Abstract
To expand our current array of safe and potent oncolytic viruses, we screened a variety of wild-type (WT) rhabdoviruses against a panel of tumor cell lines. Our screen identified a number of viruses with varying degrees of killing activity. Maraba virus was the most potent of these strains. We built a recombinant system for the Maraba virus platform, engineered a series of attenuating mutations to expand its therapeutic index, and tested their potency in vitro and in vivo. A double mutant (MG1) strain containing both G protein (Q242R) and M protein (L123W) mutations attenuated Maraba virus in normal diploid cell lines, yet appeared to be hypervirulent in cancer cells. This selective attenuation was mediated through interferon (IFN)-dependent and -independent mechanisms. Finally, the Maraba MG1 strain had a 100-fold greater maximum tolerable dose (MTD) than WT Maraba in vivo and resulted in durable cures when systemically administered in syngeneic and xenograft models. In summary, we report a potent new oncolytic rhabdovirus platform with unique tumor-selective attenuating mutations.
Introduction
Replicating viruses are being evaluated in the lab and the clinic as therapies against cancer (reviewed in ref. 1). Our interest was to identify new oncolytic agents that could be used to complement our existing arsenal of viruses, and to use as a tool to better understand the oncolytic effect with relation to immunology, host and viral genetics.
There are >250 known rhabdoviruses currently divided into six genera;2 all classically identified by their bullet shaped virion. The archetypal rhabdoviruses are rabies and vesicular stomatitis virus (VSV), the most studied of the virus family. Although these viruses share similar morphologies, they are very different in their life cycle, host range, and pathology. In fact members of the rhabdovirus family have been shown to infect all organisms except bacteria (i.e., mammals, reptiles, fish, insects, fungi, and plants).2 However, with the exception of lyssaviruses such as rabies, rhabdoviruses are rarely associated with disease in humans. These relatively simple viruses have a number of properties that make them attractive as potential oncolytic agents. For example, antibodies to these rhabdoviruses will be rare in most populations of the world. Humoral immunity is known to be a limiting factor in administering viral therapeutics.3 Thus a lack of pre-existing antibody may improve the efficacy of the initial dosing, as well as allow for serial dosing with antigenically distinct strains. Many rhabdoviruses replicate very rapidly and to high titer in mammalian cells. Their small virion size is amenable to filtration through 0.2-µm pore. These two properties together significantly ease production and purification challenges commonly faced with virus-based therapeutics. Rhabdoviruses are amenable to genetic manipulation allowing for transgene insertion, structure/function guided mutagenesis to engineer improvements, or introduction of reporter genes to help track virus in vivo. Finally, the rhabdovirus life cycle occurs entirely within the cytoplasm of infected cells and never proceeds via a DNA intermediate so there is no opportunity for genotoxicity. Based on these properties, we propose that the Rhabdoviridae are a compelling oncolytic virus platform.
We therefore embarked on a search through the rhabdovirus family for strains with favorable safety and efficacy profiles. We have tested a subset of the rhabdoviruses available from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch, for their ability to kill tumor cells of the NCI 60 cell panel. Our findings demonstrate that several of these wild-type (WT) rhabdoviruses are efficient at killing cancer cell lines. These data also demonstrate that not all rhabdoviruses are effective at tumor cell killing, and that significant variation in killing activity is evident across the members of the rhabdovirus family of viruses. We subsequently engineered mutations into our best candidate virus and demonstrated tumor selectivity in vitro, and safety and efficacy in syngeneic and xenograft mouse models of cancer.
Results
Maraba virus demonstrates potent oncolytic properties in vitro
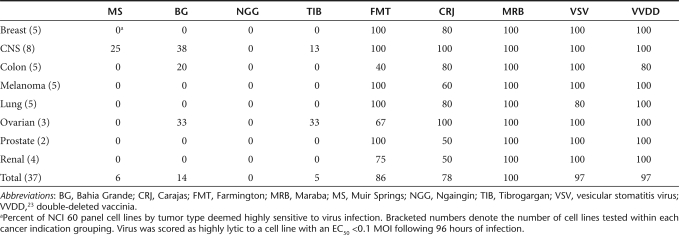
The rhabdovirus family is vast and genetically and geographically diverse. To initiate our screen, we selected a panel of 20 rhabdoviruses with a previously documented capacity to replicate in mammalian cells. In preliminary experiments using a handful of malignant and normal cell lines, we defined a subset of seven rhabdoviruses with extreme phenotypes, either highly lytic on tumors cells or profoundly attenuated on normal cells for further evaluation. These seven candidates were tested in cell killing assays in 96-well format on cell lines from the NCI 60 tumor cell panel and an assortment of mouse tumor lines (Table 1 and Supplementary Figure S1). Several species were highly lytic on human tumor lines with EC50 scores of <0.1 multiplicities of infection (MOIs) by plaque-forming units (pfu) for the majority of cell lines tested. In particular, Maraba4, Carajas,4 and Farmington virus5 appeared to be very effective at killing human tumor lines from all cancer indications represented in the cell panel (with the exception of Farmington virus on colon tumor lines). Interestingly, not all rhabdoviruses possess the capacity to efficiently kill cancer cells. Viruses such as Muir Springs,6 Bahia Grande,6 Ngaingin,7 and Tibrogargan8 showed activity in a very small proportion of tumor cells. Presently, the mechanism(s) governing the restriction of these viruses remains unknown.
Table 1. Strains of uncharacterized rhabdoviruses are highly lytic on NCI 60 cell panel.
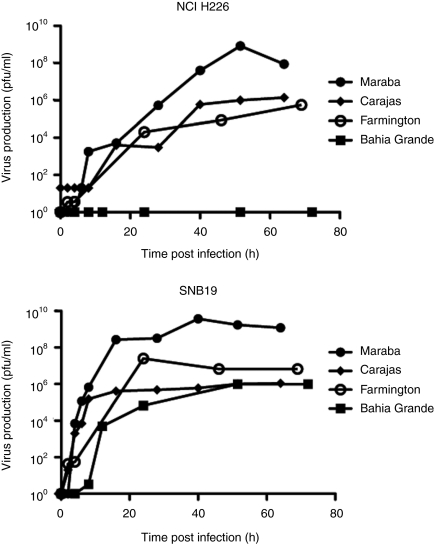
To further characterize these lytic viruses, we performed single-step growth curves on both a susceptible cell line (SNB19), as well as a relatively resistant cell line (NCI H226), to monitor the kinetics of replication and to quantify virus burst sizes. We were unable to detect virus following infection of NCI H226 cells with Bahia Grande virus, which is consistent with our observation that Bahia Grande is only poorly cytolytic on this cell line. However, Bahia Grande was able to replicate to a similar degree as Farmington and Carajas on the SNB19 cells, again correlating with its cytolytic capacity. Both Farmington and Carajas produced progeny with similar kinetics and with equivalent burst sizes when assayed on NCI H226 cells. Farmington appeared to replicate to higher titers than Carajas on SNB19 cells although both clearly produced sufficient progeny to result in rapid killing of this susceptible cell line (Figure 1). Maraba produced virus with equal or faster kinetics than the other three strains, and to a much higher titer than the other viruses on both SNB19 and NCI H226 cells.
