Abstract
DNA vaccines have undergone important enhancements in their design, formulation, and delivery process. Past literature supports that DNA vaccines are not as immunogenic in nonhuman primates as live vector systems. The most potent recombinant vector system for induction of cellular immune responses in macaques and humans is adenovirus serotype 5 (Ad5), an important benchmark for new vaccine development. Here, we performed a head-to-head evaluation of the Merck Ad5 SIV vaccine and an optimized electroporation (EP) delivered SIV DNA vaccine in macaques. Animals receiving the Ad5 vaccine were immunized three times, whereas the DNA-vaccinated animals were immunized up to four times based on optimized protocols. We observed significant differences in the quantity of IFNγ responses by enzyme-linked immunosorbent spot (ELISpot), greater proliferative capacity of CD8+ T cells, and increased polyfunctionality of both CD4+ and CD8+ T cells in the DNA-vaccinated group. Importantly, Ad5 immunizations failed to boost following the first immunization, whereas DNA responses were continually boosted with all four immunizations demonstrating a major advantage of these improved DNA vaccines. These optimized DNA vaccines induce very different immune phenotypes than traditional Ad5 vaccines, suggesting that they could play an important role in vaccine research and development.
Introduction
One of the most well-studied vaccine approaches for generating a cell-mediated response to HIV-1 is the recombinant adenovirus serotype 5 (Ad5) vector. Preclinical data in rhesus macaques demonstrated that Ad5-vectored immunogens induced the strongest cellular immune responses when compared to other viral vectors.1 They were also shown to be safe, well tolerated, and immunogenic in phase I clinical trials.2,3 The Merck trivalent Ad5 vaccine, a HIV-1 gag/pol/nef vaccine, was among the most immunogenic. It was shown to lower viral replication in a SHIV89.6P challenge study.4 The preclinical data as well as the phase I clinical data in 2004 led to the advancement of the MrkAd5 vaccine to a phase II, test-of-concept, clinical trial. With primary end points of reduction in infection rates and/or reduction of set-point viremia, the study was halted in September 2007 due to a lack of efficacy.5,6
The results of phase I trials and the Step trial highlighted the issue of pre-existing immune responses to recombinant viral vectors, which can dampen the potency of the vaccine. In the Step trial, there appeared to be more HIV-1 infections in the vaccine group over the placebo group in vaccinees that had high baseline Ad5 titers 5. Although several reports later indicated that there was no causative relationship between baseline Ad5-specific CD4+ T-cell responses and an increased rate of HIV-1 acquisition,7,8 pre-existing antibody and T-cell responses to viral vectors remain an important consideration. In this respect, plasmid DNA offers a distinct advantage for vaccine development. DNA vaccines circumvent pre-existing serology issues and have been shown to continually boost immune responses after three immunizations with homologous constructs.9 However, traditional DNA immunizations have not been as immunogenic as the Ad5 platform and have been relegated to a priming role in heterologous prime-boost strategies for recombinant viral vectors.10,11,12 Surprisingly, in contrast to small animal studies, prior DNA vaccines were biased, in particular, toward induction of CD4 rather than CD8 T-cell responses in nonhuman primates and human studies.
The immunogenicity of DNA vaccines has dramatically increased in the past 5 years. This increase in potency of plasmid DNA has been, in part, a result of enhancements in antigen expression through gene optimization by codon13,14 and RNA15 optimization. Higher concentration formulations have further improved the delivery of plasmid DNA.16 However, the use of in vivo electroporation (EP) to enhance transfection efficiency has contributed significantly to the increase in DNA-induced immune responses to levels that were previously not achievable.9,16,17,18,19,20 However, it is widely believed that DNA as a vaccine platform does not perform as well as live vector systems for the induction of immune responses. It would be a distinct advantage for vaccine design for a nonlive, nonreplicating system to be able to produce immune responses in a similar fashion to live vaccine platforms. It is also important to understand the immune phenotypes induced by the newer DNA vaccines.
Given the advances in DNA vaccine technology, a head-to-head comparison with the well-established Ad5 vaccination platform was performed to study the immune phenotype of each modality. In this study, we vaccinated groups of rhesus macaques with either an optimized DNA vaccine consisting of consensus macSIVgag, env, and pol immunogens21 or with the Merck Ad5 vaccine consisting of SIVmac239gag, nef, and pol immunogens. We assessed the magnitude and the quality of the cellular responses induced by each vaccination strategy. Following vaccination, we observed clear differences in immune magnitude and boosting induced by the two vectors. Compared to Ad5, DNA vaccination resulted in a higher number of polyfunctional T cells and a particular ability to expand T-cell proliferative responses. These responses were long-lasting and maintained at high levels. Taken together, these data demonstrate the potency of DNA vaccines and functional differences of the immune responses induced by Ad5 and DNA vaccines suggest that these platforms are unique. Furthermore, the new generation of DNA vaccines improves considerably on the induction of CD8+ T-cell immunity.
