Abstract
The prevalence of different HIV-1 subtypes in Spain varies by geographic region. In the present study isolates were collected from 72 newly diagnosed individuals in western Andalucia from 2004 to 2006. Viral sequences were amplified and the subtype diversity and prevalence of resistance mutations in the reverse transcriptase and protease genes were determined. The results presented here demonstrate that subtype B virus predominates in this region (88.9%), with the non-B subtypes CRF02_AG (9.7%) and B/G (1.4%) also present. Only two isolates (2.9%) carried resistance mutations in the reverse transcriptase gene and none of the isolates had major resistance mutations in the protease gene. Minor mutations in the protease gene were more prevalent with 86.1% of isolates containing at least one minor mutation. These results elucidate the subtype diversity present in this region and suggest that the transmission of highly resistant virus variants does not occur at a high frequency in this population.
Variants of HIV-1 have been classified into M, N, and O groups based on sequence homology. Group M viruses have been further subdivided into nine subtypes, A, B, C, D, F, G, H, J, and K, in addition to many circulating recombinant forms (CRFs) and unique recombinant forms (URFs) that have arisen via intersubtype recombination. The prevalence of HIV-1 subtypes varies greatly depending on the geographic region, with subtype B predominating in western Europe and North America, whereas multiple non-B subtypes account for the majority of infections in sub-Saharan Africa and India. Characterization of HIV variants endemic to distinct geographic regions is necessary since variation between the different subtypes of HIV-1 may affect antiretroviral treatment and future vaccination.
The use of antiretroviral therapy (ART) in developed countries has resulted in populations harboring resistant viruses. The transmission of resistant viruses has been previously reported, and in some populations the rates of transmitted resistance have increased as the use of ART becomes more widespread.1,2 This is of particular concern since it has been demonstrated that more time is required to achieve virologic suppression with ART in patients infected with resistant viruses than in patients infected with viruses free of resistance mutations.1,2 In addition, the duration of virologic suppression before virologic failure is shorter in patients infected with resistant viruses.2 Resistance mutations present in the protease gene (PR) have been classified as either major mutations or minor mutations (also called polymorphisms). Major mutations are those that occur within the active site of PR and directly confer phenotypic resistance to protease inhibitors, and it has been well established that the presence of major mutations in PR is predictive of virologic failure in patients receiving ART.3 However, the role played by minor mutations in PR in conferring resistance to PIs is less clear. It has been suggested that minor mutations in PR are compensatory mutations that increase the fitness of viruses harboring major mutations,4,5 and some studies have indicated that these mutations can affect treatment outcome.3,6
HIV surveillance studies have shown that the geographic regions of Spain are subject to unique migration patterns, resulting in populations that harbor different HIV subtype diversities.7–9 The goal of the present study is to characterize the subtype diversity and determine the prevalence of resistance mutations in the reverse transcriptase (RT) and PR regions of pol in treatment-naive individuals at a tertiary medical center in western Andalucia from 2004 to 2006. For this purpose, 72 HIV-infected, treatment-naive patients receiving care at Virgen del Rocío University Hospital between 2004 and 2006 were included in the study. Table 1 shows the characteristics of individuals included in the study. Of the 72 patients, 67 were Spanish, two were from Nigeria (NV25 and NV28), one was from Argentina (NV12), one was from India (NV35), and one was from Morocco (NV42). Patient age, sex, risk factor, coinfection status (hepatitis B and C), viral load, and CD4+ T cell counts were determined at the time of diagnosis. Plasma HIV RNA was quantified using the Amplicor HIV-1 Monitor Test 1.5 (Roche Diagnostics, Alamida, CA) with a limit of detection of 50 copies/ml. Patients were categorized as either recently or chronically infected using criteria similar to what has been used previously.12 Recently infected patients were defined as patients with documented seroconversion in the preceding 12 months or clinical evidence of acute HIV infection within 12 months of the first documented seroconversion. Chronically infected patients were defined as patients for whom there was no documented seroconversion or clinical evidence of acute infection in the preceding 12 months. For quantitative measurements, data sets were compared parametrically using the Student's t test or nonparametrically using the Mann–Whitney U test. Categorical data were analyzed using the χ2 test. For all statistical tests a p value ≤ 0.05 was considered significant. All statistical analyses were performed using SPSS version 14.0 (SPSS Inc., Chicago, IL).
