Abstract
Inorganic ions may direct the self-assembly of biomacromolecules into nanostructures which can further be used as a reactant and matrix for nanomaterials synthesis and assembly. Here we use bone mineral and filamentous bacteriophage as a model to demonstrate this concept. Divalent calcium ions are found to trigger the electrostatic self-assembly of anionic nanofiber-like bacteriophages into bundle structures where calcium ions are pre-organized between bacteriophage nanofibers. The resultant Ca2+-bacteriophage bundles can be separated and purified from the aqueous solution. The nanostructures of the bundles are verified by zeta potential analysis, small angle x-ray scattering and transmission electron microscopy. Because of the transcription of the bacteriophage chiral surface to the periodic alignment of pre-loaded Ca2+, the Ca2+-bacteriphage bundles can serve as both Ca sources and biotemplates to initiate the oriented nucleation and growth of nanocrystalline hydroxyapatite in phosphate solution or in simulated body fluid. This work provides new insights into biomineralization and represents a new approach to the fabrication of biomolecular-inorganic hybrid layered nanostructures.
Biomolecule-directed mineralization can lead to the formation of biomineralized hybrid structures with improved performance in nature. For example, bone is a hierarchically structured composite material, containing multiple levels of hierarchical organization.1 The lowest level of hierarchy is the organization of collagen fibrils mineralized with plate-like hydroxyapatite (HAP) nanocrystals. The collagen fibrils are self-assembled from collagen molecules with three intertwined polypeptide chains, and HAP crystals are nucleated and grown within these fibrils with their c-axis preferentially parallel to the collagen fibrils.2–4 Therefore, to fabricate a biomaterial mimicking some features of bone structures, it is highly desired to mimic both the chemical composition and the orientation of HAP crystals with respect to bio/organic scaffolds. Previous studies have demonstrated that the oriented biominerals in mineralized tissues might originate from organized assemblies of biomacromolecules.5–8 Bio/organic macromolecules or scaffolds such as collagen,9, 10 biopolymers,11, 12 synthetic polymers,13–16 and spider silks,17, 18 have been investigated to nucleate HAP or promote HAP crystal growth. These bio/organic macromolecules are involved in directing HAP mineralization by acting as templates through self-assembly.2, 19 However, these systems lack one dimensionally (1D) nanostructures and precise organization of functional groups or amino acids. To the best of our knowledge, except for HAP mineralization on synthetic self-assembling peptide amphiphiles reported by Stupp et al,20 no naturally occurring systems that posses 1D nanostructure and periodically assembled amino acids have been reported for HAP biomineralization. Also, investigation of macrobiomolecule-initiated mineralization has revealed that nucleation occurs on the surface of macromolecules with anionic groups (carboxylic acid, phosphates) exposed, which function as ion chelators to concentrate cations such as calcium ions, creating local supersaturation followed by oriented nucleation and growth of mineral crystals.21–23 But this generally accepted interpretation has never been directly demonstrated by experiments because the unavailability of Ca2+-bio/orgmolecule complex with pre-loaded Ca2+ (local supersaturating of Ca2+) that can be separated and characterized.
Herein we report the formation and characterization of Ca2+-phage complex, which is used as both a scaffold and a Ca source for oriented nucleation of HAP crystal along the long axis of filamentous phage (Figure S1) in phosphate solution or in simulated body fluid (SBF). The coordination of calcium ions on the surface of bacteriophage and the packing of the Ca2+-phage fibrils (Scheme 1) are investigated by zeta potential measurement and small angle X-ray scattering (SAXS), respectively. Due to the helically chiral surface of bacteriophage with periodically and precisely aligned anionic Glu and Asp residues (Figure S1 and Table S1), the pre-loaded calcium ions, which are chelated by the anionic amino acid residues, are expected to be aligned precisely on the bacteriophage surface. This precise control allows us to have a better understanding of the apatite crystallization process and reorganization mechanism at the interface between the protein and biomineral, which are poorly understood currently.24, 25 Furthermore, because the supersaturated calcium ions are pre-organized on the surface of bacteriophages, faster nucleation of HAP within the Ca2+-phage bundle-like scaffold in SBF than most reported bio/organic macromolecules is anticipated. This will provide a new scaffold with pre-loaded calcium ions for bone biomaterials fabrication. In addition, the divalent cations can trigger the self-assembly of bacteriophages and then function as a cation source. Therefore, the preloading of cations on anionic bacteriophages is expected to be extended to the production of other 1D and 2D nanomaterials.
Scheme 1.