Results
Study design
A group of five rhesus macaques (DNA) were immunized at weeks 0, 6, 12, and 18 with 1.0 mg each of SIVgag, SIVenv, and SIVpol (Table 1). Another group of five macaques were immunized at weeks 0, 4, and 24 with 1 × 1010 pfu each of recombinant Ad5-vectored SIVmac239 gag, nef, and pol.
Table 1. Study design.
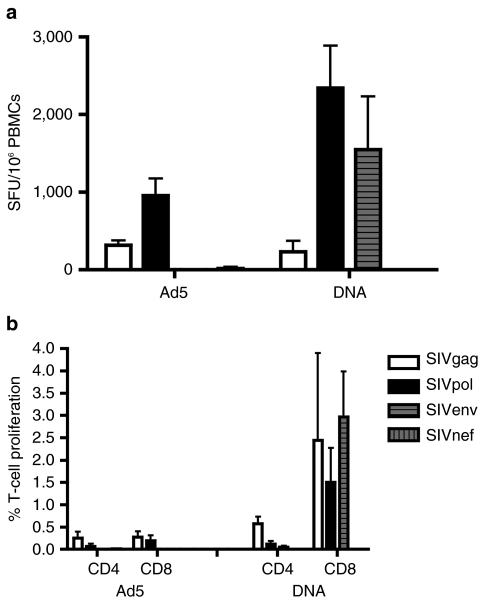
Enhanced IFNγ production following plasmid DNA vaccination compared to Ad5 vaccination
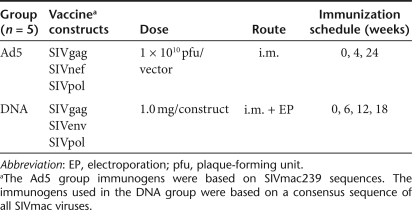
We first sought to compare the induction of cellular responses following DNA and Ad5 immunization by the standard, quantitative IFNγ enzyme-linked immunosorbent spot (ELISpot). Cellular responses to SIVgag, which was included in both vaccines, averaged 591 ± 267 spot-forming unit (SFU)/106 peripheral blood mononuclear cells (PBMCs) after the first Ad5 immunization (Figure 1a). Although this response was significantly higher, more than sevenfold (P = 0.016, Mann–Whitney), than the 75 ± 51 SFU/106 PBMCs observed following the first DNA immunization, subsequent Ad5 immunizations were much less effective in boosting this initial response. In contrast, the DNA-vaccinated animals showed boosting of SIVgag responses with each immunization, culminating in a robust 1,672 ± 1,076 SFU/106 PBMCs after the fourth immunization, a 2.5-fold enhancement compared to the highest Ad5 response.
Figure 1.
Enhanced IFNγ production following DNA vaccination. Fresh PBMCs isolated prior to immunization and 2 weeks following each immunization were assessed for IFNγ production by a standard enzyme-linked immunosorbent spot assay. (a) SIVgag- and (b) SIVpol-specific responses were measured in both the DNA and Ad5 groups. Cellular responses to (c) SIVnef were measured in the Ad5 immunized group, and (d) SIVenv responses were measured in the DNA-immunized group. Responses are shown as group averages ± SEM. PBMC, peripheral blood mononuclear cell; SFU, spot-forming unit.
Compared to SIVgag, the responses to SIVpol were higher in both immunization groups (Figure 1b). However, the trends in responses to vaccination that we observed for the SIVgag antigen were also seen for the SIVpol antigen. Immunization with Ad5 resulted in a SIVpol response of 1,217 ± 363 SFU/106 PBMCs. Four DNA immunizations resulted in a significantly higher SIVpol response compared to Ad5 immunization (6,813 ± 955 SFU/106 PBMCs, P = 0.008 Mann–Whitney). Again, Ad5 induced a stronger response right out of the box but failed to boost the induced response.
The Ad5 vaccine also included a SIVnef antigen. However, it produced a weak cellular response that was just above background (52 ± 33 SFU/106 PBMCs) (Figure 1c). A SIVenv plasmid construct was included in the DNA vaccine, and like the SIVpol antigen, it induced a very large IFNγ response (4,006 ± 1,408 SFU/106 PBMCs) (Figure 1d). In all cases, the DNA exhibited a strong ability to continue to expand T-cell ELISpot number with subsequent boosts.
