Abstract
Unique viral variants and resistance mutations may occur in the genital tract of HIV-2 ARV-naive infected women. We sequenced and phylogenetically analyzed protease (PR), reverse transcriptase (RT), and envelope (ENV) from PBMC and genital tract samples from four ARV-naive women in Senegal. HIV-2 protease polymorphisms that predict HIV-1 protease inhibitor (PI) resistance were common. Two subjects had protease mutations (T77I and I64V) in genital tract samples that were not found in PBMCs. One subject had the HIV-2 reverse transcriptase M184I mutation in CVL DNA (but not PBMCs) that is known to confer 3TC/FTC resistance in HIV-2. In another subject, the reverse transcriptase A62V mutation was also found in CVL-RNA but not PBMCs. We found no significant difference in ENV variants between PBMCs and the genital tract. HIV-2 RT and PR mutations in the genital tract of ARV-naive females may have implications for transmitted HIV-2 resistance and ARV therapy.
HIV-2 is endemic in West Africa but unlike HIV-1 has had limited spread to other locales.1 Compared to HIV-1 infection, HIV-2 is characterized by a much longer asymptomatic stage, lower plasma viral loads, slower decline in CD4 count, decreased mortality rate due to AIDS, lower rates of mother-to-child transmission, and lower rates of sexual transmission.2–5 Nonetheless, a significant proportion of HIV-2-infected individuals eventually progress to AIDS.3
Antiretroviral therapy (ART) has become increasingly available in West Africa where HIV-2 infects up to 1–2 million people. However, HIV-2 is intrinsically resistant to the nonnucleoside reverse transcriptase inhibitors (NNRTI) and T-20 (enfurvirtide) and reports suggest that HIV-2 may be partially resistant to some protease inhibitors (PI) (e.g., amprenavir and nelfinavir) and have a low genetic barrier to nucleoside reverse transcriptase inhibitors (NRTI) resistance.6,7 Recent reports of HIV-2 polymorphisms in ARV-naive subjects suggest that outright resistance to NRTIs and PIs is rare in viruses genotyped from blood specimens, although “minor” mutations that often confer low level resistance in HIV-1 have been found.6,8 However, at least one recent report from Burkina Faso suggests that the major NRTI mutations M184V and Q151M may rarely be detected in ARV-naive individuals.8 Nevertheless, transmitted HIV-2 resistance has not been noted. Furthermore, it remains to be determined whether resistance mutations can occur in the genital tract of ARV-naive women infected with HIV-2. The genital tract, as a potential reservoir of such mutations could serve as a source of virologic failure once ART in instituted or lead to transmission of resistant virus.
To assess the HIV-2 viral variation and reverse transcriptase (RT) and protease (PR) polymorphisms that might lead to ARV resistance, we sequenced RT, PR, and/or envelope (Env) genes from peripheral blood mononuclear cells (PBMCs) and genital tract samples from four ARV-naive women in Senegal.
As part of ongoing HIV cohort studies in Dakar, Senegal, women 16 years or older presenting to the University of Dakar, Fann Hospital Infectious Disease Clinic were screened for HIV-1 and HIV-2 by serologic testing. Sera were tested using a microwell plate HIV-1/HIV-2 enzyme immunoassay (Genetic Systems), with positive sera confirmed using a rapid synthetic peptide-based membrane immunoassay (Multispot; Sanofi Diagnostic Pasteur) that classifies subjects as HIV-1, HIV-2, or dually seropositive. At enrollment and subsequent (follow-up) visits, subjects underwent a physical examination and completed an interview with questions concerning demographic characteristics and sexual and other behaviors. A routine medical history was taken and recorded on a standardized form by a clinician who also performed external genital and bimanual pelvic examinations as previously described.5 Cervicovaginal specimens for detection of genital tract HIV were collected as follows. With a speculum in place, a lavage of the cervix and vagina was performed using a plastic mixing cannula connected to a 10-ml syringe. Five milliliters of sterile physiologic saline was directed toward the cervix and the washings were aspirated (noting the volume aspirated) from the posterior vaginal fornix using the same syringe and mixing cannula. The sample was tested for occult blood using a Hemastix test stick, and if positive, was not used for purposes of HIV detection. Ten milliliters of peripheral blood was collected into tubes containing EDTA and analyzed using the FACS Count analyzer (Becton Dickinson Biosciences, San Jose, CA) to determine the number of CD4, CD8, and CD3 cells/μl of blood and for HIV-2 qualitative and quantitative RNA (plasma) and DNA (PBMC) assays.