Strategy for oriented nucleation and growth of HAP nanocrystals within aligned phage nanofibers. Ca2+ ions are first added to anionic phage solution to induce the formation of Ca2+-phage assembly (bundle). Then the Ca2+-phage bundles are purified by centrifugation and incubated in a phosphate solution to induce the oriented nucleation and growth of HAP within the channels of the bundles, resulting in the formation of HAP-phage assemblies.
Filamentous bacteriophage (also called phage) (Figure S1) is a filamentous and semi-flexible virus (~1 µm long and ~7 nm wide) that specifically infects bacteria and non-toxic to human beings. It can be pictured as a core-shell nanofiber with DNA as a core and an ordered array of coat protein as a sheath. We choose bacteriophage for the following reasons. First, bacteriophage is a monodisperse, one dimensional self-assembling nanofiber (Figures S1 and S2). This resembles the shape of collagen fibrils constituting the fibrous matrices in bone, offering the potential to serve as a template for forming 1D mineralized fibers. Second, the side wall of phage is self-assembled from several thousand copies of a major coat protein called pVIII (Figure S1 and Table S1). The periodically aligned peptides form a helically chiral surface. Third, the surface chemistry of the phage can be modified by genetically fusing a foreign peptide to each of the coat protein, providing the possibility to systematically study the mineralization on the scaffolds presenting different peptides. Lastly, phage is a new workforce in biomedicine as evidenced by two facts: (1) Recent human clinical trial discovered that bacteriophage will not induce obvious toxicity and immune response in human beings.26 (2) Recent animal study also suggests that the bacteriophage can carry drugs that are chemically tethered to its side wall, penetrate brain-blood barrier and then deliver the drug to the brain.27, 28 These facts suggest the non-toxicity of bacteriophage and it will be highly desirable to use phage-inorganic composites to fabricate biomaterials.
Here wild type fd and M13 bacteriophage (Figure S1 and S2) were used as examples to demonstrate the capability of phage to direct the HAP nucleation. Fd phage is a polyelectrolyte with a contour length of L = 880 nm and a diameter of ϕ = 6.6 nm, consisting of a single-stranded DNA molecule packed in a sheath of ~4000 identical coat proteins (pVIII). The solution-exposed domain of pVIII of fd phage includes negatively charged amino acids (1 Glu and 3 Asp), making wild-type fd phage anionic (Table S1).29, 30 M13 is almost identical to fd. Its pVIII is different from that of fd in one amino acid (the 12th negatively charged residue (Asp) is replaced by a polar one (Asn); See Table S1 in the Supporting Information). In addition, M13 phage has ~2700 copies of pVIII.29 Thus, the negative charge of wild-type M13 phage is fewer than that of wild-type fd phage. Both fd and M13 phage can be amplified by infecting bacteria and purified by several rounds of centrifugation and precipitation.31
Scheme 1 illustrates the methodology adopted for the preparation of Ca2+-phage complex and nucleation of oriented HAP on the scaffold of Ca2+-phage bundles. First, addition of divalent calcium ions to a solution of anionic bacteriophage induced the assembly of bacteriophages into bundles because divalent Ca2+ ions served as a bridge to cross-link the anionic bacteriophages. The assembled Ca2+-phage bundles are less water soluble as the surface charges were neutralized through the electrostatic interaction between the calcium ions and the anionic groups on bacteriophage surface. Thus the Ca2+-phage assemblies can be separated out from the aqueous solution and purified by washing with water. In addition, because of the helical assembly of uniform coat protein32 and large-length-scale chirality33 of filamentous bacteriophages (Figure S1), calcium ions are expected to orderly coordinate with carboxylic acid residues on phage surface. The site-specific nucleation and growth of HAP crystals on bacteriophage surface are initiated when the purified Ca2+-phage bundles are incubated in a phosphate solution or in SBF. Oriented nucleation and growth of HAP nanocrystals is anticipated since the exposed peptide segment that chelates with calcium ions is flexible and at a small angle with the long axis of phage, and calcium ions are orderly pre-organized on the bacteriophage surface in the Ca2+-phage scaffold.
