Abstract
The human large bowel is colonized by complex and diverse bacterial communities. However, the relationship between commensal bowel bacteria and adenomas (colorectal cancer precursors) is unclear. This study aimed to characterize adherent bacteria in normal colon and evaluate differences in community composition associated with colorectal adenomas. We evaluated adherent bacteria in normal colonic mucosa of 21 adenoma and 23 non-adenoma subjects enrolled in a cross sectional study. Terminal restriction fragment length polymorphism, clone sequencing and fluorescent in-situ hybridization analysis of the 16S rRNA genes were used to characterize adherent bacteria. A total of 335 clones were sequenced and processed for phylogenetic and taxonomic analysis. Differences in bacterial composition between cases and controls were evaluated by UniFrac and analysis of similarity matrix. Overall, Firmicutes (62%), Bacteroidetes (26%) and Proteobacteria (11%) were the most dominant phyla. The bacterial composition differed significantly between cases and controls (UniFrac p < 0.001). We observed significantly higher abundance of Proteobacteria (p < 0.05) and lower abundance of Bacteroidetes (p < 0.05) in cases compared to controls. At the genus level, case subjects showed increased abundance of Dorea spp. (p < 0.005), Faecalibacterium spp. (p < 0.05) and lower proportions of Bacteroides spp. (p < 0.03) and Coprococcus spp. (p < 0.05) than controls. Cases had higher bacterial diversity and richness than controls. These findings reveal that alterations in bacterial community composition associated with adenomas may contribute to the etiology of colorectal cancer. Extension of these findings could lead to strategies to manipulate the microbiota to prevent colorectal adenomas and cancer as well as to identify individuals at high risk.
Key words: colorectal adenoma, mucosa adherent bacteria, cancer
Introduction
The human gastrointestinal (GI) tract is colonized by complex and diverse communities of commensal microorganisms.1 The number of bacterial cells in the adult human GI tract is an order of magnitude greater than number of human cells.2 About 1011 to 1014 microbial cells reside in the human GI tract with most of the species being uncharacterized because 60–80% of them cannot be cultivated.3 Evidence from animal and human studies suggests that intestinal commensal bacteria are not innocent bystanders but rather active participants in health. These commensal bacteria play important roles in nutrient absorption, fermentation of dietary fiber, metabolism of xenobiotics, development of the mucosal immune system and protection against foreign microbes.3,4 They also contribute to regulation of cell proliferation, differentiation and gene expression in host epithelial cells.5 Abnormalities or changes in the composition of commensal bacterial communities are associated with the pathogenesis of a range of diseases including inflammatory bowel disease (IBD),6,7 colon cancer,8,9 obesity10 and multi-system organ failure.11,12 However, the bacteria in the GI tract are largely understudied in relation to adenomas (intermediate precursors) and colorectal cancer (CRC).
Commensal bacteria reside in two compartments within the large bowel, namely the luminal compartment, which consists mostly of transients, and the mucosa-adherent compartment, which consists of entrenched residents.2,13 It is important to distinguish between these two compartments because the transient and mucosa-adherent microbial communities may relate differently to the etiology of adenomas. The fecal compartment is influenced by diet,14 while the more stable adherent compartment consists of dense cohesive microbial communities that adhere to surface-associated polysaccharide matrices, resist hydrodynamic shear forces, interact with the mucosal immune system, and persist despite the rapid turnover of cells and propulsion of debris and water through the gut.13
We hypothesized that mucosal adherent bacterial community profiles would differ between adenoma patients and non-adenoma controls and that specific bacterial species might contribute to the etiology of adenomas. Given that 60–80% of the gut bacteria are uncharacterized because they cannot be cultivated ex vivo, we relied on culture-free molecular techniques to analyze these bacterial communities. Specifically, we used terminal restriction fragment length polymorphism (T-RFLP), clone sequence analysis and fluorescence in-situ hybridization (FISH) that are based on the phylogenetically conserved 16S rRNA gene to detect and identify adherent bacteria from normal colonic biopsies. By characterizing the adherent bacterial communities in the colon, we sought to understand how the presence of adenomas affects the colonic microbial ecosystem, and elucidate the relationship between these commensal bacteria and etiology of colorectal adenomas. This new knowledge might provide targets for prevention or intervention strategies that lead to reduction of CRC risk.
Results
T-RFLP analysis reveals differences in bacterial community profiles associated with adenoma cases and non-adenoma controls.