This study was conducted according to procedures approved by the Institutional Review Boards of the Universities of Washington and Dakar and the Senegalese National AIDS Committee. All subjects provided informed consent for study participation.
Quantitative and qualitative assays for HIV-2 plasma and cervicovaginal RNA and PBMC and cervicovaginal DNA were performed using PCR-based assays developed at Roche Molecular Systems as previously described.5
Whole blood was collected in EDTA (purple top) tubes. Purified PBMC DNA was obtained using the QIAamp DNA Blood Mini Kit (Qiagen Inc., Valencia, CA). Cervicovaginal lavage (CVL) DNA was purified from the cellular fraction using the QIAamp DNA Blood Mini Kit (Qiagen Inc., Valencia, CA). HIV-2 viral RNA was purified from CVL supernatants using the QIAamp Viral RNA Mini Kit (Qiagen Inc., Valencia, CA). HIV-2-specific PCR was performed to amplify PR, RT, and Env using endpoint limiting dilution to avoid resampling as previously described.9 The HIV-2 protease-specific first round primers used were PR1 (5′-GGG AAA GAA GCC CCG CAA CTT C-3′) and PR2 (5′-GG-GTATTATAAGGATTAGTTGG-3′) and the second round primers were PR3 (5′-GCTGCACCTCAATTCTCTCTT-3′) and DP27 (5′-TAGATTTAATGACATGCCTAA-3′).10 The HIV-2 reverse transcriptase-specific first round primers used were RTC (5′-ATGACAGGGGATCCCCCAATCAATATTT-TTG-3′, 2309–2339) and RT2 (5′ -GAAGTCCCAGTCTG-GGATCCATGTCACTTGCCA-3′, 3593–3526) and second round primers were RT3 (5′-GAGGCATTAAAAGAGATCTGTGAAAAAATGG-3′, 2474–2504) and RT4 (5′-TCCCCAAATGACTAGTGCTTCTTTTTCCTAT-3′, 3500–3529).11 The HIV-2 Env-specific first round primers used were GG1 (5′ CCATACAGTGCTTGCCTGACAATGATGATTA 3′, 6298–6328 outer forward, primer numbering based on HIV-2 ROD accession no. M15390) and GG2 (5′ GGAGGT-TTTTCGTTCCCCAGACGGTCAGTCG 3′, 7872–7842 outer reverse) and the second round PCR with primers were GG3 (5′ TAATGGCACTAGAGCAGAGAATAGAACATA 3′, 6959–6988 inner forward) and GG4 (5′ TTTCTTTTGTAGGTGCGAAGCTAATTGGTG 3′, 7645–7615 inner reverse).9 PCR products were cloned and sequenced using standard methods as described.9
All PCR amplifications were done with procedural safeguards, including aliquoting of all reagents and physical separation of sample processing and post-PCR handling steps. Replicate control amplifications with no template were included in every PCR experiment to test for carryover contamination, and amplifications with 10 genome equivalents of HIV-2 (pROD10) were used as an internal control to monitor the efficiency of the PCR.