Table 1.
Characteristics of 72 HIV-Infected, Treatment-Naive Patients Receiving Care at Virgen del Rocío University Hospitala
| Accession number | Patient ID | Subtype | Type of infection | Risk factor | Age | Sex | Country | Year of diagnosis |
|---|---|---|---|---|---|---|---|---|
| EU288087 | NV01 | B | Chr | HO/BI | 28 | M | Spain | 2004 |
| EU288088 | NV02 | B | Chr | HO/BI | 39 | M | Spain | 2004 |
| EU288089 | NV03 | B | Chr | HO/BI | 19 | M | Spain | 2004 |
| EU288090 | NV04 | B | Chr | HO/BI | 57 | M | Spain | 2004 |
| EU288091 | NV05 | B | Recent | HO/BI | 27 | M | Spain | 2004 |
| EU288092 | NV06 | B | Chr | HO/BI | 39 | M | Spain | 2004 |
| EU288093 | NV07 | B | Chr | HO/BI | 26 | M | Spain | 2004 |
| EU288094 | NV08 | B | Recent | HO/BI | 30 | M | Spain | 2004 |
| EU288095 | NV09 | B | Chr | HTSX | 41 | M | Spain | 2004 |
| EU288096 | NV10 | B | Chr | HO/BI | 25 | M | Spain | 2005 |
| EU288097 | NV11 | B | Chr | HO/BI | 46 | M | Spain | 2005 |
| EU288098 | NV12 | B | Recent | HO/BI | 29 | M | Argentina | 2005 |
| EU288099 | NV13 | B | Chr | HO/BI | 23 | M | Spain | 2005 |
| EU288100 | NV14 | B | Chr | HTSX | 36 | F | Spain | 2005 |
| EU288101 | NV15 | B | Chr | HO/BI | 23 | M | Spain | 2005 |
| EU288102 | NV16 | B | Chr | ND | 47 | M | Spain | 2005 |
| EU288103 | NV17 | B | Recent | HO/BI | 39 | M | Spain | 2005 |
| EU288104 | NV18 | CRF02_AG | Chr | HTSX | 34 | M | Spain | 2005 |
| EU288105 | NV19 | B | Chr | HTSX | 46 | M | Spain | 2005 |
| EU288106 | NV20 | B | Chr | HTSX | 63 | M | Spain | 2005 |
| EU288107 | NV21 | CRF02_AG | Recent | HTSX | 26 | F | Spain | 2005 |
| EU288108 | NV22 | B | Recent | HO/BI | 36 | M | Spain | 2005 |
| EU288109 | NV23 | B | Chr | IVDU | 45 | M | Spain | 2005 |
| EU288110 | NV24 | B | Chr | HO/BI | 28 | M | Spain | 2005 |
| EU288111 | NV25 | CRF02_AG | Chr | ND | 30 | F | Nigeria | 2005 |
| EU288112 | NV26 | B | Chr | HTSX | 50 | M | Spain | 2005 |
| EU288113 | NV27 | B | Chr | HO/BI | 24 | M | Spain | 2005 |
| EU288114 | NV28 | CRF02_AG | Chr | HTSX | 25 | F | Nigeria | 2005 |
| EU288115 | NV29 | B | Chr | HO/BI | 39 | M | Spain | 2005 |
| EU288116 | NV30 | B | Chr | HO/BI | 28 | M | Spain | 2005 |
| EU288117 | NV31 | B | Recent | HO/BI | 38 | M | Spain | 2005 |
| EU288118 | NV32 | B | Chr | HO/BI | 45 | M | Spain | 2005 |
| EU288119 | NV33 | B-G | Chr | IVDU | 43 | M | Spain | 2005 |
| EU288120 | NV34 | B | Chr | HTSX | 46 | M | Spain | 2005 |
| EU288121 | NV35 | B | Chr | HO/BI | 45 | M | India | 2005 |