DNA vaccination induces CD4+ and CD8+ T-cell responses with greater proliferative capacity
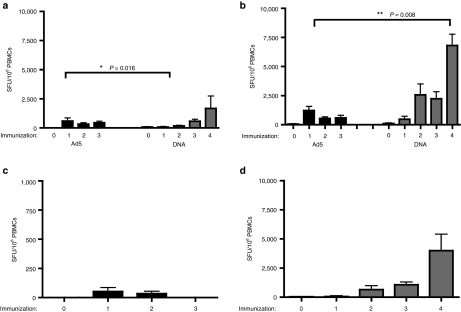
To determine the proliferative capacity of the vaccine-induced T-cell response, we stimulated PBMCs ex vivo with SIV peptides and measured proliferation by a CFSE dilution assay (Figure 2a). We measured responses 4–6 weeks after the second immunization in both the Ad5 and DNA groups. In general, we observed less CD4+ T-cell proliferation compared to CD8+ T cells in both vaccine groups (Figure 2b,c). In fact, the ratio between the CD4 and CD8 responses in both groups were biased similarly. The Ad5 group, however, supported a very weak SIVgag-specific CD4+ T-cell response that was increased only slightly over background proliferation. Stimulation with SIVnef induced CD4+ T-cell proliferation in one animal (0.17%). However, 0.24 ± 0.17% of CD4+ T cells proliferated in response to SIVpol in the Ad5 group. Similarly, we detected modest proliferation of CD4+ T cells to SIVgag and pol (0.16 ± 0.07% and 0.18 ± 0.06%, respectively), following DNA immunization. In addition, SIVenv stimulation resulted in 0.38 ± 0.24% CD4+ T-cell proliferation in the DNA group suggesting more of an antigenic difference than platform for responses to this antigen.
Figure 2.
DNA vaccination induces CD4+ and CD8+ T-cell responses with greater proliferative capacity. Fresh peripheral blood mononuclear cells isolated either 4–6 weeks following the second Ad5 and DNA immunization were stained with CFSE and stimulated with SIV peptide libraries for 5 days. Proliferation was assessed by dilution of the CFSE dye in antigen-specific T cells. (a) Dot plots of proliferating (CFSElo) CD4+ and CD4− T cells from representative animals are shown. Group average (b) CD4+ and (c) CD8+ T-cell proliferative responses to the two shared vaccine antigens, SIVgag and SIVpol, are shown as white and black bars, respectively. SIVnef responses in the Ad5 group and the SIVenv responses in the DNA group are shown as gray-vertical and horizontal bars, respectively.
As seen in the CD4+ T-cell compartment, CD8+ T-cell proliferation in the Ad5 group was predominantly to SIVpol (0.42 ± 0.13%) with one animal having a proliferative response to SIVnef stimulation (0.25%). The DNA-immunized group exhibited a very strong sixfold increase in SIVpol-specific CD8+ T-cell responses (2.84 ± 1.17%). Unlike the Ad5 group, we were able to detect SIVgag-specific CD8+ T-cell proliferative responses (1.27 ± 0.43%, P = 0.032 Mann–Whitney). SIVenv stimulation resulted in proliferation of 1.91 ± 0.79% of CD8+ T cells in the DNA group. These data suggest that DNA immunization of SIVgag and pol antigens induces highly proliferative CD8 T-cell responses that maintain proliferative capacity after expansion. It is interesting that the CD8 T cells expand without similar expansion of CD4 T cells.
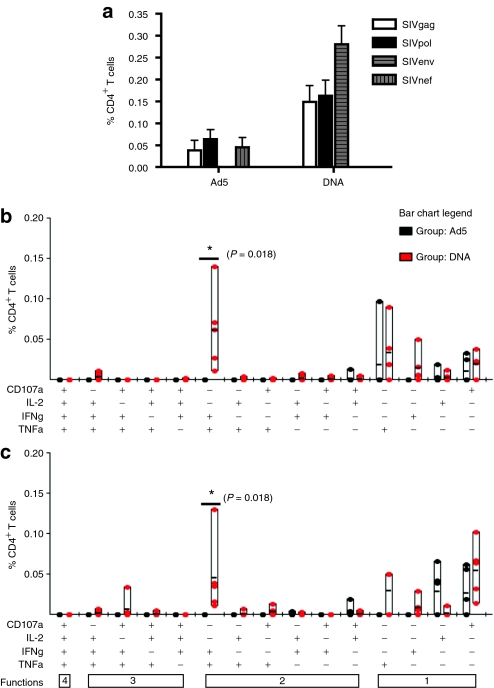
Polyfunctional profile of CD8+ T cells demonstrates greater magnitude and functionality of DNA vaccine–induced responses
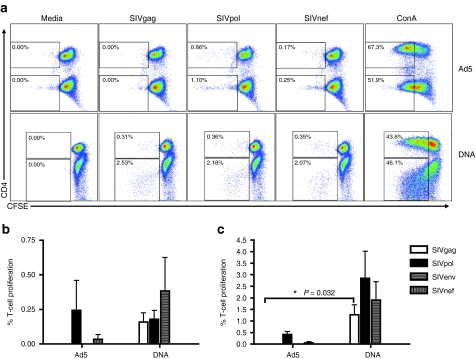
To better assess potential differences in the quality of the immune response following Ad5 or DNA vaccination, we performed polyfunctional analysis by multicolor flow cytometry. PBMCs isolated 2 weeks following the final Ad5 or DNA immunization were stained for IFNγ, IL-2, and TNFα production. Additionally, the ability for cells to degranulate in response to antigen was assessed by CD107a mobilization (Figure 3a). We were first interested in the overall magnitude of the SIV-specific responses induced by each vaccination platform (Figure 3b). Examining the total responses to SIVgag and pol, the two immunogens common to the two vaccines, DNA vaccination lead to a potent frequency of SIVgag and pol CD8+ T-cell responses, and these were higher those induced by Ad5 vaccination (1.15 ± 0.53% and 0.23 ± 0.05%, respectively). CD8+ T-cell responses to SIVgag were not significantly different between the two vaccination groups (Figure 3c). However, there was a significant increase in the overall CD8+ T-cell response (P = 0.032 Mann–Whitney) as well as the IFNγ+ TNFα+ population in the DNA group compared to the Ad5 group following SIVpol stimulation (P = 0.033, Wilcoxon) (Figure 3d). Ad5 immunization resulted in 0.05 ± 0.02% of CD8+ T cells responding to SIVnef stimulation. SIVenv responses in the DNA group were similar to SIVpol responses with 1.01 ± 0.48% of CD8+ T cells responding to stimulation.