All sequences were assessed for potential sample mix-up and contamination as previously described. HIV sequences were aligned with reference sequences from the Los Alamos National Laboratory HIV Database (http://hiv-web.lanl.gov/) using CLUSTALW followed by manual adjustment using MacClade (version 4). Pairwise nucleotide distances (excluding gaps in the pairwise alignment) were estimated using both distance-based and maximum likelihood methods. Neighbor-joining and maximum likelihood methods were used to estimate evolutionary models and phylogenetic trees (PAUP* v4.0 b10). HIV-2 sequences were assigned to clades based on the above phylogenetic analyses. Pairwise nucleotide distances were estimated using distance-based methods and evolutionary models HKY85 or GTR + Γ + I under maximum likelihood (ML) criteria and implemented in PAUP*. Viral diversity was measured by determining the ML pairwise genetic distances between all sequences obtained at a given time point in PAUP*. Viral divergence was measured by estimating, using ML criteria, a most recent common ancestor (MRCA) sequence at the root node of each subject's clade of sequences, using reference sequence (HIV-2 ROD accession no. M15390) as the outgroup as previously described. Slatkin–Maddison tests were performed to assess for evidence of compartmentalization between sequences (env, pr, rt) from the PBMCs and genital tract.12
Protease and RT nucleotide sequences were translated into amino acid sequences with open reading frames in MacClade (version 4). Amino acid positions known or suspected to confer antiretroviral resistance in HIV-2 or the corresponding positions HIV-1 were assessed using the Stanford HIV drug resistance (http://hivdb.stanford.edu/) and the International AIDS Society-USA (http://www.iasusa.org) databases.
The demographic, clinical, and virologic characteristics of the four women reported in this study are shown on Table 1. We analyzed a total of 257 contemporaneous PR, RT, and Env sequences (163 genital tract and 94 PBMC) (Table 2).
Table 1.
Demographic, Clinical, and Virological Characteristics of Subjects
| Subject ID | Age | WHO | CD4 count | Plasma HIV-2 RNA viral load (copies/ml) | CVL HIV-2 RNA viral load (copies/ml) | PBMC HIV-2 DNA viral load (copies/106 cells) | CVL HIV-2 DNA viral load (copies/106 cells) |
|---|---|---|---|---|---|---|---|
| MIN 652 | 50 | 3 | 79 | 143,116 | 2027 | 777 | 14 |
| MIN 1974 | 39 | 3 | 505 | 7,267 | 270 | 78 | UDa |
| MIN 3473 | 33 | 3 | 125 | 100,610 | 1788 | 160 | UD |
| MIN 4199 | 48 | 1 | 111 | 36,536 | +c | 84 | NDb |
UD, undetectable (<10 copies/106).
ND, not done.
Positive but not quantifiable.
Table 2.
Protease, RT, Env Clones from PBMC and Cervicovaginal Lavage
| Subject ID | Sample date | PBMC | CVL DNA | CVL RNA |
|---|---|---|---|---|
| MIN 652 | 4/28/98 | PR-12 | PR-8 | PR-13 |
| RT-14 | RT-25 | RT-15 | ||
| ENV-15 | ENV-7 | ENV-10 | ||
| MIN 1974 | 3/8/99 | PR-0 | PR-1 | PR-0 |
| RT-6 | RT-0 | RT-10 | ||
| ENV-3 | ENV-0 | ENV-2 | ||
| MIN 3473 | 5/25/99 | PR-6 | PR-5 | PR-12 |
| RT-10 | RT-8 | RT-16 | ||
| ENV-8 | ENV-9 | ENV-10 | ||
| MIN 4199 | 10/6/97 | PR-6 | PR-0 | PR-0 |
| RT-9 | RT-4 | RT-5 | ||
| ENV-5 | ENV-0 | ENV-3 |
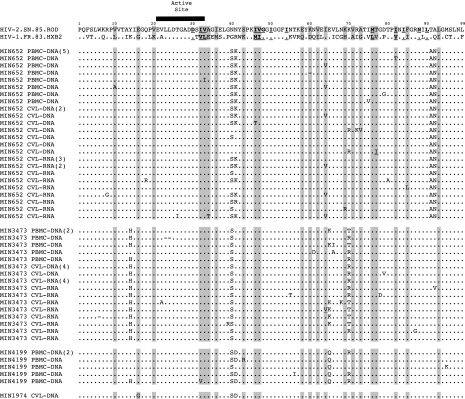