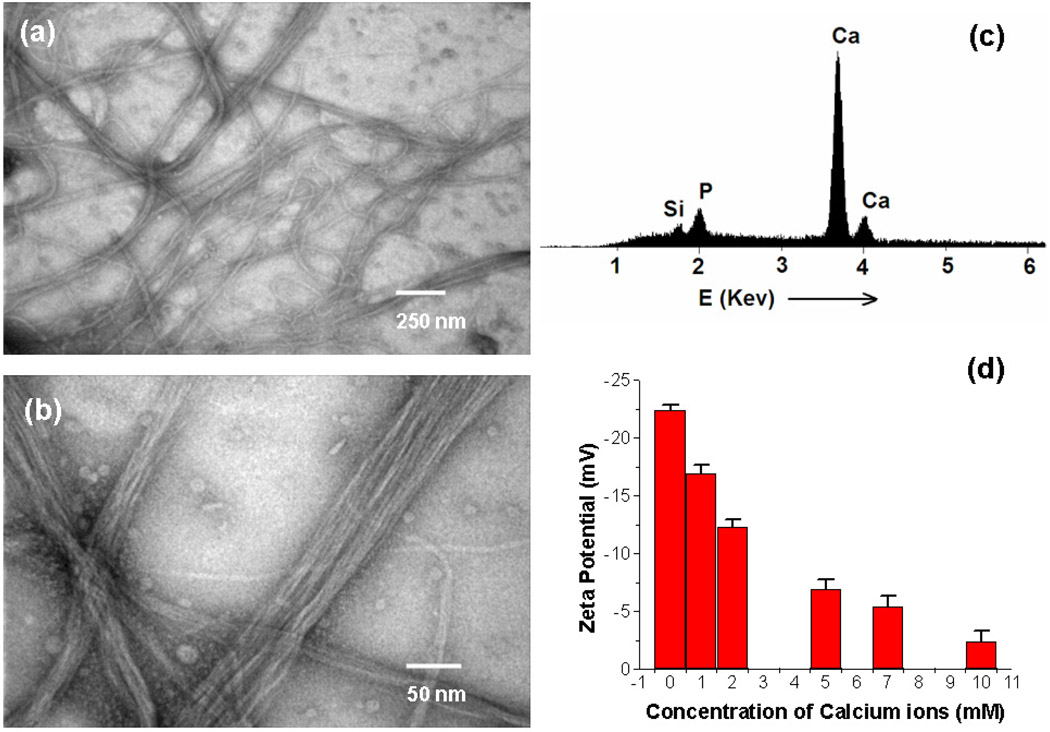
In a typical experiment, the fd bacteriophage solution (~ 2 × 1011 cfu/ml) was treated with calcium chloride (0.2 M) at room temperature. Bacteriophage solution became cloudy immediately, suggesting the aggregation of bacteriophages induced by calcium ions.34 The obtained precipitates can be purified by low-speed centrifugation (3,000 g), indicating the formation of large Ca2+-phage aggregates. The precipitates were collected and stained with 1% uranyl acetate for TEM characterization. Examination of the precipitates by TEM shows the formation of bacteriophage bundle structures, with the length up to tens micrometers (Figure 1a). Higher-magnification TEM images show parallel aligned bacteriophage fibers in these bundles (Figure 1b). In fact, the obtained bacteriophage bundles can also be visualized under TEM without uranyl stain, which was attributed to the binding of heavier calcium atoms on the surface of phage.35 The formation of bacteriophage bundles induced by Ca2+ was found to be concentration-dependent: the precipitation occurred only at a higher concentration of bacteriophages (> 5 × 1010 cfu/ml) and calcium ions (> 50 mM). At a lower concentration of Ca2+ or bacteriophage, no precipitates were observed. As will be discussed in the zeta potential measurement, although there are calcium ions coordinated to the surface of bacteriophages at lower concentration, the low molecular weight of single bacteriophage makes it difficult to precipitate out from the solution. Precipitation occurs only when tens or hundreds of bacteriophage fibers were assembled together to form a larger bundle-like aggregate. Examination of the obtained phage bundles by Fourier transform infrared spectroscopy (FTIR) revealed a broadening and shift of the −C=O stretching peaks, indicating that the carboxyl groups were ionized by the coordination of Ca2+ on the anionic bacteriophage surface (Figure S3).36 To further confirm the binding of Ca2+ ions to bacteriophage surface, the formed Ca2+-phage bundles were purified by several rounds of washing (with deionized H2O) and centrifugation, then characterized by energy-dispersive X-ray spectroscopy (EDS). The EDS analysis of the phage bundles clearly shows the presence of calcium (Figure 1c). The presence of P signal in Figure 1c is due to the existence of phosphate in bacteriophage DNA and silicon comes from the sample substrate.
Figure 1.
a) Low-magnification TEM images of fd-phage bundles (Ca2+-phage) induced by addition of CaCl2 solution (0.2 M) to phage solution. b) High-magnification TEM images of the Ca2+-(fd)phage bundles. c) EDS analysis of the formed Ca2+-(fd)phage bundles, which shows the presence of Ca and P. d) ζ potential of fd phage in aqueous solution under different concentrations of Ca2+, which shows a rapid decrease of ζ potential with increasing concentration of Ca2+.