| EU288122 | NV36 | B | Recent | HO/BI | 36 | M | Spain | 2005 |
| EU288123 | NV37 | CRF02_AG | Chr | HTSX | 27 | F | Spain | 2005 |
| EU288124 | NV38 | B | Recent | HO/BI | 31 | M | Spain | 2005 |
| EU288125 | NV39 | B | Chr | HTSX | 45 | M | Spain | 2005 |
| EU288126 | NV40 | B | Chr | HO/BI | 66 | M | Spain | 2005 |
| EU288127 | NV41 | B | Recent | HO/BI | 27 | M | Spain | 2004 |
| EU288128 | NV42 | B | Chr | HO/BI | 49 | M | Morocco | 2004 |
| EU288129 | NV43 | B | Recent | HO/BI | 24 | M | Spain | 2004 |
| EU288130 | NV44 | B | Chr | HO/BI | 32 | M | Spain | 2005 |
| EU288131 | NV45 | B | Recent | HO/BI | 30 | M | Spain | 2005 |
| EU288132 | NV46 | B | Chr | HO/BI | 28 | M | Spain | 2005 |
| EU288133 | NV47 | B | Chr | HO/BI | 27 | M | Spain | 2005 |
| EU288134 | NV48 | B | Chr | IVDU | 40 | F | Spain | 2006 |
| EU288135 | NV49 | B | Chr | HTSX | 43 | M | Spain | 2006 |
| EU288136 | NV50 | B | Chr | HO/BI | 33 | M | Spain | 2006 |
| EU288137 | NV51 | B | Chr | ND | 38 | F | Spain | 2006 |
| EU288138 | NV52 | B | Recent | HO/BI | 29 | M | Spain | 2006 |
| EU288139 | NV53 | B | Chr | HO/BI | 37 | M | Spain | 2006 |
| EU288140 | NV54 | B | Chr | HO/BI | 41 | M | Spain | 2006 |
| EU288141 | NV55 | B | Chr | HTSX | 57 | F | Spain | 2006 |
| EU288142 | NV56 | B | Chr | HO/BI | 34 | M | Spain | 2006 |
| EU288143 | NV57 | B | Chr | HO/BI | 33 | M | Spain | 2006 |
| EU288144 | NV58 | B | Recent | HO/BI | 36 | M | Spain | 2006 |
| EU288145 | NV59 | B | Chr | HTSX | 66 | M | Spain | 2006 |
| EU288146 | NV60 | B | Chr | ND | 65 | M | Spain | 2006 |
| EU288147 | NV61 | B | Chr | HO/BI | 42 | M | Spain | 2006 |
| EU288148 | NV62 | CRF02_AG | Chr | HTSX | 39 | F | Spain | 2006 |
| EU288149 | NV63 | B | Chr | HTSX | 63 | F | Spain | 2006 |
| EU288150 | NV64 | B | Chr | ND | 38 | M | Spain | 2006 |
| EU288151 | NV65 | B | Chr | HTSX | 51 | M | Spain | 2006 |
| EU288152 | NV66 | B | Chr | IVDU | 38 | M | Spain | 2006 |
| EU288153 | NV67 | B | Chr | HTSX | 58 | M | Spain | 2006 |
| EU288154 | NV68 | B | Chr | HTSX | 50 | F | Spain | 2006 |
| EU288155 | NV69 | B | Chr | HO/BI | 24 | M | Spain | 2006 |
| EU288156 | NV70 | B | Recent | HO/BI | 24 | M | Spain | 2006 |
| EU288157 | NV71 | B | Chr | HTSX | 37 | F | Spain | 2006 |
| EU288158 | NV72 | CRF02_AG | Chr | HO/BI | 40 | M | Spain | 2006 |
Recent, recently infected patients; Chr, chronically infected patients; HO/BI, homosexual/bisexual; HTSX, heterosexual; IVDU, intravenous drug use; ND, not determined; F, female; M, male.