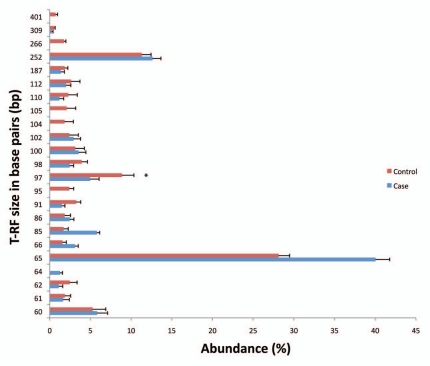
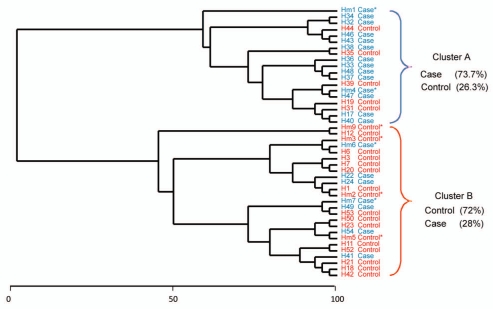
We evaluated adherent bacterial communities in normal mucosal biopsies from 44 patients (21 cases and 23 controls). The descriptive characteristics of the subjects are described in Table 1. The mean age for cases was 55.9 ± 5.8 years for cases and 53.8 ± 3.0 years for controls. We assessed several dietary factors that are relevant to colorectal cancer, however there were no statistically significant differences between cases and controls for dairy intake alcohol (g) total calcium (mg) and caloric intake (kcal). Vegetable consumption was lower in cases than controls while total fat (g) intake was higher in cases than controls. However, these results did not reach statistical significance. T-RFLP is a community fingerprinting approach that provides reliable preliminary analysis of bacteria community profiles rapidly and at low cost, but does not reveal identity of specific community members. T-RFLP analysis of adherent bacterial communities in normal colonic mucosa of adenoma and non-adenoma controls revealed 110 terminal restriction fragments (T-RFs) that ranged from 60–500 base pairs (bp), of which 23 T-RFs were predominant. T-RFs 60, 65, 97 and 252 bp were the most frequently observed (Fig. 1). The relative abundance of several T-RFs was significantly different in cases compared to controls. The abundance of T-RF 65 was 40% in cases compared to 28.1% in controls (p < 0.01) while the distribution of T-RF 97 was 4.9% for cases and 8.9% for controls, (p < 0.05). T-RFs 95, 104, 105, 266 and 401 were absent in cases but present in controls while T-RF 64 was absent in controls but present in cases. To identify patterns in T-RF bacteria community profile, we subjected the T-RF peak data to cluster analysis using T-RFLP Stats.15 We observed two main clusters (A and B) of bacterial TR-F profiles in which 73.7% of cases and 26.3% of controls were represented in one cluster and 72% of controls and 28% of cases in the second cluster (Fig. 2). This TRFLP data suggests that adenomas are associated with consistent differences in colonic adherent bacterial community profiles.
Table 1.
Descriptive characteristics of study participants
| Characteristic | Case (n = 21) | Control (n = 23) | p* |
| Age (mean, se) | 55.9 ± 5.8 | 53.8 ± 3.0 | 0.23 |
| Male (%) | 50 | 61.1 | 0.53 |
| BMI (kg/m2) (%) | |||
| <25 | 40 | 27.9 | 0.33 |
| 25–29.9 | 20 | 44.4 | |
| ≥30 | 40 | 27.8 | |
| **Constipation (%) | 0 | 13.3 | 0.21 |
| Caloric intake (kcal) (mean, se) | 2058.39 ± 178 | 1994.20 ± 177.8 | 0.80 |
| Alcohol (g) (mean, se) | 10.76 ± 2.7 | 13.76 ± 5.9 | 0.65 |
| Total calcium (mg) (mean, se) | 894.7 ± 108.9 | 1994.20 ± 177.8 | 0.91 |
| Total fat (g) (mean, se) | 82.06 ± 5.1 | 71.29 ± 8.2 | 0.27 |
| Vegetable intake (mean servings, se) | 4.31 ± 0.43 | 5.21 ± 0.86 | 0.36 |
| Dairy intake (mean, se) | 1.68 ± 0.29 | 1.57 ± 0.23 | 0.78 |
p values are based on t-test for continuous variables and chi-square statistics for categorical variables.
Constipation, defined as three bowel movements or less/week.
Figure 1.
Distribution of T-RFs in adenoma cases and non-adenoma controls. The contribution of each TRF was calculated as a percent of the total TRFs in an individual's sample. Each bar represents the average abundance (y-axis) of each TRF grouped by adenoma case or non-adenoma control status. Error bars represent standard error of the mean. Asterisks-represent TRFs that are significantly different between cases and controls. T-RF, terminal restriction fragment; size in base pairs (bp).
Figure 2.
Cluster analysis of T-RF profiles from adenoma cases and non-adenoma controls. Analysis of similarity matrix (ANOSIM) suggests modest differences between the two groups (R = 0.137, p < 0.019). Samples were standardized by calculating each TRF as a percent of the total TRFs in an individual's sample. This was followed by square root transformation and Bray-Curtis similarity matrix was used to generate a dendrogram based on group average. Sample number is presented as H##. Samples from cases and controls are indicated by color code (blue-case; red-control) and those selected for clone library analysis are marked with an asterisk.
16S rRNA sequencing reveals that adherent bacteria community members differ between adenoma cases and non-adenoma controls.
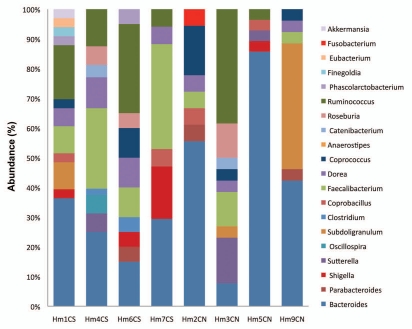
Our TRFLP analysis predicts that the adherent microbial communities in cases and controls will be relatively enriched for specific bacterial types. To test this prediction, we randomly selected eight patients (four cases and four controls) from the initial cohort of 44 patients for 16S rRNA sequence-based phylogenetic analysis. The phylogenetic distribution of the bacteria phylotypes identified among 142 clones from four controls and 200 clones from four cases are shown in Figure 3A and B. Consistent with previous reports, the adherent mucosal microbiota was dominated by few bacterial phyla (deep phylogenetic lineages, also called divisions): Firmicutes (62%), Bacteroidetes (26%) and Proteobacteria (11%) predominated, while Fusobacteria, Cyanobacteria and Verrucomicrobia comprised about 1% of all clones pooled across cases and controls.