The coordination of Ca2+ to the surface of bacteriophage was also verified by zeta potential (ζ) measurement. For example, the ζ potential was −22.4 mV for fd phages in aqueous solution with a concentration of 2 × 1010 cfu/ml. However, ζ potential of fd phages decreases with the increase of the Ca2+ concentration (Figure 1d), in the range from 1 mM to 10 mM that will not induce phage precipitation (low concentration of bacteriophage and Ca2+ was used in ζ potential measurement to avoid serious aggregation). The decrease of ζ potential suggests a strong electrostatic binding of Ca2+ to the negatively charged bacteriophage surface, which leads to a lower negative ζ potential. However, the ζ potential didn’t turn to be positive with further increase of calcium concentration, suggesting that a single layer of calcium ions is absorbed on bacteriophage surface (Figure 2). It is also interesting to note that the ζ potential changes as a function of time, indicating a kinetic process of the coordination of Ca2+ to the bacteriophage surface.
Figure 2.
Left: Comparison of SAXS profiles of pure fd phage (bottom) and Ca2+-(fd)phage bundles (top) in water. The amplified version of the SAXS profile of pure fd phage (bottom) is shown in Figure S4. Right: diagram illustrating a Ca2+-(fd)phage bundle where phage and Ca2+ layer are organized into an ordered layer structure, which explains the new peak in the SAXS profile of the Ca2+-phage bundles (see text).
To gain insight into the packing of calcium and bacteriophage in the resulting bundle structures, small angle X-ray scattering (SAXS) measurement of the phage bundles dispersed in water was carried out. Representative SAXS profiles of pure fd virus and Ca2+-(fd)phage bundles in aqueous solution are shown in Figure 2, where the scattering vector q is given by q = (4π/λ)sin(θ/2). The scattering intensity of phage bundles is significantly larger than that of pure fd phages, which was due to the scattering of heavy atom (calcium) in the bundles. For pure fd phage, three sharp peaks are available at q = 0.0425 Å−1, q = 0.0803 Å−1 and q = 0.114 Å−1 (Figure S4), which were considered to be arising from the dense lateral packing of the phage nanofibers.37 While for Ca2+-phage bundles, a very strong peak was observed at q = 0. 081 Å−1. The periodic length (d = 7.75 nm) is obtained by applying Bragg’s law to the peak position of the SAXS profile. The spacing d deduced from the peak is consistent with d = ϕfd + ϕCa, where ϕfd is the diameter of fd phage (~ 6.6 nm) and ϕCa is the hydrated ion diameter of calcium ion (~ 1 nm) (Figure 2).38 These results unambiguously show that the phage bundles adopt the parallel close-packing of the Ca2+ and rod-like fd bacteriophage as shown in Figure 2.