For genotypic analysis, viral RNA was isolated from 1 ml of plasma using the MagNA Pure LC instrument (Roche Applied Science, Indianapolis, IN) and sequenced using the TruGene Genotyping System (Bayer Healthcare), which sequences codons 10–99 of PR and 38–247 of RT. Virus subtypes were determined by analyzing sequence data using the Stanford University Drug Resistance Database (http://hivdb.standford.edu). Polymorphisms were determined by comparison with the HIV-1 reference strain LAV-1bru (accession number K02013). Resistance conferring mutations in RT and PR were defined according to the recommendations of the International AIDS Society-USA.10
Phylogenetic analysis of the pol region sequences was performed using the Vector NTI software package (version 13.3.0, Invitrogen Corporation). Multiple nucleotide sequence alignment was performed using the AlignX program, an algorithm based on a modified ClustalW algorithm in which pairwise alignments are performed between selected sequences and HIV-1 group M reference sequences. A phylogenetic tree was constructed using CLC Free workbench (version 4.0.2, CLC bio A/S) through the neighbor-joining method in which a matrix of distances between all pairs of sequence was calculated after the sequences were aligned. Branch reproducibility was assessed using bootstrap analysis (500 replicates) and the results are presented as percentages. The tree was rooted with the simian immunodeficiency virus strain SIVcpz (accession number AF115393). Reference sequences for subtype B (accession numbers M19921 and NC001802), subtype B/G (accession number AY900576), subtype F (accession number DQ358812), subtype A (accession number EF545108), subtype C (accession number AY968312), subtype E (accession number AB052995), and CRF02_AG (accession numbers AF447832, DQ845388) viruses were included in the phylogenetic analysis.
Baseline characteristics at the time of diagnosis for the 72 individuals included in the study are presented in Table 2. Seventeen individuals were identified as recently infected, whereas 55 were characterized as chronically infected due to evidence of infection for greater than 1 year or no evidence of recent infection. Significant differences between recently infected and chronically infected individuals included the following: a higher proportion of the recently infected individuals identified homosexual contact as their primary risk factor, recently infected individuals were significantly younger than chronically infected patients, and CD4+ T cell levels were lower in chronically infected individuals than in those who were recently infected.
Table 2.
Baseline Characteristics of Patients Included in the Studya
| Recently infected | Chronically infected | Total | |
|---|---|---|---|
| Participants, No. (%) | 17 (23.6) | 55 (76.4) | 72 (100.0) |
| Sex, No. (%) | |||
| Male | 16 (94.1) | 43 (78.2) | 59 (81.9) |
| Female | 1 (5.9) | 12 (21.8) | 13 (18.1) |
| Age, mean (SD) | 32.0 (5.8) | 40.0 (12.2)b | 38.1 (11.5) |
| Risk factor, No. (%) | |||
| Homosexual contact | 15 (88.2)c | 28 (50.9) | 43 (59.7) |
| Heterosexual contact | 1 (5.9)c | 19 (34.5) | 20 (27.8) |
| Intravenous drug user | 1 (5.9) | 3 (5.5) | 4 (5.6) |
| Other/unknown | 0 (0.0) | 5 (9.1) | 5 (6.9) |
| Coinfection, No. (%) | |||
| HBV | 1 (5.6) | 14 (20.9) | 15 (17.6) |
| HCV | 1 (5.6) | 9 (13.4) | 10 (11.8) |
| CD4+/μl, median (IQR) | 510 (354–719) | 333 (168–574)d | 395 (226–617) |
| HIV-RNA log10 copies/ml, median (IQR) | 4.6 (4.3–5.3) | 4.7 (4.1–5.1) | 4.6 (4.1–5.1) |
No, number; SD, standard deviation; IQR, interquartile range.
p < 0.05 compared to recently infected; Student's t test.
p < 0.05; chi-squared test.
p < 0.05 compared to recently infected; Mann–Whitney U test.