Figure 3A.
Distribution of genus level adherent bacterial phylotypes obtained from clone libraries. Each bar represents the percent contribution of genus-level profiles grouped by case-control status (A) or for each individual (B). The colors representing the different genera are shown on the side of the figures.
Comparison of the phylogenetic composition of clone libraries using UNIFRAC analysis16,17 showed significant differences between cases and controls (p < 0.001). Significant differences were observed for bacteria communities among individuals, and between the cases and controls (p < 0.001 in both p-test and UNIFRAC significance test, corrected for multiple testing using the Bonferoni correction). Analysis of similarity matrix (ANOSIM) also suggested that bacterial composition varied by case control status (R = 0.427, p = 0.02). The UNIFRAC tree also suggests that adenoma cases have reduced inter-individual variability (Suppl. Fig. 1).
Species richness and diversity.
Phylotype was based on classification at the genus level. We assessed diversity based on measures of richness and evenness for cases and controls using 16S rRNA gene sequences from clone libraries. The mean diversity results for cases and controls are shown in Table 2. Cases tended to have higher diversity and richness compared to controls but the differences were only borderline significant. We observed that the total number of individuals was also different between cases and controls (p = 0.08). Overall, the Good's coverage for the different samples ranged from 63.6 to 86.1%. This level of coverage indicated that the 16S rDNA sequences identified represent the majority of bacterial sequences present in the human colonic mucosa from the samples under study, though the sample size was small.
Table 2.
Comparison of bacterial diversity measures between cases and controls
| Measure | Case | Control | p |
| Number of clones (mean) | 54 | 36 | |
| 1Number of Phylotypes* (mean, se) | 9.25 (1.25) | 6.75 (0.85) | 0.14 |
| 2Total number of individuals (mean, se) | 27.64 (1.8) | 22.13 (2.25) | 0.08 |
| 3Richness (mean, se) | 2.46 (0.33) | 1.84 (0.21) | 0.25 |
| 4Evenness (mean, se) | 0.96 (0.009) | 0.9 (0.03) | 0.08 |
| 5Shannon-Weiner index (mean, se) | 2.11 (0.14) | 1.71 (0.16) | 0.08 |
| 6Good's coverage | 84.26 | 75.00 | - |
Phylotype number was determined on the genus level (≥97% sequence identity).
# of phylotypes in each sample with non zero counts.
Total # individuals in each sample.
Species Richness, Margaleaf's species richness, measure of the number of phylotypes present.
Evenness-measure of how evenly the individuals are distributed among different species.
Shannon Index, measure of diversity.
Good's coverage-estimate of clone library coverage.
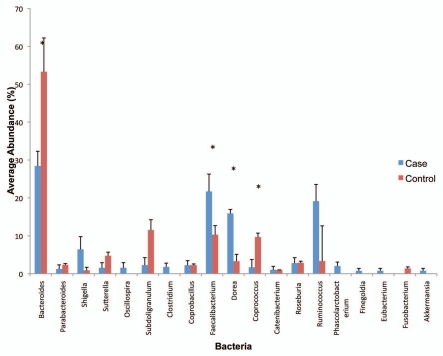
Comparison of the proportions of 16S rRNA sequences derived from adenoma cases and controls (similarity percentage analysis SIMPER, PRIMER-E,18) at the phylum and genus levels revealed the contribution of specific phylotypes to the differences between cases and controls. At the phylum level, Proteobacteria members were more abundant in cases (12.9%) compared to controls (4.85%, p < 0.05), while Bacteroidetes members were less abundant in cases (29.14%) compared to controls (37.24%, p < 0.05). Abundance of Firmicutes phylum members was similar among cases and controls, however the specific groups within the Firmicutes such as Faecalibacterium spp., unclassified Clostridiales and unclassified Lachnospiraceae differed by case-control status (data not shown). The phylotype distribution at the phylum level for individual patients is shown in Supplemental Figure 2S. The distribution of the proportions of bacterial phylotypes identified at the genus level is presented in Figure 3A and B. Compared to controls, cases were characterized by lower proportions of Bacteroides spp. (case 28.3% vs. control 53.3%, p < 0.03) Coprococcus spp., (case 1.7% vs. control 9.7%, p = 0.05) higher levels of, Dorea spp. (Case 15.9% vs. control 3.3%, p < 0.005), Faecalibacterium spp. (Case 21.7% vs. control 10.3%, p < 0.05), Shigella spp. (case 5.9% vs. control 0.89%, p = 0.16) and Ruminococcus spp. (case 19.1% vs. control 3.4%, p = 0.24). Although the following genera were not highly abundant, Oscillospira spp., Clostridium spp., Finegoldia spp., Eubacterium spp., Akkermansia spp. and Phascolarctobacterium spp. were observed only in cases but not in controls, while Fusobacterium spp. was present only in controls (Fig. 3A). The phylotype distribution at the genus level for individual patients in this study is shown in Figure 3B.
Figure 3B.
Distribution of genus level adherent bacterial phylotypes obtained from clone libraries. Each bar represents the percent contribution of genus-level profiles grouped by case-control status (A) or for each individual (B). The colors representing the different genera are shown on the side of the figures.
Identification of mucosa adherent bacteria by fluorescence in-situ hybridization (FISH).
