV173 cells. miR-34a is encoded by its own transcription unit containing p53 binding sites within the first exon (Figure 5A). ChIP analysis revealed that Nutlin-3 induces a significant increase in chromatin-bound p53 and histone acetylation at the 5’ end of the gene in both cell types (Figure 5A). Although basal levels of total RNAP II are significantly higher in HCT116 cells, Nutlin-3 treatment leads to equivalent levels of total RNAP II in both cell lines. Most importantly, similar levels of S2P-CTD are detected at the 3’ region in Nutlin-3-treated HCT116 and BV173 cells. These results mimic those obtained for the p21 locus, and prompted us to investigate if differences in miR-34a levels could be due to post-transcriptional events. To test this, we designed Q-PCR amplicons to measure total pri-miR-34a (both unprocessed and processed, labeled α in Figure 5B) or only the unprocessed form (labeled β). We also employed Taqman assays to measure the abundance of mature, fully processed miR-34a (γ). Analysis of total pri-miR-34a supports the notion that the miR-34a locus is transcriptionally activated in both cell types, as BV173 cells accumulate significant amounts of this RNA species upon Nutlin-3 treatment. Interestingly, although BV173 cells accumulate somewhat less total pri-miR-34a than HCT116 cells, they carry significantly more of the unprocessed pri-miRNA. This suggests that pri-miRNA processing is impaired in BV173 cells, and, accordingly, they express much less of the mature form of miR-34a (γ).
Figure 5. Unequal pri-miR-34a processing among cancer cell types.
A. ChIP analysis demonstrates effective RNAP II activation at the miR-34a locus in both HCT116 and BV173 cells. B. Q-PCR analysis of pri-miR-34a expression reveals its reduced processing in BV173 cells. RNA was extracted before and 16 h after Nutlin-3 treatment and analyzed by standard Q-RT-PCR with amplicons recognizing total pri-miR-34a (α) or unprocessed pri-miR-34a (β). Levels of mature miR-34a were analyzed by Taqman assays (γ). Error bars indicate standard deviations over three independent experiments.
Little is known about the regulation of miRNA expression and activity. Most pri-miRNAs are transcribed by RNAP II and are likely to be regulated by combinatorial usage of transcription factors as observed for protein-coding genes. Here, we extend the finding that miR-34a is a p53-inducible miRNA by showing that p53 binds to the miR-34a locus, mediates histone acetylation and stimulates RNAP II activity (none of this was observed in HCT116 −/− cells, data not shown). Interestingly, our analysis indicates that cell type-specific differences in miR-34a abundance can be attributed to unequal processing of its pri-miRNA. Recent reports reveal the prevalence of regulated pri-miRNA processing.25, 26 For example, the levels of several mature miRNAs are upregulated >1000-fold during mouse development without increase in the levels of the corresponding pri-miRNAs.26 Regulated pri-miRNA processing could involve editing of the pri-miRNA by adenosine deaminases27, or even blockade of Drosha-mediated cleavage by RNA binding proteins.28 Our results indicate that studies aimed at elucidating the precise mechanism(s) mediating cell type-specific processing of miR-34a, and perhaps other p53-inducible miRNAs, could lead to the identification of additional factors affecting cell fate choice to p53 activation.
Final Remarks
As our understanding of the mechanisms driving cellular responses to various stimuli increases, so does our capacity to artificially manipulate specific molecular machineries to provoke a desired outcome. This is particularly evident in cancer biology, where the identification of precise molecular perturbations driving cancerous growth has led to effective therapies, as in the treatment of CML patients with Bcr-Abl inhibitors. Finding ways to activate p53-dependent apoptosis in cancer cells for their selective elimination is a top priority in cancer research. However, these efforts are hampered by our ignorance about the mechanisms that determine the cellular response to p53 activation. Most efforts have focused on mechanisms of gene selectivity within the p53 transcriptional program (reviewed in 29). One view proposes that p53 binds selectively to target genes as instructed by p53 post-translational modifications and/or p53-interacting proteins. An alternative model postulates that gene selectivity is achieved at regulatory steps subsequent to p53 binding to chromatin by gene- and/or stimulus-specific transcriptional coregulators. Here, we demonstrate that the p53 transcriptional response can be strongly qualified by yet a different set of p53-independent mechanisms acting at the epigenetic and post-transcriptional levels. As we understand more about this enormous regulatory diversity, we will be able to envision new therapeutic strategies to harness the power of p53 in the clinic.
MATERIALS AND METHODS
Cell culture, FACS analysis, Western Blots, ChIP assays and Q-RT-PCR reactions
were performed as previously described.17, 30 Information about antibodies and oligonucleotides can be found in Supplementary Materials.