HIV-2 protease polymorphisms that correspond to HIV-1 mutations known to confer PI resistance in HIV-1 or have been described in HIV-2-infected individuals treated with PIs are shown in Fig. 1. There were HIV-2 polymorphisms at sites of predicted HIV-1 PI major resistance: V32V/I, L33V/I, M46I/T, I47V, l76M, and V82I/T, as well as polymorphisms at other associated HIV-1 resistance sites: L10V/A, G16G/E, K20V, E34A/T, M36I, K43S/R, Q58E, D60K, I62V, I64I/V, H69K/R, A71V, G73A, V77T/I, V82I, I85F, and I93L. Two subjects had PR mutations (T77I and I64V) found in genital tract samples that were not found in PBMCs (Fig. 1). One subject (MIN 3473) had the HIV-2 reverse transcriptase M184I mutation in CVL DNA (1 of 8 clones) but not CVL RNA (16 clones) or PBMCs (10 clones); although the clone containing the M184I resistance clustered with other RT sequences from MIN3473, it was truncated at amino acid position 238, unusually divergent, but without significant evidence of APOBEC-induced hypermutation using Hypermute 2.0 (http://www.hiv.lanl.gov/content/hivdb/HYPERMUT/hypermut.html) (see Fig. 2). Mutation M184I is known to confer 3TC/FTC resistance in HIV-1 and HIV-2. In another subject (MIN 652), the RT A62V mutation was also found in CVL-RNA (1 of 15 clones) but not CVL-DNA (25 clones) or PBMCs (14 clones) (Fig. 2). A62V is associated with multinucleoside resistance caused by Q151M and the 69-insertion complex in HIV-1; its effect alone and in HIV-2 is unknown.
FIG. 1.
HIV protease sequences (amino acids 1–99) from HIV-1 (HXB2), HIV-2 (ROD), and subject's PBMCs (HIV-2 DNA) and genital tract [cervicovaginal lavage (CVL) cell-associated HIV-2 DNA (CVL-DNA) and genital secretion-associated HIV-2 RNA (CVL-RNA)]. IAS-USA and Stanford HIV database predicted protease resistance mutations known to be associated with PI resistance in HIV-1 are shown (sites of predicted HIV-1 PI major resistance mutations are bolded and underlined and sites of potential HIV-1 resistance that are polymorphic between HIV-1 and HIV-2 are shaded). Note: Numbers in parentheses represent the number of identical sequences from that source; (−) represents the presence of a stop codon in the original nucleotide sequence.
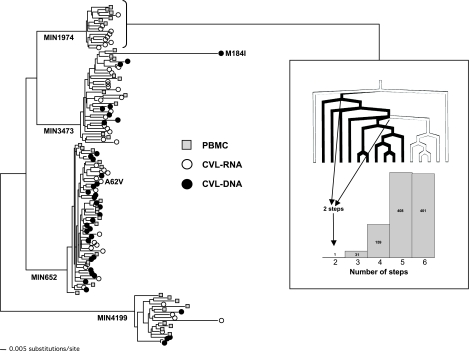
FIG. 2.
Maximum-likelihood phylogenetic tree of HIV-2 RT sequences from PBMCs, CVL-RNA, and CVL-DNA from four ARV-naive females. Potential ARV resistance mutations (M184I, A62V) found in CVL of two subjects are shown. Inset: Example of Slatkin–Maddison test for compartmentalization of CVL versus PBMC sequences for subject MIN1974. The tree shows traced character states corresponding to CVL (black lines) and PBMCs (white lines). There are two steps required to move along the tree between CVL and PBMCs. The histogram shows the of number of steps found during 1000 random character (taxa) shuffles on the ML tree.
To further assess potential differences between viral variants in the PBMCs and genital tract we used phylogenetic analyses and Slatkin–Maddison tests to quantify potential compartmentalization in HIV-2 sequences [PR, RT and Env (C2-V5)] obtained from blood (PBMCs) and the genital tract [CVL-DNA (cell-associated virus) and CVL-RNA (cell-free virus) (Table 3)]. Using the Slatkin–Maddison test to assess for compartmentalization only one subject (MIN1974) had evidence for segregation of sequences (RT) between the PBMCs and genital tract (see Fig. 2 and Table 3).
Table 3.