Figure 3.
Polyfunctional profile of CD8+ T cells demonstrates greater magnitude and functionality of DNA vaccine-induced responses. Peripheral blood mononuclear cells isolated 2 weeks after the final DNA and Ad5 immunization were stimulated in vitro with a SIVgag, nef, and pol peptide pool mix for 5 hours. Cells were stained for intracellular production of IFNγ, TNFα, and IL-2, and degranulation by CD107a. Representative gating analysis for intracellular cytokine staining is shown. The singlet population was discriminated by the use of forward scatter height (FSC-H) and forward scatter area (FSC-A). Subsequently, we isolated the lymphocyte population using side scatter area (SSC-A) by FSC-A. Live T cells were identified as negatively staining for a Pacific Blue dump gate, which includes the following: Violet Vivid Dye for viability, CD14, CD16, and CD19, and positively staining for CD3. Following this, events are sequentially gated on CD8+ and CD4+ events. In this example, the gating for identification of CD8+ T cells is shown. First, CD3+ CD8+ T cells are identified by positive CD8 staining and negative CD4 staining, to exclude any CD4+ T cells that may have upregulated CD8 following activation. (a) The resulting CD8+ T cells were then gated for each of the four functions in our panel: CD107a, IL-2, IFNγ, and TNFα. (b) The bar graph depicts the frequency of the total CD8+ T-cell responses for SIVgag, nef, env, and pol as white, gray-vertical, gray-horizontal, and black bars ± SEM, respectively. These total responses were then broken down to show the frequency of each of the 15 combinations of functional populations to (c) SIVgag and (d) SIVpol for the Ad5 (black dots) and DNA (red dots) groups.
Polyfunctional profile of SIV-specific CD4+ T-cell responses following vaccination
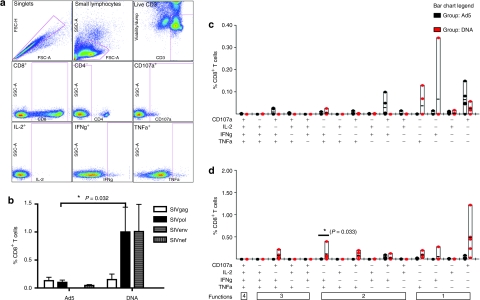
Having seen clear differences in the frequencies and functionality of the CD8+ T-cell response between the two vaccination groups, we next assessed the CD4+ T-cell response. The overall magnitude of the SIV-specific responses induced by each vaccination platform was much higher in the CD8+ T-cell compartment compared to the CD4+ T-cell compartment (Figure 4a) as was observed in the proliferation assays. As seen in the CD8+ T-cell response, we observed a significantly higher induction of total SIVgag and pol functional CD4+ T-cell responses in the DNA group compared to the Ad5 group (0.31 ± 0.07% and 0.10 ± 0.08%, P = 0.032 Mann–Whitney). The CD4+ T-cell response to SIVgag in the Ad5 group was predominantly monofunctional consisting of either IL-2 production or CD107a staining (Figure 4b). The DNA group had a much more polyfunctional response with all five animals having an IFNγ+ TNFα+ population, which was significantly higher than the Ad5 group (P = 0.018, Wilcoxon). The same trends were observed following SIVpol stimulation (Figure 4c). The Ad5 group had a SIVnef response of 0.05 ± 0.02%, whereas SIVenv was much more immunogenic in the DNA group, resulting in 0.28 ± 0.04% of CD4+ T cells making a functional response. It is clear that the phenotype of T cells induced by the two regimes varied based on the vaccine platforms.
Figure 4.