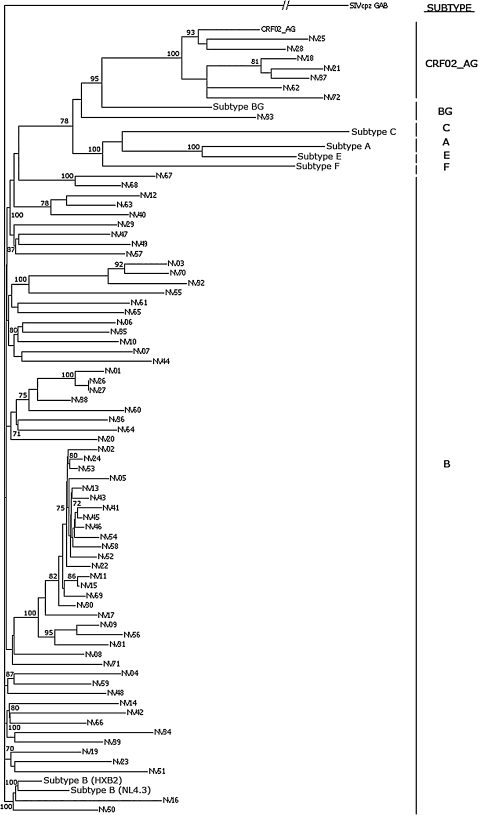
A phylogenetic tree constructed using sequences from the pol region of isolates included in the study is shown in Fig. 1. Our analysis resulted in one major cluster of 21 sequences (subtype B, NV02–NV31) and two minor clusters, each with three sequences (subtype B, NV01–NV27, and CRF02_AG, NV18–NV37). The 21 sequences comprising the major cluster were isolated from Spanish males between 23 and 46 years of age, with 20 of the 21 identifying homosexual/bisexual contact as their primary risk factor. This cluster may therefore represent an epidemiologically linked group of Spanish homosexual/bisexual males in this region of Spain. The minor cluster including the CRF02_AG sequences from NV18, NV21, and NV37 were isolated from one Spanish male and two Spanish females between the ages of 27 and 34 years, all of whom were diagnosed within 1 month of each other and identified heterosexual contact as their primary risk factor. The minor cluster including the subtype B sequences from NV01, NV26, and NV27 consist of three Spanish males two of whom identified homosexual/bisexual contact as their primary risk factor. Interestingly, sequences from NV26 and NV27 showed almost 100% sequence identity, although no social connection could be established between the two individuals.
FIG. 1.
Phylogenetic tree derived using pol sequences from the 72 HIV-1 isolates from western Andalucia. The tree was rooted using the simian immunodeficiency virus strain SIVcpz as an outgroup. Bootstrap values for selected branches are shown as percentages of 500 replicates. Reference sequences for subtype B, CRF02_AG, subtype B/G, subtype A, subtype C, subtype E, and subtype F viruses are included.
Subtype analysis revealed that the majority of the 72 individuals included in the study were infected with subtype B virus (64 of 72; 88.9%). Of those infected with non-B subtypes, seven (9.7%) were infected with the circulating recombinant form CRF_02AG and one (1.4%) was infected with the subtype recombinant B/G. Of 17 newly infected patients 15 (88.2%) were infected with subtype B, one (5.9%) was infected with CRF02_AG, and one (5.9%) with B/G. Of 55 chronically infected patients, 49 (89.1%) were infected with subtype B and six (10.9%) were infected with CRF_02AG. Interestingly, six of the 20 individuals (30%) identifying heterosexual contact as their primary risk factor were infected with CRF02_AG, whereas only one of 43 individuals (2.3%) identifying homosexual contact as their primary risk factor were infected with CRF02_AG (p < 0.01, chi-squared).
In order to characterize resistance mutations present in this population all mutations in the RT gene and both major and minor resistance mutations in the PR gene were considered.10 Table 3 shows the prevalence of mutations identified in each viral subtype at the time of diagnosis. Of the 72 isolates analyzed, 62 (86.1%) had at least one resistance-associated mutations in either the RT or PR genes. Sixty of the 72 isolates (83.3%) contained resistance-associated mutations to one class of antiretroviral drug, whereas 2 (2.9%) contained resistance-associated mutations to two classes of antiretroviral drug. No isolates had mutations associated with resistance to all three drug classes.
Table 3.