Nuclear run-on assays
Cell cultures were treated with 10 µM Nutlin-3 (Cayman Biochemicals) or left untreated for 8 h before nuclei isolation as described previously.31 Nuclei were resuspended in 100 µl of Reaction Buffer31 and incubated at 30 °C for 15 min. Labeled RNA was isolated and hybridized to nitrocellulose filters onto which the p21 cDNA had been blotted. Filters were washed as described and analyzed by PhosphoImager.
mRNA half-life studies
Cell cultures were treated with 10 µM Nutlin-3 or left untreated for 8 h followed by the addition of the transcription inhibitor actinomycin D (ActD, Sigma) at 7.5 µg/ml. Total RNA was isolated at several time points after the addition of ActD and subjected to Q-RT–PCR reactions as described.30
Methylation-specific PCR
Genomic DNA was isolated and modified by sodium bisulfite treatment as previously described.32 Methylation-specific PCR analysis was done using alternative primer sets covering CpG dinucleotides in the 14-3-3σ promoter region (see Supplementary Table S4) and 50 ng of bisulfite-treated gDNA. For gel electrophoresis analysis, PCR reactions were stopped at 37 cycles and run on EtBr-stained agarose gels. For Q-PCR quantification, reactions were run along standard curves with known amounts of PCR products obtained from either HCT116 cells (unmethylated) or LNCaP cells (methylated) and analyzed by SYBR green incorporation.
Analysis of miR-34a expression
Levels of mature miR-34a were measured using Taqman MicroRNA Assays (Applied Biosystems). Small RNAs were purified using the miRVana miRNA Isolation Kit (Ambion). Expression was normalized to that of U6 snRNA. Levels of total and unprocessed pri-miR-34a were measured by standard Q-RT-PCR using primer pairs against sequences in Exon 1 and around Drosha cleavage sites in Exon 2, respectively, and normalized to 18s rRNA.
Knock-down of miR-34a activity
miR-34a activity was inhibited in cultures of HCT116 p21−/− 14-3-3σ−/− using a specific Anti-miR™ oligonucleotide (Ambion) targeting mature miR-34a. Control or anti-miR-34a oligonucleotides were transfected with siPORT™ NeoFX™ transfection reagent (Ambion) following manufacturer’s instructions. Media was replenished at 24 h post-transfection −/+ 10 µM Nutlin-3 and cells were harvested 96 h later for apoptotic index assays.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tom Blumenthal for critiques on the manuscript and the Vogelstein lab for cell lines. This work was supported by grants from NIH (RO1- CA117907), DOD-CDMRP (CM05054), SPORE in Lung Cancer and March of Dimes (5-FY05-1217).
Footnotes
The authors have no financial conflict of interest.
REFERENCES
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Poyurovsky MV, Prives C. Unleashing the power of p53: lessons from mice and men. Genes Dev. 2006;20:125–131. doi: 10.1101/gad.1397506. [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 4.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 5.Schuler M, Green DR. Mechanisms of p53-dependent apoptosis. Biochem Soc Trans. 2001;29:684–688. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- 6.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 9.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 10.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, Myklebost O, Heimbrook DC, Vassilev LT. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan TA, Hwang PM, Hermeking H, Kinzler KW, Vogelstein B. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- 12.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 13.Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- 14.Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–33044. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 15.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–185. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 17.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 Is a Stimulus-Specific Positive Coregulator of p53 Target Genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donner AJ, Hoover JM, Szostek SA, Espinosa JM. Stimulus-Specific Transcriptional Regulation Within the p53 Network. Cell Cycle. 2007;6 doi: 10.4161/cc.6.21.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 20.Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. Embo J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henrique R, Jeronimo C, Hoque MO, Carvalho AL, Oliveira J, Teixeira MR, Lopes C, Sidransky D. Frequent 14-3-3 sigma promoter methylation in benign and malignant prostate lesions. DNA Cell Biol. 2005;24:264–269. doi: 10.1089/dna.2005.24.264. [DOI] [PubMed] [Google Scholar]
- 23.Kunze E, Wendt M, Schlott T. Promoter hypermethylation of the 14-3-3 sigma, SYK and CAGE-1 genes is related to the various phenotypes of urinary bladder carcinomas and associated with progression of transitional cell carcinomas. Int J Mol Med. 2006;18:547–57. [PubMed] [Google Scholar]
- 24.Zhu F, Xia X, Liu B, Shen J, Hu Y, Person M, Hu Y. IKKalpha shields 14-3-3sigma, a G(2)/M cell cycle checkpoint gene, from hypermethylation, preventing its silencing. Mol Cell. 2007;27:214–227. doi: 10.1016/j.molcel.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 25.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. Rna. 2006;12:1161–1117. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viswanathan SR, Daley GQ, Gregory RI. Selective Blockade of MicroRNA Processing by Lin-28. Science. 2008 doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008 doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjorge JD, Paterson AJ, Kudlow JE. Phorbol ester or epidermal growth factor (EGF) stimulates the concurrent accumulation of mRNA for the EGF receptor and its ligand transforming growth factor-alpha in a breast cancer cell line. J Biol Chem. 1989;264:4021–4027. [PubMed] [Google Scholar]
- 32.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.