Figure 1.
Maraba demonstrates high viral productivity in cancer cells. One-step growth curve to assess the viral productivity of Maraba, Carajas, Farmington, and Bahia Grande in (a) NCI H226 and (b) SNB19 cells.
Maraba virus demonstrated good cytolytic activity against tumor lines, rapid virus production, and large burst size. These are all properties we speculate may contribute to good oncolytic activity. We therefore elected to proceed with Maraba as a potential oncolytic candidate.
Maraba virus is a vesiculovirus related to VSV
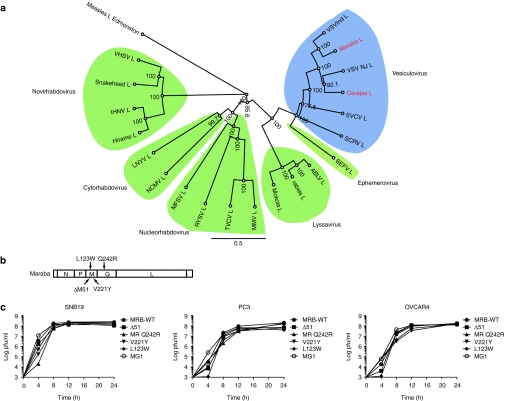
As a prelude to genetically manipulating Maraba virus, we employed a “shot gun” sequencing approach to obtain the full-length genomic sequence for this strain. Subsequent phylogenetic analysis was performed by aligning the amino acid sequence of the Maraba L protein to members of the six known genera of the rhabdovirus family (Figure 2a). This finding showed Maraba virus to indeed be a vesiculovirus as previously predicted from complement fixation serology.4 The virus has the expected genomic structure common to other vesiculoviruses, with five discrete cistrons separated by transcriptional stop/start sequences responsible for delineating the virus's N, P, M, G, and L genes (Figure 2b; sequences submitted to National Center for Biotechnology Information).
Figure 2.
Recombinant Maraba virus variants. (a) Based on the amino acid sequence of their L protein, Maraba and Carajas are characterized as species in the genus: vesiculovirus. Line lengths are proportional to genetic distance. (b) Illustration of the Maraba genome outlining the various mutation sites for our single mutants (L123W, V221Y, Q242R) and double mutant (Q242R L123W). (c) One-step growth curves were used to characterize viral productivity of our engineered Maraba mutants.
Engineered Maraba virus mutants show improved cancer cell selectivity
We developed a recombinant system to genetically manipulate Maraba virus using a strategy that has been successful in developing reverse genetic systems for several negative strand RNA viruses.9,10,11,12 We next sought to introduce mutations into our recombinant Maraba backbone (rMaraba WT) that might improve the tumor-selective killing properties of WT Maraba virus. One strategy was to engineer mutations that had previously been demonstrated to be attenuating for VSV, a close relative of Maraba, with the expectation that they may be similarly attenuating for Maraba virus. For example, we had reported that a deletion of methionine 51 in the M protein of VSV rendered the virus defective for blocking the interferon (IFN) response in infected cells.13 Similarly, we had shown that a double mutation in VSV M protein at amino acids V221F and S226R also rendered the virus unable to block nuclear cytoplasmic transport of host mRNAs and thereby allowed the host cell to propagate an IFN response.13 Considering the Glasgow strain of VSV also has an S226R variation in its matrix protein, we hypothesized that the attenuating phenotype for the V221F S226R double mutant may arise from the mutation at V221F alone. Thus, we constructed and rescued the ΔM51 Maraba recombinant virus, and V221Y Maraba mutant strain as possible attenuated variants (Figure 2b).
Alternatively, we explored two previously identified mutations that reportedly improved the replication of VSV on BHK-21 cells (M protein L123W and L protein H242R).14 We anticipated that these mutations, once engineered into Maraba virus either alone or together, might improve the virus's ability to replicate in tumor cells. Aligning the Maraba sequence to VSV, we identified the corresponding mutations to be L123W and Q242R in the Maraba sequence of the M and G proteins, respectively, and constructed recombinant Maraba viruses with the M protein L123W or the G protein Q242R single mutations, or both L123W and Q242R (hereafter referred to as Maraba MG1) (Figure 2b). All mutant viruses were subsequently fully sequenced from rescued virus stocks to confirm these engineered mutations as well as the absence of any unexpected sequence alterations.
Single-step growth assays were performed for these new Maraba mutants to assess their growth kinetics and burst size in several different cell cancer cell lines (Figure 2c). In general, the growth kinetics and burst size were similar for all recombinant Maraba variants. However, we did notice that Maraba MG1 appeared to replicate more quickly during the early phase of infection, producing 1–3 logs more progeny within the first 4 hours of infection when compared to the other mutant strains (Figure 2c).
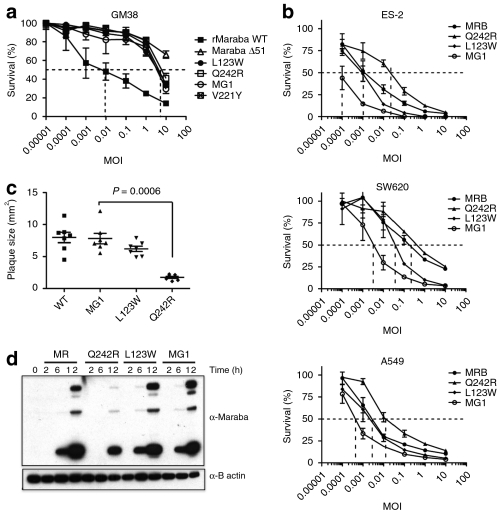
We next tested the cytotoxicity of our rMaraba WT and mutant strains on primary human skin fibroblasts (GM38 cells) to detect and quantify any attenuation resulting from our engineered mutations (Figure 3a). As expected, the Maraba ΔM51 virus is greatly attenuated on these primary cells (EC50 >> 10 MOI) compared to rMaraba WT (EC50 = 0.01 MOI). The V221Y was also attenuated, although to a slightly lesser degree than the ΔM51 (EC50 = 3 MOI). Surprisingly, both the L123W and the Q242R single mutants were also highly attenuated (EC50 = 3 MOI). Furthermore, the double mutant combining both L123W and Q242R mutations was equally attenuated as compared to the single mutants, resulting in a 100-fold increase in EC50 after 72-hour infection of primary human fibroblasts (EC50 = 3 MOI). These results were surprising given that both mutations were expected to improve replication, not attenuate the virus. These phenotypes correlated with plaque formation as well. Following infection of GM38 fibroblasts, detectable plaques became visible 1 week after infection with rMaraba WT. However, no plaques were visible over the same time frame for the various Maraba single mutants or Maraba MG1. This again highlighted the severely attenuated nature of V221Y, L123W, and Maraba MG1 on normal primary fibroblasts. In contrast, large plaques formed on tumor cells (SNB19) following just 24 hours of infection with either rMaraba WT, V221Y, L123W, or Maraba MG1 (Figure 3c). The Q242R mutant, however, produced smaller plaques as compared to the other strains, suggesting that this mutation may slightly impair replication of this strain in tumor cells. Interestingly, however, the double mutant that contains the Q242R mutation clearly demonstrated no such impairment on malignant cells (Figure 3c).