Prevalence of Resistance Mutations in RT and PR Genesa
| Drug class | Mutations |
Subtype B (n = 64) |
CRF_02AG (n = 7) |
B/G (n = 1) |
Total (n = 72) |
|---|---|---|---|---|---|
| Protease inhibitors | L10I | 2 (3.1) | 2 (2.8) | ||
| L10V | 1 (1.6) | 1 (1.4) | |||
| K20R | 4 (6.3) | 4 (5.6) | |||
| K20I | 7 (100.0) | 7 (9.7) | |||
| M36I | 24 (37.5) | 5 (71.4) | 1 (100.0) | 30 (41.7) | |
| M36P | 2 (3.1) | 1 (14.3) | 3 (4.2) | ||
| M36L | 1 (1.6) | 1 (1.4) | |||
| L63P | 32 (50.0) | 2 (28.6) | 1 (100.0) | 35 (48.6) | |
| A71V | 3 (4.7) | 3 (4.2) | |||
| A71T | 3 (4.7) | 3 (4.2) | |||
| V77I | 1 (1.6) | 1 (1.4) | |||
| Reverse transcriptase inhibitors | K103N | 1 (1.6) | 1 (14.3) | 2 (2.8) | |
| Y188C | 1 (1.6) | 1 (1.4) | |||
| No resistance mutations | 10 (15.6) | 0 | 0 | 10 (13.9) | |
| Mutations to one or more classes | 54 (84.4) | 7 (100.0) | 1 (100.0) | 62 (86.1) | |
| Mutations to > 1 class | 1 (1.6) | 1 (14.3) | 0 | 2 (2.8) | |
| Mutations to > 2 classes | 0 | 0 | 0 | 0 |
Numbers in parentheses indicate percentages.
None of the patient isolates contained major resistance mutations within the PR gene. However, 62 of the 72 isolates (86.1%) contained at least one minor mutation within the PR gene that is associated with resistance to protease inhibitors.10 Of the mutations identified, M36I and L63P were the most common, present at frequencies of 41.7% and 48.6%, respectively. Mutations in the RT region were less prevalent than those in the PR region. Of the 72 isolates analyzed, only two had mutations in RT that are associated with resistance to nucleoside or nonnucleoside reverse transcriptase inhibitors (NRTIs or NNRTIs). The K103N mutation, which confers resistance to all NNRTIs, was present in two isolates (2.8%), while Y188C, which has been associated with resistance to nevirapine, was present in one isolate (1.4%). When data from newly infected patients were analyzed alone in order to determine the level of transmitted resistance, none of the newly infected individuals carried viruses with major mutations, whereas 13 of 17 carried minor mutations in PR.
In the present study we have evaluated the prevalence of HIV-1 subtypes and resistance mutations in RT and PR in ART-naive patients at a tertiary medical center in Seville, Spain. Our data show that subtype B virus predominates in this region with a frequency similar to what has been reported for other regions of mainland Spain7,8 and France.11 However, the prevalence of subtype B virus is higher in western Andalucia than has been reported in neighboring Portugal12 and the Spanish Canary Islands,9 likely due to the differing immigration patterns in these regions. The data presented here also demonstrate a relatively high prevalence of the CRF02_AG recombinant form in infected individuals identifying heterosexual contact as their primary risk factor compared to those identifying homosexual contact as their primary risk factor. These results may indicate that distinct virus subtype diversities exist within the different subpopulations of this geographic region, similar to what has been reported in other regions of Europe.13
Our analysis also indicates that there are few major resistance mutations present in either RT or PR in this population. Of the 72 isolates analyzed, only two had resistance mutations in RT and none had major resistance mutations in PR. While the prevalence of primary mutations in this population was low, the frequency of isolates with at least one minor mutation in the PR gene was 86.1%. The effect that these mutations have on ART in the absence of major resistance mutations is unclear. These findings are in agreement with previous reports evaluating the presence of resistance mutations in drug-naive individuals in which the prevalence of major resistance mutations is low.14,15 The low prevalence of major resistance mutations in the chronically infected group may simply reflect the persistence of virus in the absence of selective pressure from antiretrovirals. However, the low prevalence of major resistance mutations in the recently infected group likely reflects a low level of transmission of resistant strains in this population. There are reports describing higher levels of major resistance mutations in some ART-naive cohorts due to the transmission of resistant strains.1,2 These differences may be due to either the virus variants circulating in that population, or the availability of antiretroviral drugs in that region.