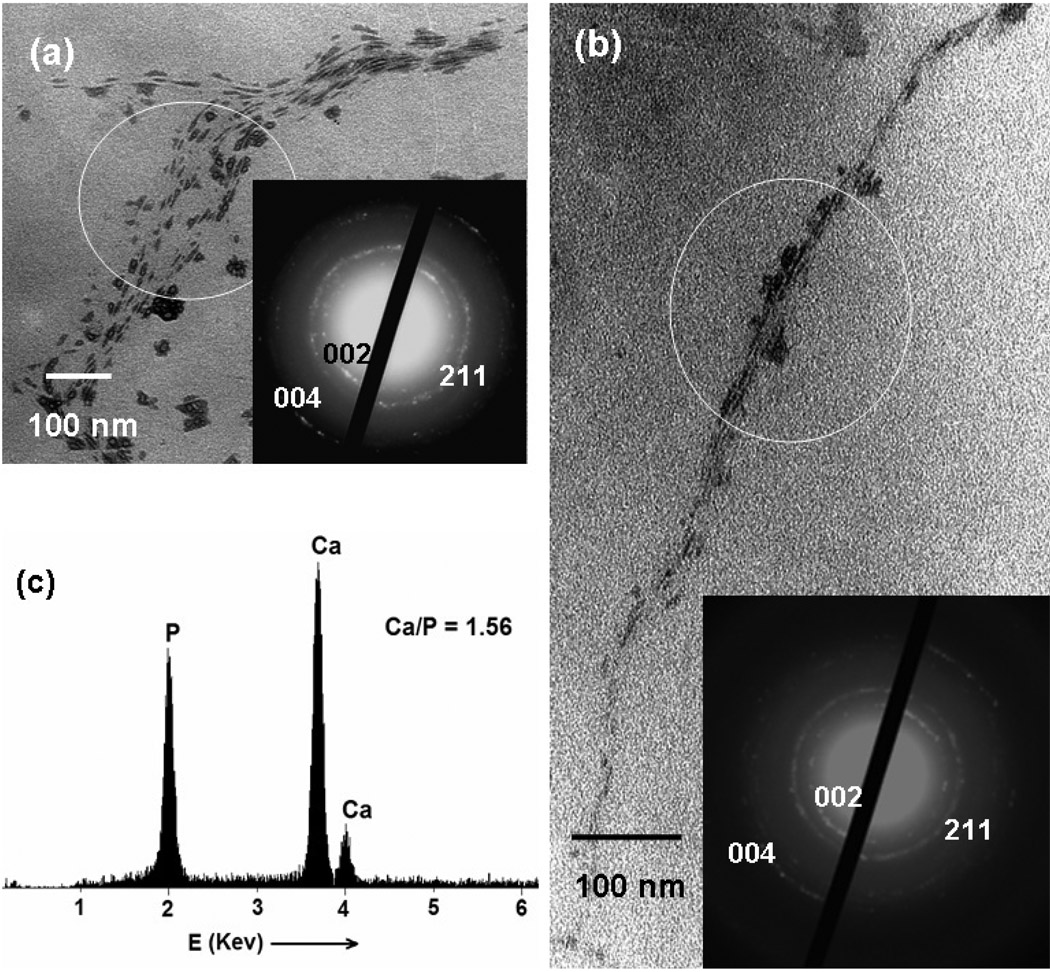
The obtained Ca2+-phage bundles formed a bulky gel after centrifugation and are slightly soluble in water. They were soaked in the phosphate solution (Na2HPO4 or Na3PO4, 0.01M, pH = 7, adjusted by ammonia) or in SBF to initiate the HAP nucleation within Ca2+-phage bundles. To get small bundles for better TEM characterization and to accelerate the penetration of other HAP precursor ions (PO43− and OH−) into the channels of the Ca2+-phage bundles, ultrasonication was applied occasionally during incubation time. After 12 h incubation at room temperature, the obtained white precipitates were collected by centrifugation and washed with DI water for several times before characterization. For fd phage, mineralized HAP-phage bundles (Figure 3a) were frequently observed. On the same TEM sample, occasionally, narrower mineralized phage bundles were found (Figure 3b). In both cases, HAP is oriented with its c-axis almost parallel to the phage, as evidenced by the corresponding selected area diffraction (SAED) analysis (Figure 3a and b, inset). In the SAED patterns, the presence of the diffraction arcs corresponding to the 002 and 004 plane indicates the c-axis preferred orientation of HAP nanocrystals. This c-axis preferred orientation is consistently found both along the individual HAP-phage bundles and among different bundles we imaged. The formation of crystalline HAP was also confirmed by XRD measurement, which shows a typical 211 Bragg peak at 31.7° (Figure S5). The mineralized bundles were analyzed by EDS, which revealed a Ca/P ratio of 1.56 ± 0.05 (Figure 3c). The ratio is consistent with the formation of HAP with a formula of Ca10(PO4)6(OH)2, if the phosphate groups in phage DNA are taken into consideration. Our results indicate that the preferred orientation of HAP results from the organized carboxyl groups on the phage surface due to the natural self-assembled structure of phage (Figure S1). The c-axis preferred orientation of HAP is similar to that in bone where the c axis of HAP nanocrystals is preferentially parallel to the long axis of the collagen fibrils.39
Figure 3.
TEM images and corresponding SAED patterns of HAP-phage nanocomposites. (a) wider and (b) narrower HAP-fd phage bundles. Insets are the SAED patterns taken from the highlighted sections. (c) EDS analysis of the HAP-phage bundle in (a). It should be noted that phages in these images are not visible under TEM because they are not stained.