Figure 3.
Maraba mutants retain their killing potency in cancer cells yet are attenuated on normal cells. (a) Mutations in the M and G protein attenuate the ability of Maraba to kill GM38 cells. Viability assays were performed in GM38 cells at 72 hours postinfection with Maraba, and the indicated Maraba mutants. (b) Maraba and Maraba variants are highly lytic in a variety of tumor cell lines. Cells infected with rMaraba WT and the Maraba variants were assayed for viability using resazurin. (c) Engineering L123W mutation into Q242R Maraba mutant reverts plaque sizes to a wild-type phenotype on tumor cells. (d) Time course of viral protein induction. Immunoblots of OVCAR4 mock-infected and MR, rMaraba wild type, Q242R-, L123W-, and MG1-infected cells. WT, wild type.
In contrast to our observations on normal fibroblasts, all of the mutant strains remained highly lytic when assayed on a panel of malignant cell lines (Figure 3b). The L123W strain appeared to be as cytolytic as the rMaraba WT on tumor cells and thereby demonstrated an improved in vitro therapeutic index compared to the WT strain. Maraba Q242R was very cytolytic on all three tumor lines, albeit less so than its parental WT virus and in line with our plaque size observations. The double mutant, however, demonstrated an interesting reversal of this phenotype as it showed no impairment in cytotoxicity due to the Q242R mutation it harbors. In fact, Maraba MG1 consistently appeared to be the most lytic strain on cancer cell lines (Figure 3b), even more cytolytic than rMaraba WT. It appears that the combination of L123W and Q242R gives rise to a Maraba strain that is selectively hypervirulent only on cancer cells yet remains attenuated on normal fibroblasts (compare Figure 3a to Figure 3b). This was also evident when viral protein production was assayed over time in OVCAR4 human ovarian carcinoma cells (Figure 3d). rMaraba WT and the L123W strains showed rapid viral protein induction, whereas the Q242R mutant lagged behind. Here, again the Q242R L123W double mutant Maraba showed no impairment in viral protein kinetics.
Having established Maraba MG1 as a promising new oncolytic virus candidate, we were interested in seeing how it compared to the well-characterized oncolytic rhabdovirus ΔM51 VSV by comparing their cytolytic activity in normal human fibroblast and cancer cell lines. Although both viruses were equally attenuated on GM38 cells, Maraba MG1 was significantly more virulent in several cancer cell lines when compared to ΔM51 VSV (*P < 0.05, Supplementary Figure S2). These results suggest that the in vitro therapeutic index of Maraba MG1 may be greater than that of our previously developed ΔM51 VSV virus.
Maraba mutants are variably defective in blocking host IFN antiviral responses
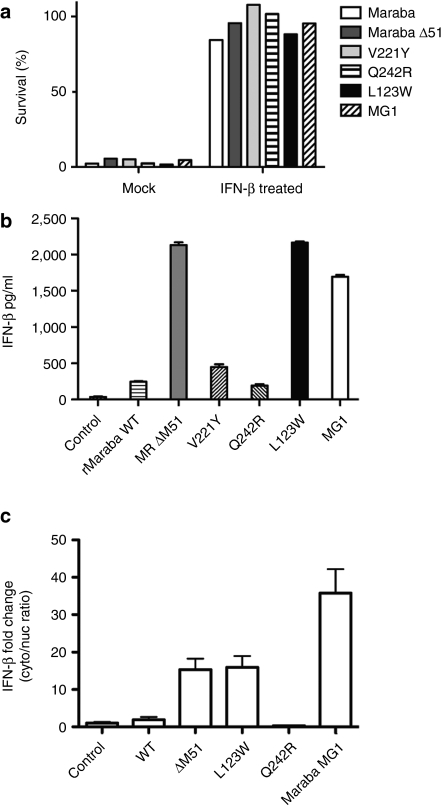
To examine the interaction of our Maraba variant viruses with host IFN signaling, we first established that Maraba virus and each of the engineered mutants were susceptible to IFN-mediated antiviral responses by observing robust protection of Vero cells following pretreatment with IFN-β (Figure 4a). Because Maraba mutant strains are selectively attenuated in normal primary fibroblasts, we sought to understand whether this attenuation was due to defects in innate immune blockade. For example, ΔM51 and V221 mutations had previously been shown in VSV to render the virus unable to block nuclear/cytoplasmic mRNA transport, thereby inhibiting the host IFN transcriptional cascade.13 We anticipated these mutations to function in the same way when introduced into Maraba virus. When PC3 cells were either mock-infected, or infected with rMaraba WT, we could detect very little IFN-β production, consistent with the ability of the parental virus to block innate immune responses (Figure 4b). As expected, the ΔM51 and V221Y mutants did show defects in the ability to block IFN production as measured in our IFN-β enzyme-linked immunosorbent assay (Figure 4b). The L123W mutant also demonstrated a defect in its ability to block IFN production (Figure 4b). The Q242R mutant, however, was similar to the WT virus in its ability to block cytokine production in PC3 cells, indicating that this mutant has no defect in IFN blockade. Therefore, the profound attenuation of the Q242R mutant appears to be unrelated host IFN responses. When the two single mutations are combined in the Maraba MG1 variant, the resulting virus was similarly defective in blocking IFN production as the L123W single mutant (Figure 4b).
Figure 4.
Maraba mutants vary in their ability to block interferon production. (a) IFN-β protects Vero cells from Maraba-mediated cell killing. Cells were infected with the rMaraba WT or the Maraba variants (MOI = 10) or pretreated with IFN-β (100 U) followed by infection. Viability was assessed by crystal violet assay. (b) The L123W mutant, V221Y mutant, Maraba ΔM51, and the double mutant L123W-M/Q242R-G allow IFN to be produced following infection of PC3 cells. Enzyme-linked immunosorbent assay was used to assess human IFN-β production 48 hours postinfection. (c) rMaraba WT blocks nuclear/cytoplasmic transport of IFN-β. IFN-β mRNA induction is not detected in the cytoplasmic fraction after infection with rMaraba WT or Maraba Q242R as determined by qRT-PCR. Cells infected with Maraba Δ51M, L123W, and MG1 show IFN-β mRNA induction in the cytoplasm following infection. IFN, interferon; MOI, multiplicity of infection; qRT-PCR, quantitative reverse transcriptase-PCR.