Slatkin–Maddison Test for Compartmentalization between PBMC and Genital Tracta
| Subject ID | Sample date | ENV (p) | PR (p) | RT (p) |
|---|---|---|---|---|
| MIN 652 | 4/28/98 | 0.19 all | 0.74 all | 0.72 all |
| 0.31 DNA | 0.08 DNA | 0.99 DNA | ||
| 0.59 RNA | 0.71 RNA | 0.97 RNA | ||
| 0.77 CVL DNA/RNA | 0.25 CVL DNA/RNA | 0.71 CVL DNA/RNA | ||
| MIN 1974 | 3/8/99 | ND | ND | 0.001 RNA |
| MIN 3473 | 5/25/99 | 0.54 all | 0.19 all | 1.0 all |
| 0.64 DNA | 0.98 DNA | 1.0 DNA | ||
| 0.50 RNA | 0.28 RNA | 0.72 RNA | ||
| 0.98 CVL DNA/RNA | 1.0 CVL DNA/RNA | 0.73 CVL DNA/RNA | ||
| MIN 4199 | 10/6/97 | 0.35 RNA | ND | 0.57 all |
| 0.13 DNA | ||||
| 0.08 RNA | ||||
| 0.75 CVL DNA/RNA |
Slatkin–Maddison test implemented in MacClade, p-value < 0.05/30 = 0.002 (Bonferroni correction) considered evidence for site segregation.
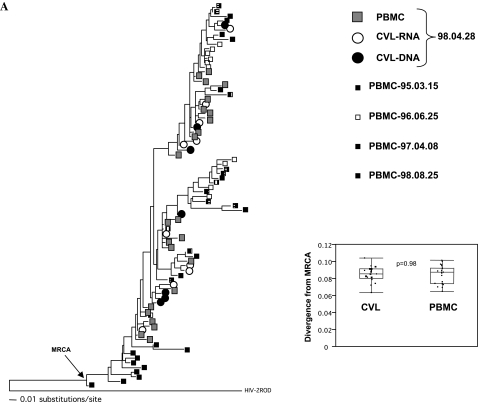
To further assess potential differences between viral variants in the PBMCs and genital tract we intensively sampled one subject's (MIN652) envelope sequences and performed phylogenetic analyses (Fig. 3). Longitudinal sampling of viral Env sequences in PBMCs over 3.5 years showed evidence of time-ordered evolution (p values: p = 0.0001, p < 0.0001, p = 0.26, and p = 0.0002 for SM test between 1995–1998, 1995–1996, 1996–1997, and 1997–1998 sequences, respectively; see Fig. 3). However, we found no evidence of compartmentalization between PBMCs and genital tract sequences or differences in the levels of diversity or divergence from the estimated MRCA (see Fig. 3).
FIG. 3.
Maximum-likelihood phylogenetic trees of HIV-2 Env sequences from PBMC, CVL-RNA and CVL-DNA from subject MIN652. (A) Boxes represent longitudinal HIV-2 Env PBMC sequences between March, 1995 and August, 1998. Open and closed circles represent HIV-2 Env sequences from April, 1998 CVL-RNA and CVL-DNA, respectively. Estimated most recent common ancestor (MRCA) is shown, as is HIV-2 ROD (M15390) used as an outgroup. Inset shows box-plots of the levels of divergence of CVL and PBMC sequences from MRCA. (B) Maximum-likelihood phylogenetic tree of HIV-2 Env sequences from PBMC, CVL-RNA, and CVL-DNA from April, 1998. Inset shows box-plots of the levels of diversity of CVL and PBMC sequences. P-values are nonparametric (Wilcoxon) between groups, PBMC vs. CVL or CVL-DNA vs. CVL-RNA. Box-plots show median, 25–75% IQR and range.