The orientation control may be gained by oriented arrangement of acidic residues (Glu and Asp) on bacteriophage surface (Figure S1), which chelate Ca2+ orderly and promote oriented nucleation and growth of HAP by an epitaxial mechanism.7, 35, 40 It is well known that acidic moieties with carboxyl groups play an important role in biomineralization process.35, 41, 42 For fd phage, the viral capsid is composed of several thousand copies of helically arranged coat protein (pVIII) with a uniform tilt angle (13° < θ < 20°) between the α-helix of pVIII and the long axis of bacteriophage (Figure S1), forming a large-length-scale chiral filamentous virion.33 At the solution-exposed N-terminal end of each pVIII, 4 acidic residues (1 Glu and 3 Asp) are protruding from the phage outer surface. Therefore, the chiral ordering of pVIII on the landscape of phage will be imparted to Ca2+ alignment when Ca2+ ions are coordinated with the carboxylic groups on the bacteriophage surface. When the Ca2+-phage bundles with pre-aligned Ca2+ ions were soaked in a phosphate solution or in SBF, PO43− will be electrostatically attracted to the bacteriophage surface. This leads to an increase in the degree of supersaturation of HAP at the phage/solution interface. The high degree supersaturation and hydrogen bonding between amino acids and hydroxide ions can result in the heterogeneous nucleation of HAP on the bacteriophage surface.43 Once HAP nuclei are formed, HAP crystals will grow preferentially along bacteriophage surface from those oriented nuclei due to the chiral ordering and dense packing of pVIII (Figure S1). To confirm the transcription of the chirality of phage surface to the resulting HAP crystals, we carefully measured the angle of the connection line (i.e. average c-axis of HAP) between the middle point of (002) and (002) diffraction arc in the ED patterns and the long axis of the corresponding phage fibres derived from TEM images. Interestingly, we found the angles (10° < θ < 20°) are in a narrow range at different positions or at different area of samples on the TEM grid. This angle is also consistent with the tilt angle between the α-helix of major coat protein (pVIII) and the long axis of bacteriophage (Figure S1). Thus, we believe that the helical arrangement of the major coat protein (pVIII) leads to a highly ordered arrangement of calcium ions on phage surface, and this was transcribed to the oriented nucleation and growth of HAP crystals. Meanwhile, HAP tends to nucleate within and grow along the channels (between two neighboring phage fibers) in the bundles because the Ca2+ ions are enriched within the channels. This process will result in the oriented nucleation and growth of plate-shaped HAP nanocrystals along the bacteriophage.
To verify the above mechanism, a control experiment was carried out by the addition of phosphate and Ca2+ ions (with a molar ratio of 1.67 with [Ca2+] = 0.01M) to a fd phage solution, instead of using Ca2+-phage bundles as scaffold and Ca2+ source. The control experiment shows that no HAP crystals were found to be aligned along the phage nanofibrils (Figure S6). SAED pattern analysis showed the amorphous nature of the material, indicating that there is no orientated nucleation and growth of HAP on the surface of bacteriophage (Figure S6, inset). In another control experiment, wild type M13 bacteriophage (less anionic than fd phage) was used in the preparation of Ca2+-phage bundles. M13 phage formed similar bundle structures as shown in Figure 1. However, no oriented HAP crystallites were observed when Ca2+-(M13) phage bundles were soaked in phosphate solution or in SBF (Figure S7), which might be ascribed to the unfavorable spacing and a smaller number of anionic groups on the surface of M13 phage for HAP mineralization. One possible mechanism is that the orderly assembled acidic group of Asp12 in fd phage might not only bind calcium cations, but also regulate the orientation of the growing crystals (Table S1). These results suggest that the ordering and spacing of anionic groups within phage bundles, which leads to the pre-organization of calcium ions prior to nucleation, plays an important role in the oriented nucleation and growth of HAP on the surface of biomacromolecules.
Our experiments also show that the oriented nucleation and growth of HAP crystals on the bacteriophage can be completed in about 12 h, while most references reported that HAP growth on the bio/organic substrates take up to several days or even weeks.44, 45 The faster mineralization of HAP within the Ca2+-phage scaffolds may be due to the local supersaturation of calcium ions on bacteriophage surface by those pre-loaded calcium ions, avoiding the time-consuming kinetic process of calcium ions enrichment as in the reported macromolecules.
In addition, the divalent cation induced assembly of the bacteriophages and the mineralization of the assemblies in a counter-ion solution can be generally applied to prepare other nanoscale composites by using bacteriophage as templates. For example, Zn2+-phage bundles can be separated and purified similarly as that of Ca2+-phage bundles. After the Zn2+-phage bundles were soaked in the solution of thioacetamide (60 mM), ZnS-phage bundles were formed as shown in the supporting information (Figure S8).