Finally, we observed that IFN-β mRNA transport from the nuclear compartment to the cytoplasm was blocked following infection with either rMaraba WT or the Q242R mutant (Figure 4c). These results are consistent with previous reports that indicate vesiculoviruses rely on their matrix proteins to inhibit the IFN transcriptional cascade by several mechanisms including blocking mRNA transport to the cytoplasm.13,15,16 These data suggest Maraba virus may employ this same strategy. In contrast, Maraba ΔM51, the L123W strain, and Maraba MG1 all showed a “leak” of IFN-β mRNA detectable in the cytoplasm following virus infection, and this deficiency in mRNA blockade correlated with the viruses' ability to block IFN responses as measured by enzyme-linked immunosorbent assay (Figure 4b).
Maraba MG1 in less toxic in vivo
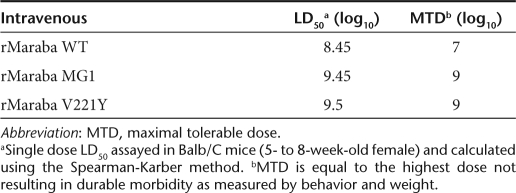
We determined the LD50 and maximum tolerable doses (MTDs) for Maraba WT and several attenuated strains. Because our desired therapeutic route of administration to treat disseminated tumors would likely be intravenous, we treated mice at a range of doses intravenously with either WT virus, or two of our most promising mutant strains. We observed that Maraba virus is well-tolerated following intravenous injection of immunocompetent Balb/C mice. As predicted from the in vitro data (Figure 3a), Maraba MG1 was attenuated in vivo with an MTD 2 logs higher than the parental Maraba WT strain (Table 2). Animals that received lethal doses of rMaraba WT, V221Y, or Maraba MG1 display signs of central nervous system infection and had significant titers of virus in their brains (data not shown). At doses below the MTD, mice generally showed transient weight loss, dehydration, and piloerection consistent with a virus infection. These symptoms resolved within 3–4 days postinfection and no virus was detected in the brains of these mice killed 12 days postinfection.
Table 2. Intravenous single-dose toxicity of rMaraba virus strains.
Maraba MG1 is efficacious in syngeneic and xenograft tumor models
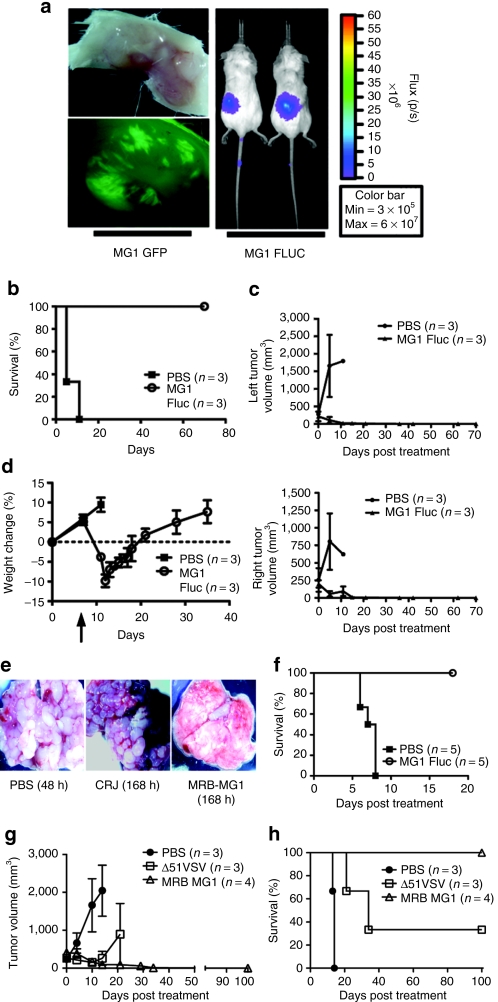
We engineered Maraba MG1 strains expressing GFP or firefly luciferase and examined their replication in subcutaneous CT26 tumors following systemic administration. We observed Maraba MG1 virus to be delivered to tumor beds and to replicate in tumor tissue using both bioluminescent imaging in whole animals, and fluorescent microscopy in tumor explants (Figure 5a). We then examined the efficacy of Maraba MG1 on a bilateral CT26 subcutaneous tumor model (Figure 5b,c). Five days after the first treatment, control animals treated with saline reached end point with tumors reaching a size of ≥750 mm3. However, animals that received six systemic doses of Maraba MG1 responded to treatment with complete tumor regression by day 35 and durable cures in 100% of the animals (**P = 0.01, Figure 5b,c). Finally, intravenous Maraba MG1 treatment was well tolerated in the animals, with no mortality and minimal morbidity. Piloerection, mild dehydration, and transient weight loss was observed but all these symptoms resolved within 2 weeks of the first treatment (Figure 5d).
Figure 5.
Systemic delivery of Maraba MG1 is efficacious in the syngeneic CT26 mouse tumor model. (a) Maraba MG1-GFP and Maraba MG1-FLUC replicate selectively at the tumor site 24 hours after intravenous injection of 5 × 108 pfu. (b) Durable survival of Balb/C mice with bilateral subcutaneous CT26 tumors after Maraba MG1 treatment. “n” denotes number of animals in the experimental group; P = 0.0224. (c) Tumor volumes were calculated on a biweekly basis. Error bars denote SEM. (d) Mouse weights measured before and after treatment with Maraba MG1 and control. Error bars denote SEM. (e) Systemic treatment of disseminated CT26 lung tumors. Tumors were treated intravenously with either PBS, Carajas virus, or Maraba MG1 at day 10 after tumor implantation. At day 17, animals were killed and lung images were captured. (f) Effective lung tumor treatment with six intravenous doses (5 × 108 pfu/dose) of Maraba MG1. “n” denotes the number of animals in the experimental group; P = 0.0023. (g) Maraba MG1 is more effective at treating large CT26 subcutaneous tumors as compared to VSV Δ51. Error bars denote SEM. (h) Durable survival of all Balb/C animals with CT26 subcutaneous tumors after Maraba MG1 treatment (six doses of 5 × 108 pfu/dose). “n” denotes the number of animals in the experimental group, **P < 0.01, P = 0.0091. PBS, phosphate-buffered saline; VSV, vesicular stomatitis virus.
We also sought to determine the utility of Maraba MG1 to reduce tumor burden in a disseminated disease model. CT26 cells were injected intravenously into Balb/C mice to induce disseminated lung tumors. Although saline [phosphate-buffered saline (PBS)]- and Carajas-treated animals had a massive tumor burden, Maraba MG1 animals showed little to no tumor burden and exhibited a normal lung phenotype (Figure 5e). Moreover, Maraba MG1 led to a significant prolongation in survival when administered systemically thrice weekly for 2 weeks (**P = 0.0023; Figure 5f). These findings demonstrate the potency of Maraba MG1 to effectively treat an aggressive, disseminated syngeneic tumor model.