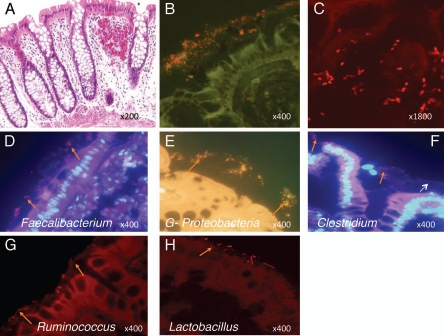
Although 16S rRNA TRFLP and clone sequencing can reveal the overall composition of a bacterial community, these methods do not provide information about the physical structure of the bacterial community or localization of specific community members. We therefore performed fluorescence in-situ hybridization to localize specific adherent bacterial types on colonic tissue sections. We employed 16S rRNA FISH probes Eubacteriaceae (EU 388) universal bacteria probe, Gammaproteobacteria (Gpro 384), Lachnospiraceae (Clostr 482), Ruminococcus (Rbro 730), Lactobacillus (Lacto 645) and Faecalibacterium (Fecali 645), to detect these mucosal adherent bacteria. The specificity of these probes was validated with corresponding cultured bacteria (data not shown). FISH analysis on colonic tissue sections from cases and controls confirmed that these respective bacterial types were present within the adherent community, and predominantly localized to the mucus layer overlying the epithelium in normal colonic tissue (Fig. 4).
Figure 4.
Fluorescence in-situ hybridization (FISH) using bacterial 16S rRNA probes showing bacteria localized to the mucus layer. (A) H&E stained section of normal colonic mucosa showing crypts and mucosal layer. Tissue sections from colonic mucosa were hybridized with general bacteria (EUB 388) probe: (B and C) show that general bacteria (include most of Eubacteria species) were distributed in the mucosal layer; (B) shows bacteria (red) at magnification, 400X while (C) shows confocal laser scanning image of bacteria (red) in mucosa at a higher magnification. Colon tissue sections were also hybridized with specific bacteria probes namely Faecalibacterium, Proteobacteria, Clostridia, Ruminococcus and Lactobacillus (D–H). (D and E) were stained with bacteria specific probe and general bacteria probe. (F–H) were stained with single bacteria specific probe. Mammalian cell nuclei are stained blue by DAPI staining (D and F). Orange arrows show different shaped bacteria localized to the mucus layer. White arrow points to the mucus layer above the colonic epithelium.
Discussion
The microbiota plays an important role in the gut but little is known about the composition of the bacteria in the mucosal compartment, especially in relation to colorectal cancer. In this study, we defined adherent bacteria in normal colonic mucosa of patients with and without adenomas and assessed whether differences in bacterial profiles or specific bacterial species were associated with adenoma status. Three predominant phyla, Firmicutes, Bacteroidetes and Proteobacteria, were identified in normal colonic mucosa. Case subjects had lower relative abundance of Bacteroidetes and higher abundance of Proteobacteria than controls. Analysis at shallower phylogenetic resolution revealed a distinct bacterial genus profile that consisted of decreased abundance of Bacteroides and Coprococcus, and increased abundance of Dorea, Faecalibacterium and Shigella was characteristic of adenoma subjects. We observed borderline statistically significant differences in bacterial diversity with cases having higher diversity than controls. We did not detect the phylum Actinobacteria and its important genus, Bifidobacterium, in the adherent colonic mucosa of our subjects. This phylum is commonly observed in infants19,20 but is less dominant in adults.21,22 Our inability to detect Bifidobacterium spp. in our subjects may relate to coverage and the number of clones evaluated in this study. Deeper sequencing using high throughput methods should clarify the detection of Actinobacteria in the adherent ecosystem.
Identifying the bacterial composition of the gastrointestinal tract and understanding the role of bacteria in health and disease is currently a focus of ecological studies.1,21,23,24 Our observation that Firmicutes, Proteobacteria and Bacteroidetes were the dominant phyla in normal colonic mucosa are also concordant with previous studies of the gut microflora.10,21,25–27 Our findings suggest that alterations in the composition and structure of adherent bacterial communities may contribute to the development of adenomas.
The mucosal biopsies used in this study were from the rectum, not from adenoma tissue or from mucosa in the surrounding area of adenomas. Previous studies have indicated that the bacterial community in an individual host is relatively stable along the distal digestive tract21,28 and over time.29,30 Therefore, the observed compositional changes in rectal mucosal bacterial communities associated with colorectal adenomas may reflect the presence of adenomas elsewhere in the colon, and may have preceded the onset of adenoma formation.
Recent human and animal studies also indicate that bacterial dysbiosis may contribute to obesity10,31 and inflammatory bowel diseases.32–35 Although, it is not clear how mucosal adherent bacteria might influence the development of adenomas, factors such as antibiotics use,29 host factors1 and diet10,30 that change the environment in the large bowel could contribute to bacterial dysbiosis by altering the normal homeostasis between the gut bacteria and the host. While antibiotic use has been shown to contribute to immediate shifts in the microbiota, most of the bacterial communities are fairly resilient and return to pretreatment stages within four weeks.21 A recent study observed that knock out of Myd88, the universal adaptor used by the host pattern-recognition Toll-like receptors (TLR) resulted in lower Firmicutes to Bacteroidetes ratio.36 Although it is not clear how loss of Myd88 contributes to the shift in microbial composition, nevertheless, these findings illustrate the importance of host factors in gut bacterial dysbiosis.