In summary, we have reported the successful purification and characterization of Ca2+-phage bundles structures. The zeta potential and SAXS measurement show the monolayer absorption of calcium ions on phage surface and close-packing of bacteriophage in the bundle structures. The obtained Ca2+-(fd)phage bundles can function both as a scaffold and calcium source in the nucleation and growth of HAP. The obtained HAP-phage composites resemble some features of the lowest level of hierarchical structure of bone, in which the c-axis of HAP crystals is preferentially oriented along the long axis of filamentous bacteriophage. The present work provides direct experimental evidence for the mechanism of protein-mediated HAP nucleation and growth, and how the periodic arrangement of surface proteins was imparted to orientation of the resulting HAP crystals. The advantage of using Ca2+-phage complex for mineralization includes faster HAP nucleation due to the pre-organized calcium ions, the periodically aligned peptides on the chiral phage surface, the easy introduction of a foreign functional peptide on the side wall of phage by phage display technique,46 and the formation of mineralized bundles that can serve as building blocks for further self-assembly. Therefore, the filamentous cation-induced phage assemblies could potentially be an ideal scaffold for the fabrication of layered nanomaterials from the bottom up. Moreover, it is easy to display a foreign peptide on the surface of phage, indicating mineralized phage bundles (or scaffolds assembled from the bundles) with different peptides can be produced. Such bundles or scaffolds may be a unique system that can be used to systematically study the response of cells (e.g., mesenchymal stem cells or osteoblasts) to different peptides.
Supplementary Material
Acknowledgement
We thank the financial support from the National Science Foundation, National Institutes of Health, Department of Defense and the Oklahoma Center for the Advancement of Science and Technology. We also thank Profs. B. Grady, K. J. Dormer and Drs. H. Lu and P. Ngweniform for their kind help. Supporting Information is available online from Wiley InterScience or from the author.
Footnotes
Supporting Information Available Detailed experimental description, protein sequences, structure and morphology of filamentous phage, FT-IR spectra of Ca2+-phage complex, SAXS profiles of pure virus, TEM images of HAP obtained from a control experiments, and TEM image of ZnS-mineralized bundles. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Weiner S, Wagner HD. Ann. Rev. Mater. Sci. 1998;28:271–298. [Google Scholar]
- 2.Mann S. Nature. 1988;332:119–124. [Google Scholar]
- 3.Berman A, Ahn DJ, Lio A, Salmeron M, Reichert A, Charych D. Science. 1995;269:515–518. doi: 10.1126/science.269.5223.515. [DOI] [PubMed] [Google Scholar]
- 4.Traub W, Arad T, Weiner S. Proc. Nat. Aca. Sci. USA. 1989;86:9822–9826. doi: 10.1073/pnas.86.24.9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauerlein E. Angew. Chem. Int. Ed. 2003;42:614–641. doi: 10.1002/anie.200390176. [DOI] [PubMed] [Google Scholar]
- 6.Snead ML. Connect. Tissue Res. 2003;44:47–51. [PubMed] [Google Scholar]
- 7.Aizenberg J, Black AJ, Whitesides GM. Nature. 1999;398:495–498. [Google Scholar]
- 8.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande AS, Beniash E. Cry. Grow. Des. 2008;8:3084–3090. doi: 10.1021/cg800252f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Liao SS, Cui FZ. Chem. Mater. 2003;15:3221–3226. [Google Scholar]
- 11.Daculsi G, Pilet P, Cottrel M, Guicheux G. J. Biomed. Mater. Res. 1999;47:228–233. doi: 10.1002/(sici)1097-4636(199911)47:2<228::aid-jbm13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 12.Leonor IB, Baran ET, Kawashita M, Reis RL, Kokubo T, Nakamura T. Acta Biomaterialia. 2008;4:1349–1359. doi: 10.1016/j.actbio.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Filmon R, Grizon F, Basle MF, Chappard D. Biomaterials. 2002;23:3053–3059. doi: 10.1016/s0142-9612(02)00069-8. [DOI] [PubMed] [Google Scholar]