Having established the effectiveness of Maraba MG1 in the previously characterized CT26 model, we next sought to determine how Maraba MG1 compared to our previous best oncolytic virus, ΔM51 VSV,13 in the same model. As both viruses are effective oncolytic agents, we made the current model more stringent by commencing treatment when tumors reached a size of 300 mm3. With a tumor of this size, we were able to better delineate the limits of these viruses with respect to efficacy. Both viruses reduced the tumor burden; however, only Maraba MG1 was able to eradicate all treated tumors (Figure 5g). Moreover, Maraba MG1 led to durable cures in 100% of the treated animals as compared to only 30% in the ΔM51 VSV–treated group (Figure 5h). This result demonstrates the enhanced nature of Maraba MG1 in successfully treating sizeable, aggressive syngeneic tumors.
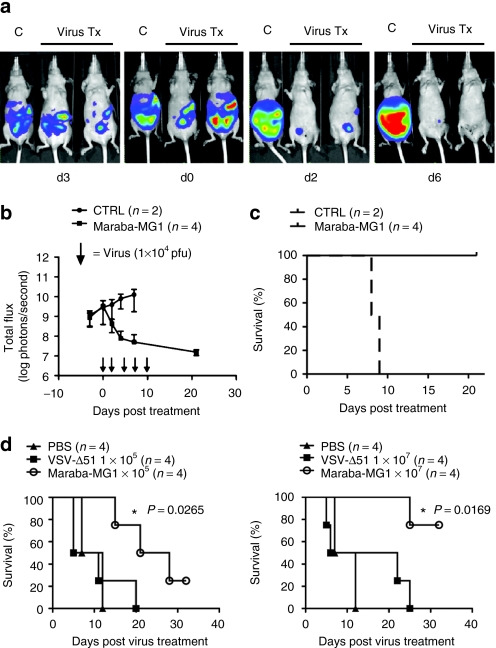
To complement these studies in immunocompetent animals, we tested Maraba MG1 using a bioluminescent human ES2 ovarian xenograft model. Even at very low doses (1 × 104 pfu), animals treated with Maraba MG1 had a significant decrease in tumor burden (*P = 0.0177, Figure 6a–c). In contrast, control-treated mice rapidly developed ascites with increasing tumor burden until reaching end point. Systemic treatment of ES2 tumor–bearing mice using low and high doses of virus demonstrated a dose-dependent tumor response (Figure 6d). Again, when tested against ΔM51 VSV, Maraba MG1 showed better efficacy at both the high (*P = 0.0169) and low doses (*P = 0.0265, Figure 6d).
Figure 6.
Maraba MG1 demonstrates enhanced efficacy in an ovarian xenograft mouse tumor model. (a) IVIS images demonstrating a rapid tumor regression following virus treatment. ES2 human ovarian tumor cells adapted for bioluminescent imaging were injected intraperitoneally into athymic mice (CD-1 nude) followed by treatment with Maraba MG1 10 days after tumor injection. (b) Bioluminescent flux plot quantifying tumor in response to virus treatment from a. (c) Maraba MG1 is efficacious in a human ovarian ES2 xenograft model (5 intraperitoneal doses of 1 × 104 pfu/dose); *P < 0.05, P = 0.0177. (d) Kaplan–Meier plot demonstrates enhanced survival after virus treatment. Maraba MG1 is superior to VSV Δ51 in treating ES2 xenograft tumors with either 5 intravenous injections of 1 × 105 (left panel) or 1 × 107 pfu/dose (right panel). “n” denotes the number of animals in each experimental group; *P = 0.0265 (1 × 105 pfu dose) and *P = 0.0169 (1 × 107 pfu dose). CTRL, control; PBS, phosphate-buffered saline; VSV, vesicular stomatitis virus.
Discussion
In this report, we sought to identify a new oncolytic virus to serve as a platform on which to build effective virus-based cancer therapies for the clinic. Several factors led us to focus our attention on the rhabdovirus family (summarized in the introduction). Our approach was to screen the Rhabdoviridae for a virus with properties we felt contributed to strong oncolytic effects. Because we anticipate systemic delivery will be an imperative in treating disseminated cancers in the clinical setting, our intent was to develop a virus that could be delivered intravenously, and initiate an infection at disparate tumor sites. We postulate that one of the in vivo limitations to effective therapy could be that virus delivery to the tumor bed may be limiting. In fact, we have previously seen dose thresholds, below which, virus is not effectively delivered to the tumor in mouse models, and these doses were not efficacious.13 Thus, we were interested in finding viruses able to kill tumor cells at low MOI, replicate quickly, and to produce large numbers of progeny to potentially maximize the probability and magnitude of tumor bed infection. These experiments identified Maraba virus as the most promising candidate in our screens.
We subsequently derived the full genome sequence of Maraba and Carajas, the first sequence data for these viruses, and performed phylogenetic analysis using the L protein amino acid sequences to align to Rhabdoviridae members. Previous serological data at the time of identification of Maraba virus predicted it to be a vesiculovirus.4 Our sequence analysis confirmed that Maraba and Carajas are indeed vesiculoviruses closely related to VSV Indiana and VSV New Jersey, respectively. Its interesting that after screening a number of disparate rhabdoviruses, that vesiculoviruses like VSV, Carajas, and Maraba appear to be most well suited to killing human cancer cells and that not all rhabdoviruses are equal in this regard. This is perhaps not surprising given the complexities that govern host range. However, because poorly oncolytic rhabdoviruses such as Ngaingin, Tibrogargan, and Bahia Grande can infect mammalian cells, seroconvert animals and can productively infect diverse permissive cells from monkey (Vero) and hamster (BHK),6,7,8 it would seem unlikely that surface receptor restrictions are at play. Perhaps instead, intracellular constituents and responses are determinants, with the most prominent candidate being IFN signaling. However, most of the tumor cell lines in the NCI60 panel display type-I IFN defects,13 yet these viruses do poorly on these cells. This finding suggests other unknown intracellular restrictions to rhabdovirus replication and/or cytolysis of cancer cells may be responsible, and warrant further exploration.