A potential mechanism by which bacterial dysbiosis might influence the development of adenomas and colorectal cancer is through inflammation and diet. Metabolites from diet could create an environment that alters the gut pH, which in turn could promote bacterial dysbiosis, inflammation and cellular transformation.37 A recent study by Vannucci et al.37 compared tumors in conventional and germ-free rats treated with azoxymethane and found that germ-free animals had fewer tumors and smaller sized tumors compared to conventional animals. In general, studies in germ-free animals as well as human studies on Crohn and ulcerative colitis support a link between the gut bacteria, inflammation and cancer.38–41
The gut bacteria may also promote adenomas through direct effects of metabolites on the colonic epithelium. Bacteria may convert procarcinogenic metabolites from diet into DNA damaging agents. For example, oxygen radicals produced by E. fecalis have been linked to DNA damage in colonic epithelial cells.42,43 Similarly, H2S, produced by sulfur reducing bacteria has been shown to modulate pathways involved in colorectal cancer.44,45 In our study, we observed Clostridium spp. in cases and not controls. Another mechanism by which bacteria may promote colorectal cancer is through production of toxic polycyclic aromatic compounds by bile acid metabolizing bacteria such as Clostridium paraputrificum.46–48 Future studies will evaluate the association between diet, inflammation, gut bacteria and colorectal adenomas in humans.
Recent advances in molecular methods that are based on the highly conserved bacterial 16S ribosomal RNA (rRNA) gene have enhanced our ability to study and characterize adherent bacteria communities in the gut. In this study, we used clone sequencing and terminal restriction fragment length polymorphism (T-RFLP), a high throughput fingerprinting technique that is widely used for bacterial community fingerprinting.49,50 Sequencing of 16S rRNA gene is the gold standard for validating T-RFLP analysis. Cluster analysis of the T-RFs obtained from mucosal samples identified subjects that shared similar profiles and eight subjects were randomly selected for generation of clone libraries. By combining T-RFLP and clone sequence analysis, we observed that cases and controls had both unique and shared phylotypes, thus this method proved to be useful in demonstrating differences in adherent bacterial community structure. A major advantage of this approach is the efficient use of resources, cost effectiveness and ability to correlate T-RFs with bacteria species identified from clone library to determine their phylogenetic affiliation.
The gut epithelium is protected by a mucus layer, which serves several functions such as keeping epithelial cells from direct contact with microorganisms as well as sustaining the physical and nutritional activities of various components of the commensal microbiota. Using bacteria specific probes and FISH analysis, we found that Clostridium, Faecalibacterium, Gammaproteobacteria, Ruminococcus and Lactobacillus were localized to the thick mucus layer above the colonic epithelium. These observations are compatible with some studies24,51,52 and but not all studies.53 The inconsistencies in detecting bacteria within the mucus layer may relate to potential differences in biopsy location, sample preparation and method of detection.
In summary, we used molecular methods to characterize adherent bacteria in the normal colon of patients with and without adenomas. Our findings suggest that adherent bacteria may be significant players in the development of adenomas and colorectal cancer. We identified a bacterial profile that characterized adenoma subjects, namely increased Proteobacteria and decreased Bacteroidetes. This finding suggests that changes in the adherent bacterial composition and structure may contribute to development of adenomas. To extend our understanding of how adherent composition changes as a function of adenoma, it will be helpful in future studies to include a larger number of subjects with longitudinal time points and utilize second-generation sequencing technology to provide deeper coverage of bacterial communities. Future studies will also utilize conventionally raised and gnotobiotic animal models to elucidate the mechanisms by which altered colonic bacterial profiles contribute to adenomas. Identifying the composition of adherent bacterial communities in the colon is an important step in our understanding of their role in large bowel cancer and development of effective prognostic, preventative or therapeutic strategies.
Materials and Methods
Biopsy sample collection.
Colorectal biopsies were collected from consenting subjects enrolled in an ongoing study, the Diet and Health Study V (DHS) who presented for routine screening colonoscopy. No patient had received antibiotic therapy within eight weeks prior to his or her colonoscopy. All the patients received polyethylene glycol for bowel cleansing. Eligible participants provided written informed consent, agreed to a telephone interview, donated rectal biopsies during the procedure. Reasons for exclusion in the study were incomplete examination (cecum not reached), age < 30 years, inability to give informed consent, polyposis (>100 polyps), previous colon resection or cancer, colitis (such as ulcerative colitis and Crohn disease) and previous colon adenoma. Normal mucosal biopsies were obtained 10–12 cm from the anal verge from consented participants and the mucosal biopsies were rinsed in sterile PBS and snap-frozen in liquid nitrogen on site and later transferred to −80°C. A pathologist reviewed all polyps according to standard pathologic criteria and classified subjects with adenomatous polyps as cases and those without adenomas as controls. The study was approved by the Institutional Review Board (IRB) at the University of North Carolina School of Medicine (Protocol #05-3138).
DNA extraction.
Bacterial genomic DNA was extracted from biopsy specimens. Mucosal biopsies (10–20 mg) were placed in lysozyme (30 mg/ml; Sigma, St. Louis MO) for 30 minutes. The biopsy-lysozyme mixture was homogenized on a bead beater (Biospec Products Inc., Bartlesville, OK) at 4,800 rpm for 3 minutes at room temperature followed by DNA extraction using the Qiagen DNA isolation kit (cat # 14123) per the manufacturer's recommended protocol.
T-RFLP analysis.