To our knowledge this study is the first to analyze HIV-2 protease and RT variants found in the PBMCs and genital tract of ARV-naive HIV-2-infected women. We found HIV-2 protease polymorphisms that predict PI resistance in HIV-1 were common in both the PBMCs and genital tract; however, their phenotypic effects on ARV resistance in HIV-2 infection remain to be further delineated. That HIV-2 PR polymorphisms are commonly found in the blood of ARV-naive HIV-2-infected individuals has been reported in previous studies;8,13 however, their presence in the genital tract is of some concern as this may be a site of lower penetration of some antiretroviral agents and consequently resistance may develop more easily. In two subjects we found evidence of potential PR resistance polymorphisms in the genital tract but not in the PBMCs. In addition, these same two subjects had evidence of NRTI mutations (M184I and A62V) in the genital tract but not the PBMCs. Despite this possible compartmentalization of ARV resistance mutations we found no evidence of more global compartmentalization between sequences in the PBMCs and genital tract in these two subjects (see Table 3). The M184I mutation predicts resistance to 3TC (lamivudine) and FTC (emtricitabine) in HIV-2. It was found in a genital tract sequence that although truncated and quite divergent, was monophyletic with other sequences from this subject ruling out contamination. It is possible that the sequence divergence occurred due to hypermutation, which was not detected using standard methods, or recombination between the predominant parental strain noted and an undetected second strain. Nevertheless, that a significant mutation NRTI such as M184I can apparently occur in the genital tract of an ARV-naive subject has implications for HIV-2 treatment (where 3TC is a first line ARV in West Africa) and transmission of HIV-2 drug resistance. Interestingly a recent report by Ruelle and colleagues from Burkina Faso found unlinked M184V and Q151M NRTI mutations in two subjects from a cohort of ARV-naive HIV-2-infected individuals.8 In their report they could not determine whether the mutations arose de novo, were transmitted from an ARV-experienced partner, or whether these two subjects had been taking unsupervised ARV. In our subjects, we cannot exclude these possibilities, but given their low frequency, we suspect that they arose de novo.
Despite a significant amount of progress in our understanding of the anatomic compartments and viral reservoirs associated with HIV-1 infection in ARV-naive and ARV-suppressed individuals, almost nothing is known about these factors in HIV-2 infection.14,15 Although we found evidence for differential detection of ARV resistance-associated mutations in the genital tract in two of four subjects studied, neither of these two subjects had statistically significant levels of compartmentalization based upon Slatkin–Maddison testing or viral diversity or divergence analyses. Only one subject (MIN1974) had evidence of statistically significant levels of compartmentalization based upon Slatkin–Maddison testing; it is noteworthy that she had a relatively high CD4 count and low plasma viral load (see Table 1). In a previous study, Sankale and colleagues assessed potential compartmentalization between PBMCs and female genital tract HIV-2 envelope C2–V3 region viral sequences; they found some evidence for differences between the two sites in two of three subjects but they could not rule out bias due to resampling in these two subjects.14
Limitations of this study include the small sample size, the lack of longitudinal genital tract sampling, our inability to amplify HIV-2 sequences in some subjects/regions, and lack of plasma HIV-2 RNA sequences available (due to sample availability/degradation) for comparison with PBMCs and genital tract sequences. Despite these limitations, this small study provides new insights into HIV-2 variants and potential ARV resistance in the female genital tract.
In summary, this study is the first to analyze HIV-2 protease and RT variants found in the PBMCs and genital tract of ARV-naive women. HIV-2 RT and PR polymorphisms/mutations may be found at low levels in the genital tract of ARV-naive females, which has implications for transmitted HIV-2 resistance and potentially could serve as a source of virologic failure once ART is instituted. Ultimately, however, ARV regimens designed specifically for HIV-2 infection, with proven benefit from randomized controlled trials, are needed in West Africa and elsewhere (Gen Bank Accession Nos. EU530819-EU531126).
Acknowledgments
We thank Deana Rich, Elise Reay-Ellers, Macoumba Toure, Dr. Mame B. Diouf, Dr. Mame A. Faye Niang, and Dr. Awa M. Coll-Seck for their invaluable coordination and supervision of study procedures in Senegal; Dr. Aissatou Diop, Dr. Pierre Ndiaye, Marie Pierre Sy, and Mame Dieumbe Mbengue-Ly for patient care; and Alison Starling for forms development and data management. In addition, we would like to thank Shirley Kwok, Rich Respess, Kelly Lagassic, Cindy Christopherson, and Jane Kuypers for their work in the development of assays of HIV-2 viral load, David C. Nickle, Laura Heath, and Gerald H. Learn for helpful discussions, and James I. Mullins for comments on the manuscript. We would like to thank the study participants without whom these studies would not be possible. These studies were supported by grants from the NIH/NIAID (to G.S.G. and N.B.K.) and the University of Washington CFAR.