Once we identified Maraba virus as a suitable genetic background, we sought to improve its tumor selectivity through engineered mutations. Two interesting mutations we tested were originally identified in an experiment to monitor RNA virus fitness in changing environments.14 Both L123W and H242R (Q242R in Maraba) were individually able to increase VSV replication in BHK-21 cells. Additionally, the authors reported that the combination of the two mutations retained this fitness phenotype. We reasoned that BHK-21 cells might be similar to many tumor cells in that they are highly permissive to virus infection and devoid of an IFN response. If these mutations could increase VSV fitness in BHK-21 cells, perhaps they might improve Maraba virus replication in tumor cells. Although we could not detect any improvement in tumor cell killing of the individual mutants engineered into Maraba virus, it appeared that the combination of L123W and Q242R mutations does improve virulence of Maraba MG1 virus on the tumor cell lines tested. Importantly, the L123W/Q242R mutations do not increase Maraba virus cytolysis of primary human fibroblast cells, thereby creating a significant therapeutic index of at least 3 logs (EC50 < 10‐3 MOI on some tumor cells; EC50 = 3 MOI on GM38 fibroblasts). To our surprise, each of the Q242R and L123W mutations were in fact attenuating on normal fibroblasts. The L123W mutation seems to function much the same as ΔM51, resulting in a deficit in the ability to block nuclear/cytoplasmic transport thereby inhibiting the host IFN transcriptional cascade. To our knowledge, this is the first demonstration of a role for this region of the matrix protein in mitigating host innate immune defenses. Previously, mutations in this region have been reported to affect translation of virus mRNA.17 Although the Q242R mutation also severely reduces Maraba virus cytolysis of normal cells, it does so in an IFN-independent manner. Presently, we do not understand how Q242R attenuates Maraba virus on GM38 fibroblasts, nor do we appreciate how the combination of two mutations that individually attenuate Maraba virus on normal cells, result in a more virulent phenotype on tumor cells. The mechanisms underlying these observations are currently being investigated. Nonetheless, these properties form the basis of a potent tumor selectivity that results in a significant increase in therapeutic index for this newly described Maraba-based oncolytic virus platform.
As predicted from our in vitro results, the Maraba MG1 variant was significantly less toxic than the WT virus when delivered intravenously into Balb/C mice. The MTD was 100-fold greater than the WT virus. This allowed us to dose well below the MTD to achieve significant tumor regressions in both tumor models. In the CT26 model for example, six doses of Maraba MG1 virus were sufficient to provide complete durable cures in all mice. Particularly important for the clinical setting, Maraba MG1 was effective at treating both human xenograft tumors and an immunocompetent syngeneic tumor model by systemic delivery. Virus replication was demonstrated at the tumor site in the CT26 tumor model following intravenous injection, consistent with viral mediated oncolysis as a contributor to efficacy. In fact, Maraba MG1 appeared to be more effective than our previous best candidate VSV ΔM51 in the CT26 syngeneic and ES2 xenograft models. This is consistent with the in vitro data demonstrating Maraba MG1 to be more effective at killing tumor cells than ΔM51 VSV or even rMaraba WT virus.
Several studies have definitively demonstrated that the host immune response plays a positive and negative role in oncolytic virus efficacy.18,19,20,21 It will be interesting to determine how Maraba MG1 interacts with the host immune response, both antiviral and antitumor, and to compare and contrast this with effects from other oncolytic viruses.
In conclusion, we describe here a newly described oncolytic platform and a recombinant system to genetically manipulate the virus. We introduce the engineered mutant, Maraba MG1, and demonstrate the strain to be safe and efficacious by systemic delivery in multiple tumor models, both immunocompetent and human xenograft.
Materials and Methods
Cell lines. Human A549 lung carcinoma, human HeLa cervical carcinoma, human ES2 ovarian carcinoma, murine CT26 colon carcinoma (American Type Tissue Collection), human GM13030 melanoma cells, human GM38 primary fibroblasts (National Institute of General Medical Sciences Mutant Cell Repository, Camden, NJ), and cell lines from the NCI 60 cell panel obtained from the Developmental Therapeutics Program, National Cancer Institute (Bethesda, MD)were propagated in Dulbecco's modified Eagle's medium (Hyclone, Logan, UT) supplemented with 10% fetal calf serum (Cansera, Etobicoke, Ontario, Canada).
In vitro cytotoxicity screen. Cells from the NCI 60 cell panel were plated in 96-well plates to a confluency of 90%. These cells were infected at log dilutions with various rhabdoviruses, as indicated. After 96 hours postinfection, the monolayers were washed, fixed, and stained with 1% crystal violet solution. Stained monolayers were subsequently solubilized in 1% sodium dodecyl sulfate in water to create homogenous lysates. Absorbance was read at 595 nm to score for viable cells.
Single-step growth curves. Cells were infected with the indicated viruses at an MOI of 5 pfu/cell for 1 hour. Cells were then washed with PBS and incubated at 37 °C. Aliquots (100 µl) were taken at time 0, 4, 8, 12, 24, and 48 hours postinfection and titers assessed with a standard plaque assay.
Viability assays. The indicated cell lines were seeded into 96-well plates (1 × 104 cells/well). The next day cells were infected with the indicated viruses at various MOIs (0.0001–10 pfu/cell). Following a 48-hour incubation, Alamar Blue (Resazurin sodium salt; Sigma-Aldrich, St Louis, MO) was added to a final concentration of 20 µg/ml. After a 6-hour incubation, the absorbance was read at a wavelength of 573 nm.
Plaque assays. GM38, SNB19, or Vero cells were seeded into 6-well plates (5 × 105 cells per/well). The next day 100 µl of serial viral dilutions were prepared and added for 1 hour to Vero cells. After viral adsorption, 2 ml of agarose overlay was added (1:1, 1% agarose:2× Dulbecco's modified Eagle's medium and 20% FCS). Plaques were counted the following day. Where applicable, diameters were measured and plaque area calculated using the following formula Area = πr2.
Phylogenetic analysis. Phylogenetic relationships between rhabdoviruses based on a muscle alignment of L protein amino acid sequences, and using the paramyxovirus Measles Edmonston strain as the outgroup. The tree was generated by the neighbor-joining method and bootstrap values (indicated for each branch node) were estimated using 1,000 tree replicas. Branch lengths are proportional to genetic distances. The scale bar corresponds to substitutions per amino acid site.
Recombinant Maraba rescue system. A549 lung carcinoma cells seeded at 3.0 × 105 cells/well in 6-well plates were infected 24 hours later with vaccinia virus (MOI = 10) expressing the T7 RNA polymerase22 in OptiMeM medium for 1.5 hours. Following removal of the vaccinia virus, each well was transfected with LC-KAN Maraba (2 µg) together with pCI-Neo constructs encoding for Maraba N (1 µg), P (1.25 µg), and L (0.25 µg) with lipofectamine 2000 (5 µl per well) according to the manufacturer's instructions. The transfection reagent was removed 5 hours later and replaced with Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. At 48 hours following the transfection, medium was collected (pooled from two plates), filtered (0.2 µm) to remove contaminating vaccinia virus, and 1 ml was used to infect SNB19 glioblastoma cells in each well of a 6-well plate. Cytopathic effects visible 24–48 hours later were indicative of a successful rescue, which was confirmed by purifying viral RNA and reverse transcriptase (RT)-PCR with Maraba-specific primers. All viruses underwent three rounds of plaque purification (on SNB19 cells), before scale up, purification on sucrose cushion, and resuspension in PBS containing 15% glucose.