Enhanced magnitude and polyfunctionality of SIV-specific CD4+ T-cell responses following DNA vaccination. Peripheral blood mononuclear cells isolated 2 weeks after the final DNA and Ad5 immunization were stimulated in vitro with a SIVgag, nef, and pol peptide pool mix for 5 hours. Cells were stained for intracellular production of IFNγ, TNFα, and IL-2, and degranulation by CD107a and analyzed as described in Figure 3. (a) The bar graph depicts the frequency of the total CD4+ T-cell responses for SIVgag, nef, env, and pol as white, gray-vertical, gray-horizontal, and black bars ± SEM, respectively. The total responses were then broken down to show the frequency of each of the 15 combinations of functional populations to (b) SIVgag and (c) SIVpol for the Ad5 (black dots) and DNA (red dots) groups.
Memory T-cell responses following DNA vaccination
Having seen the robust induction of immune responses following vaccination, we were next interested in the durability of this response. We first examined the memory response induced by each vaccine by the standard quantitative IFNγ ELISpot (Figure 5a). Interestingly, the DNA and Ad5 group had similar SIVgag responses 5 months following the final immunization (231 ± 143 and 319 ± 60 SFU/106 PBMCs, respectively). In contrast, SIVpol-specific memory responses were maintained at higher levels in the DNA group (2,343 ± 547 SFU/106 PBMCs) compared to the Ad5 group (955 ± 224 SFU/106 PBMCs). There was only one animal in the Ad5 group that had a detectable SIVnef response 5 months following the last immunization. The SIVenv-specific memory response in the DNA group was robust with ELISpot counts of 1,546 ± 686 SFU/106 PBMCs. In general, we observed memory responses in the DNA group that were greater than the peak responses in the Ad5 group, but there was an antigen-specific component to this observation.
Figure 5.
Maintenance of high-frequency memory T-cell responses following DNA vaccination. To assess the maintenance of SIV-specific T-cell responses following vaccination, PBMCs were isolated 5 months following the final Ad5 and DNA immunization. (a) Cellular immune responses were measured by IFNγ enzyme-linked immunosorbent spot. (b) The ex vivo proliferative capacity was also assessed by the CFSE dilution assay. Group mean responses for SIVgag, nef, env, and pol are shown as white, gray-vertical, gray-horizontal, and black bars ± SEM, respectively. PBMC, peripheral blood mononuclear cell; SFU, spot-forming unit.
In addition to IFNγ production, we assessed the proliferative capacity of the memory response by an in vitro CFSE dilution assay (Figure 5b). Examination of the SIVgag and pol combined response showed that the DNA and Ad5 group had similar CD4+ T-cell proliferative responses (0.35 ± 0.07 and 0.16 ± 0.10%, respectively). A much larger difference in the memory proliferative capacity was seen in the CD8+ T-cell population with an average 3.95 ± 2.39% proliferation in the DNA group compared to 0.46 ± 0.24% proliferation in the Ad5 group. These results suggest that although both vaccination strategies are able to induce and maintain robust cellular responses, DNA vaccination maintains higher frequencies of vaccine-specific responses with greater proliferative capacity most likely due to the additional benefits of boosting. Furthermore, the vector systems may have an influence specifically on specific antigens, as we observed for the gag responses versus the pol responses.
Discussion
In this study, we sought to compare the immune responses induced by an optimized DNA vaccine with an established benchmark for cellular HIV vaccines, the Merck trivalent Ad5 vaccine. We observed both modalities induced strong immune responses. This is an important distinction over prior versions of DNA vaccines where Ad5 was clearly a superior vaccine profile. Furthermore, the ability of DNA to be boosted in the absence of induced serology ultimately appeared beneficial.
It is important to note that this study was designed to test two vaccine candidates, which were previously developed and formulated individually by different groups and available for study, and was not designed to evaluate the individual vaccination platforms. Thus, there are several differences in antigens, viral gene sequences, and immunization protocols between these two vaccines. First, the Ad5 vaccine consists of the SIV antigens gag, nef, and pol. In contrast, the DNA vaccine consisted of plasmids encoding the SIV antigens gag, env, and pol. Second, the SIVpol antigen in the Ad5 vaccine lacks the sequence encoding protease. To determine the effect that this disparity in antigenic sequences might have, we excluded the IFNγ responses to the pool of peptides that spanned the protease sequence from our calculation of the SIVpol response following DNA vaccination. Despite the exclusion of the protease-specific response, the SIVpol response was significantly higher following DNA vaccination compared to the Ad5-induced response. Finally, the gene sequences used in the Ad5 vaccine were based on the SIVmac239 virus, whereas the immunogens in the DNA group were derived from a consensus sequence of all SIVmac viruses. In all of the assays reported in this study, SIVmac239 peptide libraries were used to stimulate PBMCs. These are exactly matched to the immunogen inserts contained in the Ad5 vaccine, but are not exactly matched to the consensus immunogens contained in the DNA vaccine. Thus, despite having unmatched peptide libraries, the DNA-immunized group had cellular response of strong magnitude in all assays.