The DNA extracted from mucosal biopsies served as a template for amplification of an approximately 1,500 bp region of the 16S rRNA gene. The reaction mixture contained, 100 mM Tris (pH 8.5), 2 mM MgCl2, 20 mM (NH4)2SO4, 10 µM dNTPs, 0.2 µM each fluorescently labeled universal primers 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GYT ACC TTG TTA CGA CTT–3′),54 and 1 unit of Taq polymerase (Apex). Cycling conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 30 min, 55°C for 45 sec and 72°C for 1.5 min, with a final extension period of 10 min at 72°C. Amplified PCR products were verified by electrophoresis in 1.5% agarose gel followed by purification (Qiaquick PCR purification kit, Qiagen) and restriction enzyme (RE) digestion with Hha 1 and Msp1 in separate reaction mixtures that consisted of 1X restriction buffer, 20 µg bovine serum albumin and 10 units of restriction enzyme in a final volume of 20 µl at 37°C for 3 hours. The digested DNA fragments were purified and mixed with 0.5 µl internal size standards (ROX 500 Applied Biosystems), deionized formamide and denatured for 3 minutes at 95°C followed by capillary electrophoresis (ABI 3100 genetic analyzer, Applied Biosystems). The PCR conditions and RE digests were optimized to ensure maximum reproducibility. Individual reference bacteria strains assessed by T-RFLP analysis showed a single T-RF peak for a single strain and multiple peaks for a mixture of reference strains (data not shown). The fragment peak size, area and height were determined by GeneMapper (Applied Biosystems Inc.). Peak area and fluorescence data were normalized and processed as described by Abdo et al.15 followed by cluster analysis. To account for differences in biopsy sizes, we used a standard amount of DNA for PCR, restriction digests and TRFLP analysis. The contribution of individual T-RFs was calculated as a proportion of the total T-RF peak area for each sample. For our analysis, we used these proportions rather than absolute numbers. Samples from eight subjects were randomly selected for cloning and sequencing.
Construction of 16S ribosomal RNA clone libraries.
Clone libraries were constructed from four adenoma cases and four controls. The 16S rRNA gene was amplified using unlabeled 8F and 1492R primers. The reaction mixture and cycling conditions were as described above. The resulting PCR products were gel purified with the Qiagen extraction kit according to the manufacturer's recommendations followed by sub cloning into TOPO TA pCR2.1, and transformation into Escherichia coli TOP10 competent cells (Invitrogen). Plasmid DNAs were extracted using Qiagen Plasmid DNA Purification Kit following manufacturer's protocol.
DNA sequencing, consensus alignment and taxonomic classification.
A minimum of 30 randomly selected clones were sequenced per patient using M13-F and M13-R primers (MWG-BIOTECH AG, High Point, NC). Vector sequences and ambiguous terminal sequences were trimmed and the 16S rRNA gene sequences were edited, assembled and aligned into consensus sequences using Sequencher (version 4.8, Genecode). The sequences were checked for possible chimeras using Bellerophon v.3.55 The operational taxonomic units (OTUs) were determined using >97% similarity with SeqMatch in the Ribosomal Database-II (RDP-II, http://rdp.cme.msu.edu/cgis/). The taxonomic classification were confirmed by naïve Bayesian rRNA classifier56 and the BLAST program.57 The classified genus level phylotypes were used for analysis.
Nucleotide sequence accession numbers.
All 353 gene sequences reported in this paper are available in the GenBank database under the accession numbers (EU530161-EU530519). Sixteen sequences were not registered because the similarity was <60% and they could not be matched to known bacteria.
Estimation of richness and biodiversity.
Phylotypes were estimated based on the genus level classification (≥97% sequence identity). Each OTU was considered a separate phylotype. Microbial diversity was estimated with the Shannon-Weiner diversity index (H'), which accounts for both the number of phylotypes present (richness) and the proportion of the total accounted for by each phylotype (evenness) (PRIMER E).18 We calculated Good's coverage58 to estimate if the clone library reflected the actual bacterial diversity in the samples using the following equation, G = 1 − (n1/N), where G is the coverage index, n1 is the number of phylotypes appearing only once, and N is the number of all sequences in the library.
Fluorescent in situ hybridization (FISH).
We used FISH analysis to localize and identify bacteria in the colonic mucosa. Carnoy's fixed paraffin-embedded colonic tissue sections (6 µm) on Probe-On Plus slides (Fisher Scientific, Pittsburgh, Pa.) were deparaffinized in xylene (three washes), and graded series of alcohol (100%, 95%, 70%) followed by lysozyme treatment (15 U/µl in 1% PBS) at 37°C for 20 min, two rinses with PBS and water. Slides were hybridized with FISH probes 5′ labeled with Cy3 or 6-FAM at a concentration of 5 ng/µl in hybridization buffer (20 mM Tris-HCl, 0.1% sodium dodecyl sulfate (SDS), 0.9% NaCl [pH 7.2]) and incubated at 56°C overnight. The following probes were used to detect general bacteria (EUB 338, GCT GCC TCC CGT AGG AGT) and specific bacteria (Gamma-Proteobacteria, Gpro, GAA GCC ACG CCT CAA GGG CAC AA; Clostridia Cluster Clostr482, GCT TCT TAG TCA GGTACC G; Faecalibacterium, Fecali645, CCT CTG CAC TAC TCA AGA AAA AC; Lactobacillus, Lactob645, GGT ATT AGC A(C/T)CT GTT TCC A and Ruminococcus, Rbro730, TAA AGC CCA G(C/T)AG GCC GC) in mucosal biopsies. Slides were washed in hybridization buffer without SDS at 58°C for 20 min followed by rinse in 1% PBS and distilled water. Slides were dried at 46°C for 10 min and mounted with ProLong antifade Dapi kit (Molecular Probes Inc., Eugene, OR). Sections were examined on a BX71 (Olympus America, Melville, NY) epifluorescence microscope and Zeiss LSM5 Pascal Confocal Laser Scanning Microscope (Olympus America, Melville, NY), and images were captured with Olympus DP-7 camera or Zeiss Axiocam respectively.