References
- 1.De Cock KM. Adjorlolo G. Ekpini E, et al. Epidemiology and transmission of HIV-2. Why there is no HIV-2 pandemic. JAMA. 1993;3(270):2083–2086. doi: 10.1001/jama.270.17.2083. [Published erratum appears in JAMA 1994;271(3):196; see comments.] [DOI] [PubMed] [Google Scholar]
- 2.Simon F. Matheron S. Tamalet C, et al. Cellular and plasma viral load in patients infected with HIV-2. AIDS. 1993;7:1411–1417. doi: 10.1097/00002030-199311000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Marlink R. Kanki P. Thior I, et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- 4.Kanki PJ. Travers KU. MBoup S, et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343:943–946. doi: 10.1016/s0140-6736(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb GS. Sow PS. Hawes SE, et al. Equal plasma viral loads predict a similar rate of CD4+ T cell decline in human immunodeficiency virus (HIV) type 1- and HIV-2-infected individuals from Senegal, West Africa. J Infect Dis. 2002;185:905–914. doi: 10.1086/339295. [DOI] [PubMed] [Google Scholar]
- 6.Pieniazek D. Rayfield M. Hu DJ, et al. HIV-2 protease sequences of subtypes A and B harbor multiple mutations associated with protease inhibitor resistance in HIV-1. AIDS. 2004;18:495–502. doi: 10.1097/00002030-200402200-00016. [DOI] [PubMed] [Google Scholar]
- 7.Smith RA. Gottlieb GS. Anderson DJ. Pyrak CL. Preston BD. Human immunodeficiency virus types 1 and 2 exhibit comparable sensitivity to AZT and other nucleoside analog inhibitors in vitro. Antimicrob Agents Chemother. 2007;52(1):329–332. doi: 10.1128/AAC.01004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruelle J. Sanou M. Liu HF. Vandenbroucke AT. Duquenne A. Goubau P. Genetic polymorphisms and resistance mutations of HIV type 2 in antiretroviral-naive patients in Burkina Faso. AIDS Res Hum Retroviruses. 2007;23:955–964. doi: 10.1089/aid.2007.0034. [DOI] [PubMed] [Google Scholar]
- 9.Gottlieb GS. Sow PS. Hawes SE, et al. Molecular epidemiology of dual HIV-1/HIV-2 seropositive adults from Senegal, West Africa. AIDS Res Hum Retroviruses. 2003;19:575–584. doi: 10.1089/088922203322230941. [DOI] [PubMed] [Google Scholar]
- 10.Rodes B. Holguin A. Soriano V, et al. Emergence of drug resistance mutations in human immunodeficiency virus type 2-infected subjects undergoing antiretroviral therapy. J Clin Microbiol. 2000;38:1370–1374. doi: 10.1128/jcm.38.4.1370-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao F. Yue L. Robertson DL, et al. Genetic diversity of human immunodeficiency virus type 2: Evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slatkin M. Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damond F. Brun-Vezinet F. Matheron S, et al. Polymorphism of the human immunodeficiency virus type 2 (HIV-2) protease gene and selection of drug resistance mutations in HIV-2-infected patients treated with protease inhibitors. J Clin Microbiol. 2005;43:484–487. doi: 10.1128/JCM.43.1.484-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sankale JL. Mboup S. Essex ME. Kanki PJ. Genetic characterization of viral quasispecies in blood and cervical secretions of HIV-1- and HIV-2-infected women. AIDS Res Hum Retroviruses. 1998;14:1473–1481. doi: 10.1089/aid.1998.14.1473. [DOI] [PubMed] [Google Scholar]
- 15.Seck K. Samb N. Tempesta S, et al. Prevalence and risk factors of cervicovaginal HIV shedding among HIV-1 and HIV-2 infected women in Dakar, Senegal. Sex Transm Infect. 2001;77:190–193. doi: 10.1136/sti.77.3.190. [DOI] [PMC free article] [PubMed] [Google Scholar]