As previously reported, we did not observe boosting in immune responses subsequent to the first Ad5 immunization. The attenuation of immune induction following homologous viral vector boosts by vector-specific neutralizing antibodies has been a limitation of this approach.12,22 To address this, other Ad5-based approaches have focused on heterologous viral vector boosts23,24 or the use of mixed modalities such as DNA/Ad5 prime-boost strategies.10,11,12 It is in this respect that DNA vaccines could provide the greatest advantage as there are no issues with pre-existing serology, and thus, a single vaccine formulation could be used repeatedly to induce immune responses. Furthermore, the combination of a DNA and Ad5, or other viral vector platform may be very useful and open up new possibilities for vaccination. Theoretically, if a DNA vaccine was potent enough, such a DNA should be able to boost repeatedly without interference. The results of this study have, in part, borne out this hypothesis as we observed boosting of immune responses following each of the four DNA immunizations.
The lack of efficacy observed in the Merck Step trial has made clear that evaluation of vaccine candidates by IFNγ ELISpot is not sufficient to predict the efficacy of that vaccine in a human clinical trial. Although there is an agreement that the quality of the immune response is likely to be more important than the magnitude of the vaccine response, there has been much debate on what type of cellular response would be desirable for an HIV vaccine. Studies in long-term nonprogressors suggest that polyfunctional CD4+ T cells25,26,27 and CD8+ T cells28 as well as highly proliferative CD8+ T cells29 may be better equipped to maintain viral control. In this study, we evaluated the functional phenotype of CD4+ and CD8+ T cells following DNA or Ad5 vaccination. We observed clear differences in the polyfunctional responses induced by the two vaccine regimes. Similarly, CFSE dilution assays also demonstrated clear differences in the ability of the two platforms to drive proliferative responses. It has been speculated that Ad5 responses may be handicapped in the induction of T-cell proliferative responses, and this suspicion was supported by the data obtained in this study. Furthermore, we also observed that with improved delivery, a novel shift in potency was observed between the two platforms. Specifically, it appears that Ad5 is particularly potent after a single immunization, whereas the enhanced DNA could be boosted multiple times. This suggests that these more potent platforms may be useful in reversed roles where Ad5 is used for the prime to jump-start the immune response, and DNA is then used to continue to boost this more efficient prime. More work is needed in this interesting area.
Taken together, the data in this study suggest that EP-delivered highly optimized DNA vaccines are capable of inducing cellular immune responses that are improved in magnitude and quality compared to prior generations of DNA vaccines suggesting a renewed importance of this platform. In addition, the responses induced appear to differ from AD5 studied here in magnitude and quality.30 Although these results are encouraging, further studies, including additional primate challenge studies, will need to be performed to more fully assess the potential of DNA vaccines and to study such approaches in prime-boost modalities and to understand their utility for further examination in the clinic.
Materials and Methods
Animals. Chinese-origin rhesus macaques (Macaca mulatta) were housed at BIOQUAL (Rockville, MD), in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care. Animals were allowed to acclimate for at least 30 days in quarantine prior to any experimentation.
Recombinant Ad5 vaccine. Five animals were immunized with recombinant Ad5 vectors encoding SIVmac239gag, SIVmac239nef, and SIVmac239pol. Genes coding for Gag, Pol, and Nef were synthesized based on codons frequently used in mammalian cells.31 All genes were based on reported sequences from SIVmac239 with the exception of Nef that was based on the sequence reported for SIVmac251 (ref. 32). The pol gene spanned reverse transcriptase through integrase; protease was not included. Enzymatically, active sites were inactivated by alanine substitutions at nine locations33,34,35,36,37,38,39 (reverse transcriptase Asp-112, Asp-187, and Asp-188; RNase H Asp-445, Glu-480, and Asp-500; integrase Asp-626, Asp-678, and Glu-714). The nef open-reading frame was altered by substituting the myristoylation site located at position 2 from glycine to alanine. Replication-defective Ad5 sequence was synthesized as previously described;40 modified by deleting the E1 region and replacing the expression cassettes with the early gene promoter from human cytomegalovirus,41 the coding sequence of either gag, pol, or nef as described above, and the bovine growth hormone polyadenylation signal sequence.42 The animals were immunized three times at weeks 0, 4, and 24 with a dose of 1 × 1010 viral particles per construct. The vaccine was formulated into a total volume of 1.0 ml and divided into two injections into the quadriceps muscle.