Sequence Data
GenBank accession numbers for the sequences reported here are from EU288087 to EU288158.
Acknowledgments
Financial support was provided through Plan Nacional de Investigacion Cientifica, Desarrollo e Innovacion Tecnologica (I+D+I) PI05/0226, Instituto de Salud “Carlos III”-Fondo de Investigacion Sanitaria; Consejeria de Salud, Junta de Andalucia 0183/2006; Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía P06-CTS-01915; the Spanish Ministry of Health; and Red de Investigacion en SIDA, RD06/0006 (RIS network). M.J.M was supported by T32 GM07863 and T32 GM08353 from the NIH.
References
- 1.Grant RM. Hecht FM. Warmerdam M, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 2.Little SJ. Holte S. Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 3.Zolopa AR. Shafer RW. Warford A, et al. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann Intern Med. 1999;131:813–821. doi: 10.7326/0003-4819-131-11-199912070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda Y. Venzon DJ. Mitsuya H. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J Infect Dis. 1998;177:1207–1213. doi: 10.1086/515282. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Picado J. Savara AV. Sutton L. D'Aquila RT. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perno CF. Cozzi-Lepri A. Balotta C, et al. Secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naive patients treated with protease inhibitor-based therapy. J Infect Dis. 2001;184:983–991. doi: 10.1086/323604. [DOI] [PubMed] [Google Scholar]
- 7.Casado C. Urtasun I. Martin-Walther MV, et al. Genetic analysis of HIV-1 samples from Spain. J Acquir Immune Defic Syndr. 2000;23:68–74. doi: 10.1097/00126334-200001010-00009. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez M. Garcia F. Martinez NM, et al. Introduction of HIV type 1 non-B subtypes into Eastern Andalusia through immigration. J Med Virol. 2003;70:10–13. doi: 10.1002/jmv.10368. [DOI] [PubMed] [Google Scholar]
- 9.Holguin A. Pena MJ. Troncoso F. Soriano V. Introduction of non-B subtypes among Spaniards newly diagnosed with HIV type 1 in the Canary Islands. AIDS Res Hum Retroviruses. 2007;23:498–502. doi: 10.1089/aid.2006.0191. [DOI] [PubMed] [Google Scholar]
- 10.Johnson VA. Brun-Vezinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1: Fall 2006. Top HIV Med. 2006;14:125–130. [PubMed] [Google Scholar]
- 11.Couturier E. Damond F. Roques P, et al. HIV-1 diversity in France, 1996–1998. The AC 11 laboratory network. AIDS. 2000;14:289–296. doi: 10.1097/00002030-200002180-00011. [DOI] [PubMed] [Google Scholar]
- 12.Esteves A. Parreira R. Venenno T, et al. Molecular epidemiology of HIV type 1 infection in Portugal: High prevalence of non-B subtypes. AIDS Res Hum Retroviruses. 2002;18:313–325. doi: 10.1089/088922202753519089. [DOI] [PubMed] [Google Scholar]
- 13.Casado C. Urtasun I. Saragosti S, et al. Different distribution of HIV type 1 genetic variants in European patients with distinct risk practices. AIDS Res Hum Retroviruses. 2000;16:299–304. doi: 10.1089/088922200309403. [DOI] [PubMed] [Google Scholar]
- 14.Perno CF. Cozzi-Lepri A. Balotta C, et al. Low prevalence of primary mutations associated with drug resistance in antiviral-naive patients at therapy initiation. AIDS. 2002;16:619–624. doi: 10.1097/00002030-200203080-00014. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues R. Scherer LC. Oliveira CM, et al. Low prevalence of primary antiretroviral resistance mutations and predominance of HIV-1 clade C at polymerase gene in newly diagnosed individuals from south Brazil. Virus Res. 2006;116:201–207. doi: 10.1016/j.virusres.2005.10.004. [DOI] [PubMed] [Google Scholar]