Mutagenesis and Maraba variants. Single phosphorylated mutagenic primers (45–55 bp) were used with the high-fidelity Phusion enzyme (NEB, Pickering, ON, Canada) to create the panel of LC-KAN Maraba mutants described within. Briefly, a PCR reaction was carried out with 100 ng of mutagenic primer and 100 ng DNA template with Hot Start addition of enzyme (98 °C—2 min, 80 °C hold—add enzyme) and typical PCR setup (98 °C—10 seconds, 55 °C—30 seconds, 72 °C for 7 minutes for 30 cycles). Dimethyl sulfoxide was added in the range of 0–6% in increments of 2%. The parental plasmid was digested with DpnI (NEB) (37 °C for 1 hour) and 4 µl of the 25 µl DpnI-digested PCR mixture was used to transform TOP-10 competent cells (Invitrogen, Carlsbad, CA). Positive clones were screened by introduction of noncoding change restriction site changes (adding or removing) followed by sequencing. The different attenuated mutants described here include deletion of Met-51 in the M protein (ΔM51), Leu-123 to Trp in the M protein (L123W), Val-221 to Tyr in the M protein (V221Y), Gln-242 to Arg in the G protein (Q242R), and double mutant Leu-123 to Trp in the M protein, and Gln-242 to Arg in the G protein (Maraba MG1).
IFN treatment. To assess the ability of recombinant IFN-β to protect Vero cells from infection and killing by rMaraba WT and the Maraba variants, we incubated cells with either control media or IFN-β (100 U) for 18 hours followed by viral infection (MOI = 10). Cell viability was assessed using crystal violet assay. Briefly, cells were incubated with 1% crystal violet solution, washed, dried, resuspended in 1% sodium dodecyl sulfate, and read at a wavelength of 595 nm.
IFN enzyme-linked immunosorbent assay. IFN-β levels were measured using the VeriKine Human IFN-β enzyme-linked immunosorbent assay kit (PBL Interferon Source, Piscataway, NJ). Briefly, PC3 cells were infected (MOI = 5) with ΔM51 VSV, rMaraba WT, and the Maraba mutants ΔM51, V221Y, Q242R, L123W, and MG1. Culture media (1 ml) was collected 48 hours postinfection and incubated along side standards provided by the manufacturer.
Sequencing and cloning of Maraba rhabdovirus. Maraba rhabdovirus was amplified on Vero cells and RNA was isolated from purified virus by standard techniques (Trizol+RNeasy; Invitrogen). With the exception of the 5′ and 3′ terminal ends, the virus sequence was obtained using the mRNA Complete cloning kit (Invitrogen). The 3′- and 5′-end sequencing was completed following T4 RNA ligase–mediated ligation of T7 DNA primers to either end followed by RT-PCR and cloning into pCR2.1-TOPO (Invitrogen). The viral complementary DNA was amplified in a single RT-PCR reaction (yielding a >11 kbp fragment) and cloned into a modified LC-KAN vector (Lucigen, Middleton, WI) carrying a T7 promoter upstream of the 5′-antigenomic leader sequence and immediately downstream of the 3′-terminator a modified hepatitis delta virus ribozyme and T7 polymerase termination signal sequence.
Quantitative RT-PCR to detect nuclear and cytoplasmic IFN. Nuclear and cytoplasmic RNA was separated as described previously.13 Briefly OVCAR4 cells either mock treated or infected with rMaraba WT, ΔM51, L123W, Q242R, or Maraba MG1 were harvested in PBS, pelleted, and resuspended in 200 µl of lysis buffer (25 mmol/l Tris: pH 7.4, 15 mmol/l NaCl, 12.5 mmol/l MgCl2 5% sucrose, and 1% NP-40). The lysates were incubated at 4 °C for 10 minutes with occasional vortexing. Nuclei were collected by centrifugation at 1000g for 3 minutes. The supernatant (cytoplasmic fraction) was collected while nuclear fraction was washed once with 250 µl of lysis buffer followed by total RNA extraction using the Qiagen RNeasy kit (as per manufacturer's instructions; Qiagen, Almeda, CA). Quantitative RT-PCR of IFN-β mRNA was performed using the Quantitect SYBR Green RT-PCR kit from Qiagen with previously described primers. IFN-β was assayed from nuclear and cytoplasmic fractions and normalized to hypoxanthine-guanine phosphoribosyltransferase mRNA from the same compartment. Normalized values were normalized again to values from uninfected nuclear and cytoplasmic fractions, respectively, to determine fold induction values in each compartment, following virus infection. Plotted values indicate the ratio of normalized mRNA induction from the cytoplasmic to nuclear compartments. All quantitative PCR values were calculated using the ΔCT method.
Determination of in vivo toxicity. Groups of 3–5 Balb/C mice (6–8 weeks old) were injected once intravenously in half log increments of virus ranging from 3 × 106 to 3 × 109 pfu. The animals were monitored for signs of distress including weight loss, morbidity, piloerection, hindlimb paralysis, and respiratory distress. All experiments were conducted with the approval of the University of Ottawa Animal Care and Veterinary Service in concordance with guidelines established by the Canadian Council on Animal Care.
CT26 subcutaneous tumor model. Subcutaneous tumors were established by injecting 3 × 105 murine CT26 colon cancer cells23 were injected in the right and/or left flanks of 6–8 week old female Balb/C mice. Palpable tumors (10–600 mm3) were treated with six intravenous injections (thrice weekly) of either 51VSV or Maraba MG1 (5 × 108 pfu/dose). Animals were monitored for piloerection, weight loss, morbidity, hind leg paralysis, and respiratory distress. When tumor burden exceeded a size of 750 mm3 animals were killed. Tumor volumes were calculated using (L × W2)/2, where L = tumor length and W = tumor width.
Imaging Maraba MG1 virus in a subcutaneous tumor model. Maraba MG1 was adapted for fluorescent or bioluminescent imaging by genetically engineering in eGFP or firefly luciferase (FLUC), respectively. MG1-GFP and MG1-FLUC was injected IV (1 × 108) into Balb/C animals bearing subcutaneous CT26 tumors. At 24 hours postinfection MG1-GFP-infected animals were killed and their tumors were extracted and imaged under a Nikon fluorescent microscope (Nikon Instruments, Melville, NY). Animals infected with MG1-FLUC were injected with luciferin and underwent live imaging using the IVIS Xenogen 200 system (Caliper Life Sciences, Hopkinton, MA).
CT26 lung tumor model. Lung tumors were established by a single intravenous injection of 3 × 105 CT26 colon cancer cells into 6- to 8-week-old female Balb/C animals.24 Generally, mice develop severe respiratory distress, piloerection, and hunched phenotype at day 16–18 at which point they are killed. Mice were either intravenously treated with PBS, Carajas, or Maraba MG1 (5 × 108 pfu/dose) treated at day 10, 12, 14, 17, 19, and 21. Some animals were killed at day 17 and images were captured on a Nikon dissecting microscope. The remaining animals were monitored for survival.