DNA vaccine. The plasmids used in this study express the modified proteins for SIV Gag (pSIVgag), SIVpol (pSIVpol), or SIVenv (pSIVenv), and were generated in our laboratory. Briefly, consensus sequences for macaque SIVgag, SIVenv, and SIVpol were generated with several modifications. For the SIVenv construct, the V1 and V2 regions were shortened by removing N-linked glycosylation sites, and the cytoplasmic tail was truncated to prevent envelope recycling. For SIVpol, seven mutations were introduced to deactivate the protease, reverse transcriptase, RNAse H, and integrase regions. We added an efficient IgE leader sequence to all SIV antigen sequences to improve expression. The resulting optimized SIV DNA immunogens were codon and RNA-optimized, synthesized, and cloned into the pVAX1 expression vector by GENEART (Toronto, Ontario, Canada) to create optimized expression constructs for SIVgag (pSIVgag), SIVenv (pSIVenv), and SIVpol (pSIVpol). A group of five rhesus macaques (DNA) were immunized at weeks 0, 6, 12, and 18 with 1.0 mg each of SIVgag, SIVenv, and SIVpol. The DNA at each immunization time point was delivered into a single site in the quadriceps muscle followed by in vivo EP. All EP procedures were performed using the constant current CELLECTRA device (Inovio Pharmaceuticals, formerly VGX Pharmaceuticals, Blue Bell, PA). EP conditions were 0.5 A, 3 pulses, 52 ms pulse length with 1 second between pulses.
Blood collection. Animals were bled 2 and 4 weeks following each immunization and again 5 months following the final immunization. Twenty milliliters of blood were collected in EDTA tubes. PBMCs were isolated by standard Ficoll-Hypaque centrifugation and resuspended in complete culture medium (RPMI 1640 with 2 mmol/l l-glutamine supplemented with 10% heat-inactivated fetal bovine serum, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 55 µmol/l β-mercaptoethanol). Red blood cells were lysed with ACK lysis buffer (Cambrex Bioscience, East Rutherford, NJ).
Peptides. Reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: SIVmac239 gag peptides (no. 6204), SIVmac239 env peptides (no. 6883), SIVmac239 nef peptides (no. 8762), and SIVmac239 pol peptides (no. 6443). As the SIV pol antigen contained in the Ad5 vector was deleted in the protease region, these peptide pool responses were not included in the vaccine responses to allow for comparative data.
IFNγ ELISpot. MultiScreen-IP 96-well plates (Millipore, Bedford, MA) were coated overnight at 4 °C with capture antibody diluted in phosphate-buffered saline (PBS) at a concentration of 7.5 µg/ml (anti-IFN-γ clone GZ-4; Mabtech, Cincinnati, OH). Plates were washed five times with PBS and blocked with complete culture medium for 2 hours. PBMCs were added in triplicate at an input cell number of 2 × 105/well in 100 µl complete culture medium. For third and fourth immunization, the input cell number was decreased to 1 × 105 cells due to excessive spot number for the DNA group. SIV peptide pools were diluted 1/200 in complete culture medium, and 100 µl were added per well (final 1/400 dilution). Cells resuspended in complete culture medium only served as a negative control. ConA (5 µg/ml; Sigma-Aldrich, St Louis, MO) was used as a positive control. Plates were incubated 18 hours at 37 °C, washed five times with PBS (and incubated overnight at 4 °C with 100 µl/well of biotinylated detection antibody diluted in PBS (1 µg/ml; clone 7-B6-1, Mabtech). Plates were washed five times with PBS and then incubated with 100 µl/well of streptavidin-alkaline phosphatase diluted in PBS (1:1,000) for 1 hour at room temperature. Plates were then washed five times with PBS and developed with 100 µl/well BCIP/NBT substrate solution for 5–10 minutes at room temperature. Washing the wells three times with tap water terminated the colorimetric reaction. Plates were air-dried, and the spots were counted using an automated ELISpot reader system (CTL Analyzers, Shaker Heights, OH) with the ImmunoSpot software. The mean number of spots from triplicate wells was adjusted to 1 × 106 PBMCs. The SIV-specific responses were calculated after subtraction of spots formed in response to culture medium alone. In addition, a response that was at least twofold higher than the medium control was considered positive. For comparisons in the vaccine responses to SIVpol, the peptide pool including the SIV protease encoding peptides were not included in the calculation of the DNA vaccine–induced response.
CFSE labeling and flow cytometry. Freshly isolated PBMCs were stained with 5 µmol/l CFSE (Invitrogen, Carlsbad, CA) in prewarmed PBS for 10 minutes, washed two times, and suspended in RPMI 1640 containing 2 mmol/l l-glutamine, 100 IU/ml penicillin, and 100 IU/ml streptomycin and 10% fetal bovine serum (R10 medium). Cells were stimulated using either pooled peptides from SIVgag, SIVenv, SIVnef, and SIVpol or 5 µg/ml Concanavalin A (ConA) (Sigma-Aldrich) for 5 days. Cells were then washed in PBS and stained using the LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Invitrogen) and subsequently stained for CD3, CD4, and CD8. Antibodies were incubated with cells for 30 minutes on ice. Following staining, cells were washed twice in PBS and fixed with 1% paraformaldehyde. For flow cytometry, cells were gated on singlets using forward scatter height by forward scatter area followed by gating on Pacific Bluelo CD3+ T cells to examine live T-cell populations. Pure CD4+ and CD8+ T-cell populations were determined by sequential CD4 and CD8 gating to account for upregulation of CD8 on activated CD4+ T cells. Responses are shown as stacked group mean responses ± SEM, after media subtraction, to SIVgag, nef, env, and pol.