Statistical analysis.
Terminal restriction fragments (T-RFs) analysis: Processing of T-RFs was conducted as described in the methods and the resulting matrix consisted of columns representing normalized peak areas of individual TRF-s and rows representing individual subjects, cases and controls. The T-RF data was subjected to cluster analysis using the unweighted mean pair group method with arithmetic mean (UPGMA) to establish the dendrogram type.15
Bacterial communities and structural analysis: We used complementary phylogenetic and taxon based methods to compare 16S rRNA sequences among cases and controls. Structure based alignments of the 16S rRNA gene sequences59 based on multiple-aligned gap containing sequence data from GreenGenes55 were performed in the ARB database. The tree data were subjected to UniFrac analysis,16,60 to compare bacterial communities and phylogenetic structure between adenoma cases and non-adenoma controls using the p-test which compares differences in phylogeny and community composition.61 The analysis was performed on the UNIFRAC web server using 1000 permutations and weighting according to abundance of taxa. We employed Bonferroni corrections because of multiple comparisons. Multivariate analysis (non metric multidimensional scaling, MDS) was performed on the UNIFRAC distances of the different libraries using Primer E. The UNIFRAC distances were converted to similarities in PRIMER E and the similarities between groups (case/control) and within groups were assessed by analysis of similarities (ANOSIM), a non-parametric test (PRIMER E). We computed the significance in PRIMER E by permutation of group membership with 999 replicates. The test statistic R, which measures the strength of the results ranges from −1 to 1. An R value of 1 signifies differences between groups while an R value of 0 signifies that the groups are identical. To determine the specific phylotypes that contributed to the differences in bacterial composition between cases and controls, we used SIMPER (similarity percentage, PRIMER E) to compute the proportions of phylotypes overall and for cases and controls separately. Differences in distribution of specific phylotypes between cases and controls were evaluated by Wilcoxon rank sum test (Stata).
Supplementary Material
Acknowledgements
This research was supported, in part, by grants from the National Institutes of Health P30 DK34987; R01 CA44684 and P50 CA106991, K01 CA093654, R01-CA136887, R01 DK081426, K01 DK073695 and a Pew Scholars Award to John F. Rawls. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Abbreviations
- FISH
florescence in situ hybridization
- CRC
colorectal cancer
- T-RFLP
terminal restriction fragment length polymorphism
- T-RF
terminal restriction fragment
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/12360
References
- 1.Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136:1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savage DC. Microbial ecology of the gastrointestinal tract. Ann Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 3.Van Citters GW, Lin HC. Management of small intestinal bacterial overgrowth. Curr Gastroenterol Rep. 2005;7:317–320. doi: 10.1007/s11894-005-0025-x. [DOI] [PubMed] [Google Scholar]
- 4.Mutch DM, Simmering R, Donnicola D, Fotopoulos G, Holzwarth JA, Williamson G, et al. Impact of commensal microbiota on murine gastrointestinal tract gene ontologies. Physiol Genomics. 2004;19:22–31. doi: 10.1152/physiolgenomics.00105.2004. [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa M, Kawasaki A, Yang K, Fujimoto Y, Masumoto J, Breukink E, et al. A role of lipophilic peptidoglycan-related molecules in induction of Nod1-mediated immune responses. J Biol Chem. 2007;282:11757–11764. doi: 10.1074/jbc.M700846200. [DOI] [PubMed] [Google Scholar]
- 6.Guarner F, Malagelada JR. Role of bacteria in experimental colitis. Best Pract Res Clin Gastroenterol. 2003;17:793–804. doi: 10.1016/s1521-6918(03)00068-4. [DOI] [PubMed] [Google Scholar]
- 7.Shanahan F. Gut flora in gastrointestinal disease. Eur J Surg. 2002:47–52. [PubMed] [Google Scholar]
- 8.Edmiston CE, Jr, Avant GR, Wilson FA. Anaerobic bacterial populations on normal and diseased human biopsy tissue obtained at colonoscopy. Appl Environ Microbiol. 1982;43:1173–1181. doi: 10.1128/aem.43.5.1173-1181.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scanlan PD, Shanahan F, Clune Y, Collins JK, O'Sullivan GC, O'Riordan M, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environmental microbiology. 2008;10:789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 10.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 11.Campieri M, Gionchetti P. Bacteria as the cause of ulcerative colitis. Gut. 2001;48:132–135. doi: 10.1136/gut.48.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandek A, Bauditz J, Swidsinski A, Buhner S, Weber-Eibel J, von Haehling S, et al. Altered intestinal function in patients with chronic heart failure. J Am College Cardiol. 2007;50:1561–1569. doi: 10.1016/j.jacc.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nature Immunol. 2004;5:569–73. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 14.Oswald IP. Role of intestinal epithelial cells in the innate immune defence of the pig intestine. Vet Res. 2006;37:359–368. doi: 10.1051/vetres:2006006. [DOI] [PubMed] [Google Scholar]
- 15.Abdo Z, Schuette UM, Bent SJ, Williams CJ, Forney LJ, Joyce P. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environmental Microbiol. 2006;8:929–938. doi: 10.1111/j.1462-2920.2005.00959.x. [DOI] [PubMed] [Google Scholar]
- 16.Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative b diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke K, Warwick RM. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. 2nd ed. Primer-E Ltd., Plymouth: Plymouth Routines In Multivariate Ecological Research; 2001. [Google Scholar]