Ovarian xenograft model. Human ovarian ES2 cells were adapted for bioluminescent imaging at which time 1 × 106 ES2 cells were injected intraperitoneally into 6- to 8-week-old athymic CD-1 nude mice. Untreated CD-1 animals develop ascites at about day 15–17. Intraperitoneal and intravenous injections were performed on day 8, 9, 12, 14 and 16 with 1 × 104 to 1 × 107 pfu/dose of Maraba MG1 or VSV Δ51. Tumor imaging was captured with a Xenogen 200 IVIS system (Caliper Life Sciences).
Statistics. For plaque size determinations, one-way analysis of variance was performed using the Bonferroni multiple comparison's test to derive a P value (Graphpad Prism; Graphpad Software, La Jolla, CA). For Kaplan–Meier plots, we compared survival plots using Mantel–Cox log-rank analysis (Graphpad Prism). We compared titers and viability using a two-tailed unpaired Student's T test to derive a P value (Graphpad Prism).
SUPPLEMENTARY MATERIAL Figure S1. Rhabdovirus mediated cell killing on the NCI 60 cell panel. Figure S2. Maraba-MG1 demonstrates both enhanced cytolytic capability as compared to Δ51VSV.
Acknowledgments
We thank Robert Tesh (University of Texas Medical Branch) for providing virus isolates from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch. Funding was provided by the National Cancer Institute of Canada with funds raised by the Terry Fox Foundation (D.F.S), Canadian Foundation for Innovation (D.F.S.) and a Program Project Grant from the Terry Fox Foundation (J.C.B, J.A.M, and D.F.S).
Supplementary Material
Rhabdovirus mediated cell killing on the NCI 60 cell panel.
Maraba-MG1 demonstrates both enhanced cytolytic capability as compared to Δ51VSV.
REFERENCES
- Vähä-Koskela MJ, Heikkilä JE., and, Hinkkanen AE. Oncolytic viruses in cancer therapy. Cancer Lett. 2007;254:178–216. doi: 10.1016/j.canlet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U., and, Ball LA. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier: London; 2005. [Google Scholar]
- Power AT, Wang J, Falls TJ, Paterson JM, Parato KA, Lichty BD.et al. (2007Carrier cell-based delivery of an oncolytic virus circumvents antiviral immunity Mol Ther 15123–130. [DOI] [PubMed] [Google Scholar]
- Travassos da Rosa AP, Tesh RB, Travassos da Rosa JF, Herve JP., and, Main AJ. Carajas and Maraba viruses, two new vesiculoviruses isolated from phlebotomine sand flies in Brazil. Am J Trop Med Hyg. 1984;33:999–1006. doi: 10.4269/ajtmh.1984.33.999. [DOI] [PubMed] [Google Scholar]
- Travassos da Rosa AP, Mather TN, Takeda T, Whitehouse CA, Shope RE, Popov VL, et al. Two new rhabdoviruses (Rhabdoviridae) isolated from birds during surveillance for arboviral encephalitis, northeastern United States. 2002;8:614–618. doi: 10.3201/eid0806.010384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschner JH, Calisher CH, Vorndam AV., and, Francy DB. Identification and characterization of Bahia Grande, Reed Ranch and Muir Springs viruses, related members of the family Rhabdoviridae with widespread distribution in the United States. J Gen Virol. 1986;67 (Pt 6):1081–1089. doi: 10.1099/0022-1317-67-6-1081. [DOI] [PubMed] [Google Scholar]
- Doherty RL, Carley JG, Standfast HA, Dyce AL, Kay BH., and, Snowdon WA. Isolation of arboviruses from mosquitoes, biting midges, sandflies and vertebrates collected in Queensland, 1969 and 1970. Trans R Soc Trop Med Hyg. 1973;67:536–543. doi: 10.1016/0035-9203(73)90084-9. [DOI] [PubMed] [Google Scholar]
- Cybinski DH, George St TD, Standfast HA., and, McGregor A. Isolation of Tibrogargan virus, a new Australian rhabdovirus, from Culicoides brevitarsis. Aust J Biol Sci. 1980;5:301–308. [Google Scholar]
- Nakaya T, Cros J, Park MS, Nakaya Y, Zheng H, Sagrera A, et al. Recombinant Newcastle disease virus as a vaccine vector. J Virol. 2001;75:11868–11873. doi: 10.1128/JVI.75.23.11868-11873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Stillman EA, Whitt MA., and, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Ball LA, Barr JN., and, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MJ, Mebatsion T., and, Conzelmann KK. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Sanjuán R, Moya A., and, Elena SF. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci USA. 2004;101:15376–15379. doi: 10.1073/pnas.0404125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terstegen L, Gatsios P, Ludwig S, Pleschka S, Jahnen-Dechent W, Heinrich PC, et al. The vesicular stomatitis virus matrix protein inhibits glycoprotein 130-dependent STAT activation. J Immunol. 2001;167:5209–5216. doi: 10.4049/jimmunol.167.9.5209. [DOI] [PubMed] [Google Scholar]
- Ferran MC., and, Lucas-Lenard JM. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JH, McKenzie MO., and, Lyles DS. Role of residues 121 to 124 of vesicular stomatitis virus matrix protein in virus assembly and virus-host interaction. J Virol. 2006;80:3701–3711. doi: 10.1128/JVI.80.8.3701-3711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocca EA. The host response to cancer virotherapy. Curr Opin Mol Ther. 2008;10:38–45. [PubMed] [Google Scholar]
- Endo Y, Sakai R, Ouchi M, Onimatsu H, Hioki M, Kagawa S, et al. Virus mediated oncolysis induces danger signal and stimulates cytotoxic T-lymphocyte activity via proteasome activator upregulation. Oncogene. 2008;27:2375–2381. doi: 10.1038/sj.onc.1210884. [DOI] [PubMed] [Google Scholar]
- Altomonte J, Wu L, Meseck M, Chen L, Ebert O, Garcia-Sastre A, et al. Enhanced oncolytic potency of vesicular stomatitis virus through vector-mediated inhibition of NK and NKT cells. Cancer Gene Ther. 2009;16:266–278. doi: 10.1038/cgt.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D, Spencer JF, Toth K., and, Wold WS. Effect of preexisting immunity on oncolytic adenovirus vector INGN 007 antitumor efficacy in immunocompetent and immunosuppressed Syrian hamsters. J Virol. 2009;83:2130–2139. doi: 10.1128/JVI.02127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier FW., and, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brattain MG, Strobel-Stevens J, Fine D, Webb M., and, Sarrif AM. Establishment of mouse colonic carcinoma cell lines with different metastatic properties. Cancer Res. 1980;40:2142–2146. [PubMed] [Google Scholar]
- Specht JM, Wang G, Do MT, Lam JS, Royal RE, Reeves ME, et al. Dendritic cells retrovirally transduced with a model antigen gene are therapeutically effective against established pulmonary metastases. J Exp Med. 1997;186:1213–1221. doi: 10.1084/jem.186.8.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rhabdovirus mediated cell killing on the NCI 60 cell panel.
Maraba-MG1 demonstrates both enhanced cytolytic capability as compared to Δ51VSV.