- 14.Murphy WL, Kohn DH, Mooney DJ. J. Biomed. Mater. Res. 2000;50:50–58. doi: 10.1002/(sici)1097-4636(200004)50:1<50::aid-jbm8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Verma D, Katti K, Katti D. J. Biomed. Mater. Res. A. 2006;78A:772–780. doi: 10.1002/jbm.a.30774. [DOI] [PubMed] [Google Scholar]
- 16.Boduch-Lee KACT, Petricca SE, Marra KG, Kumta P. Macromolecules. 2004;37:8959. [Google Scholar]
- 17.Kino R, Ikoma T, Monkawa A, Yunoki S, Munekata M, Tanaka J, Asakura T. J. App. Polym. Sci. 2006;99:2822–2830. [Google Scholar]
- 18.Cao B, Mao C. Langmuir. 2007;23:10701–10705. doi: 10.1021/la7014435. [DOI] [PubMed] [Google Scholar]
- 19.Addadi L, Weiner S. Proc. Nat. Aca. Sci. USA. 1985;82:4110–4114. doi: 10.1073/pnas.82.12.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 21.Veis A. New York: VCH; 1989. Biomineralization: Chemical and Biochemical Perspectives. [Google Scholar]
- 22.Albeck S, Weiner S, Addadi L. Chem. Eur. J. 1996;2:278–284. [Google Scholar]
- 23.Lakshminarayanan R, Kini RM, Valiyaveettil S. Proc. Nat. Aca. Sci. USA. 2002;99:5155–5159. doi: 10.1073/pnas.072658899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George A, Sabsay B, Simonian PAL, Veis A. J. Biol. Chem. 1993;268:12624–12630. [PubMed] [Google Scholar]
- 25.He G, Dahl T, Veis A, George A. Nat. Mater. 2003;2:552–558. doi: 10.1038/nmat945. [DOI] [PubMed] [Google Scholar]
- 26.Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 27.Carrera MRA, Kaufmann GF, Mee JM, Meijler MM, Koob GF, Janda KD. Proc. Nat. Aca. Sci. USA. 2004;101:10416–10421. doi: 10.1073/pnas.0403795101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickerson TJ, Kaufmann GF, Janda KD. Expert Opin. Biol. Th. 2005;5:773–781. doi: 10.1517/14712598.5.6.773. [DOI] [PubMed] [Google Scholar]
- 29.Dogic Z, Fraden S. Curr. Opin. Colloid In. Sci. 2006;11:47–55. [Google Scholar]
- 30.Webster RE. Display of Peptides and proteins. London: Academic Press; 1996. [Google Scholar]
- 31.J. Sambrook EFF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1989. [Google Scholar]
- 32.Overman SA, Tsuboi M, Thomas GJ. J. Mol. Biol. 1996;259:331–336. doi: 10.1006/jmbi.1996.0323. [DOI] [PubMed] [Google Scholar]
- 33.Tomar S, Green MM, Day LA. J. Am. Chem. Soc. 2007;129:3367–3375. doi: 10.1021/ja068498d. [DOI] [PubMed] [Google Scholar]
- 34.Tang JX, Janmey PA, Lyubartsev A, Nordenskiold L. Biophys. J. 2002;83:566–581. doi: 10.1016/S0006-3495(02)75192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiner S, Addadi L. J. Mater. Chem. 1997;7:689–702. [Google Scholar]
- 36.Sato K, Kumagai Y, Tanaka T. J. Biomed. Mater. Res. 2000;50:16–20. doi: 10.1002/(sici)1097-4636(200004)50:1<16::aid-jbm3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 37.Yang LH, Liang HJ, Angelini TE, Butler J, Coridan R, Tang JX, Wong GCL. Nat. Mater. 2004;3:615–619. doi: 10.1038/nmat1195. [DOI] [PubMed] [Google Scholar]
- 38.Vericat F, Grigera JR. J. Phys. Chem. 1982;86:1030–1032. [Google Scholar]
- 39.Kikuchi M, Itoh S, Ichinose S, Shinomiya K, Tanaka J. Biomater. 2001;22:1705–1711. doi: 10.1016/s0142-9612(00)00305-7. [DOI] [PubMed] [Google Scholar]
- 40.Mann S, Hannington JP, Williams RJP. Nature. 1986;324:565–567. doi: 10.1038/324565a0. [DOI] [PubMed] [Google Scholar]
- 41.Hunter GK, Goldberg HA. Biochem. J. 1994;302:175–179. doi: 10.1042/bj3020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer LC, Newcomb CJ, Kaltz SR, Spoerke ED, Stupp SI. Chem. Rev. 2008;108:4754–4783. doi: 10.1021/cr8004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li PJ, Ohtsuki C, Kokubo T, Nakanishi K, Soga N, Nakamura T, Yamamuro T. J. Am. Chem. Soc. 1992;75:2094–2097. [Google Scholar]
- 44.Boccaccini AR, Chicatun F, Cho J, Bretcanu O, Roether JA, Novak S, Chen Q. Adv. Funct. Mater. 2007;17:2815–2822. [Google Scholar]
- 45.Sato K, Kogure T, Kumagai Y, Tanaka J. J. Colloid In. Sci. 2001;240:133–138. doi: 10.1006/jcis.2001.7617. [DOI] [PubMed] [Google Scholar]
- 46.Liu A, Abbineni G, Mao CB. Adv. Mater. 2009;21:1001–1005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.