- 19.Enck P, Zimmermann K, Rusch K, Schwiertz A, Klosterhalfen S, Frick JS. The effects of maturation on the colonic microflora in infancy and childhood. Gastroenterol Res Pract. 2009;2009:752401. doi: 10.1155/2009/752401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lievin V, Peiffer I, Hudault S, Rochat F, Brassart D, Neeser JR, et al. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. 2000;47:646–652. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, et al. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009;3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 23.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 27.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepage P, Seksik P, Sutren M, de la Cochetiere MF, Jian R, Marteau P, et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis. 2005;11:473–480. doi: 10.1097/01.mib.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- 29.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komanduri S, Gillevet PM, Sikaroodi M, Mutlu E, Keshavarzian A. Dysbiosis in pouchitis: evidence of unique microfloral patterns in pouch inflammation. Clin Gastroenterol Hepatol. 2007;5:352–360. doi: 10.1016/j.cgh.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 35.Sokol H, Lay C, Seksik P, Tannock GW. Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm Bowel Dis. 2008;14:858–867. doi: 10.1002/ibd.20392. [DOI] [PubMed] [Google Scholar]
- 36.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vannucci L, Stepankova R, Kozakova H, Fiserova A, Rossmann P, Tlaskalova-Hogenova H. Colorectal carcinogenesis in germ-free and conventionally reared rats: different intestinal environments affect the systemic immunity. Int J Oncol. 2008;32:609–617. [PubMed] [Google Scholar]
- 38.Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002;160:2253–2257. doi: 10.1016/S0002-9440(10)61172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engle SJ, Ormsby I, Pawlowski S, Boivin GP, Croft J, Balish E, et al. Elimination of colon cancer in germ-free transforming growth factor β1-deficient mice. Cancer Res. 2002;62:6362–6366. [PubMed] [Google Scholar]
- 40.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de la Cochetiere MF, Piloquet H, des Robert C, Darmaun D, Galmiche JP, Roze JC. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: the putative role of Clostridium. Pediatr Res. 2004;56:366–370. doi: 10.1203/01.PDR.0000134251.45878.D5. [DOI] [PubMed] [Google Scholar]
- 42.Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 43.Huycke MM, Gaskins HR. Commensal bacteria, redox stress and colorectal cancer: mechanisms and models. Exp Biol Med. 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- 44.Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 45.Kanazawa K, Konishi F, Mitsuoka T, Terada A, Itoh K, Narushima S, et al. Factors influencing the development of sigmoid colon cancer. Bacteriologic and biochemical studies. Cancer. 1996;77:1701–1706. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1701::AID-CNCR42>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 46.Horie H, Kanazawa K, Okada M, Narushima S, Itoh K, Terada A. Effects of intestinal bacteria on the development of colonic neoplasm: an experimental study. Eur J Cancer Prev. 1999;8:237–245. doi: 10.1097/00008469-199906000-00012. [DOI] [PubMed] [Google Scholar]
- 47.Hughes R, Kurth MJ, McGilligan V, McGlynn H, Rowland I. Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro. Nutr Cancer. 2008;60:259–266. doi: 10.1080/01635580701649644. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura J, Kubota Y, Miyaoka M, Saitoh T, Mizuno F, Benno Y. Comparison of four microbial enzymes in Clostridia and Bacteroides isolated from human feces. Microbiol Immunol. 2002;46:487–490. doi: 10.1111/j.1348-0421.2002.tb02723.x. [DOI] [PubMed] [Google Scholar]
- 49.Blackwood CB, Marsh T, Kim SH, Paul EA. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl Environ Microbiol. 2003;69:926–932. doi: 10.1128/AEM.69.2.926-932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coolen MJ, Post E, Davis CC, Forney LJ. Characterization of microbial communities found in the human vagina by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Appl Environ Microbiol. 2005;71:8729–8737. doi: 10.1128/AEM.71.12.8729-8737.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035–1045. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 52.Swidsinski A, Loening-Baucke V, Theissig F, Engelhardt H, Bengmark S, Koch S, et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Waaij LA, Harmsen HJ, Madjipour M, Kroese FG, Zwiers M, van Dullemen HM, et al. Bacterial population analysis of human colon and terminal ileum biopsies with 16S rRNA-based fluorescent probes: commensal bacteria live in suspension and have no direct contact with epithelial cells. Inflamm Bowel Dis. 2005;11:865–871. doi: 10.1097/01.mib.0000179212.80778.d3. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt TM, Relman DA. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 1994;235:205–222. doi: 10.1016/0076-6879(94)35142-2. [DOI] [PubMed] [Google Scholar]
- 55.DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:394–399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Good RA, Denny FW, Smith RT, Thomas L. Factors involved in the production of tissue damage by endotoxins derived from Gram-negative micro-organisms. AMA Am J Dis Child. 1953;86:473–474. [PubMed] [Google Scholar]
- 59.Ludwig A, von Rhein C, Bauer S, Huttinger C, Goebel W. Molecular analysis of cytolysin A (ClyA) in pathogenic Escherichia coli strains. J Bacteriol. 2004;186:5311–5320. doi: 10.1128/JB.186.16.5311-5320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.