Intracellular cytokine staining: antibody reagents. Directly conjugated antibodies were obtained from the following: BD Biosciences (San Jose, CA): IL-2 (PE), CD3 (APC Cy7), CD8 (APC), IFN-γ (Alexa Fluor 700), TNF-α (PE Cy7), and CD4 (PerCP Cy5.5).
Cell stimulation and staining. PBMCs were resuspended to 1 × 106 cells/100 µl in complete RPMI and plated in 96-well plates with SIVgag and pol stimulating peptides 100 µl of 1:200 dilutions. An unstimulated and positive control (Staphylococcus enterotoxin B, 1 µg/ml; Sigma-Aldrich) was included in each assay. Cells were incubated for 5 hours at 37 °C. Following incubation, the cells were washed (PBS) and stained with surface antibodies. The cells were washed and fixed using the Cytofix/Cytoperm kit (BD Pharmingen, San Diego, CA) according to instructions. Following fixation, the cells were washed twice in the perm buffer and stained with antibodies against intracellular markers. Following staining, the cells were washed, fixed (PBS containing 1% paraformaldehyde), and stored at 4 °C until analysis.
Flow cytometry. Cells were analyzed on a modified LSR II flow cytometer (BD Immunocytometry Systems, San Jose, CA). One hundred thousand live CD3+ events were collected per sample. Data analysis was performed using FlowJo version 8.8 (TreeStar, San Carlos, CA). Initial gating used a forward scatter area versus forward scatter area height plot to remove doublets. The events were subjected to a lymphocyte gate by a forward scatter area versus SSC plot. Live T cells were identified by a live/dead versus CD3+ plot. Following this, events are sequentially gated on CD8+ and CD4– events versus IFN-γ to account for downregulation. Following identification of CD8+ and CD4+ T cells, a gate was made for each respective function using combinations that provided optimal separation. After the gates for each function were created, we used the Boolean gate platform to create the full array of possible combinations, equating to 15 response patterns when testing four functions. Data are reported after background correction.
Statistics. For comparisons of IFNγ ELISpots, T-cell proliferation, and cytokine production, Mann–Whitney tests were performed using SPSS 17.0 Statistical Software (SPSS, Chicago, IL). For comparisons of Boolean populations, Wilcoxon signed rank tests were performed by the SPICE 5.05 software. P values that were <0.05 were considered significant.
Acknowledgments
Inovio acknowledges funding support from NIAID/DAIDS under an HVDDT contract (HHSN272200800063C) for the nonhuman primate studies and VGXI, Inc. (The Woodlands, TX) for manufacturing the DNA vaccine candidates used in the study. The laboratory notes possible commercial conflicts associated with this work, which may include the following: Wyeth, Inovio, BMS, Virxsys, Ichor, Merck, Althea, and Aldevron, and possibly others.
REFERENCES
- Casimiro DR, Chen L, Fu TM, Evans RK, Caulfield MJ, Davies ME, et al. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J Virol. 2003;77:6305–6313. doi: 10.1128/JVI.77.11.6305-6313.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priddy FH, Brown D, Kublin J, Monahan K, Wright DP, Lalezari J, et al. Merck V520-016 Study Group. (2008Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults Clin Infect Dis 461769–1781. [DOI] [PubMed] [Google Scholar]
- Catanzaro AT, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, et al. Vaccine Research Center 006 Study Team. (2006Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector J Infect Dis 1941638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiver JW, Fu TM, Chen L, Casimiro DR, Davies ME, Evans RK, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002;415:331–335. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, et al. Step Study Protocol Team. (2008Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial Lancet 3721881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, et al. Step Study Protocol Team. (2008HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis Lancet 3721894–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick NA, Carnathan DG, Dubey SA, Makedonas G, Cox KS, Kierstead L, et al. Baseline Ad5 serostatus does not predict Ad5 HIV vaccine-induced expansion of adenovirus-specific CD4+ T cells. Nat Med. 2009;15:876–878. doi: 10.1038/nm.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KL, Liu J, King SL, Sun YH, Schmitz JE, Lifton MA, et al. Adenovirus-specific immunity after immunization with an Ad5 HIV-1 vaccine candidate in humans. Nat Med. 2009;15:873–875. doi: 10.1038/nm.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao LA, Wu L, Khan AS, Hokey DA, Yan J, Dai A, et al. Combined effects of IL-12 and electroporation enhances the potency of DNA vaccination in macaques. Vaccine. 2008;26:3112–3120. doi: 10.1016/j.vaccine.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro DR, Wang F, Schleif WA, Liang X, Zhang ZQ, Tobery TW, et al. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J Virol. 2005;79:15547–15555. doi: 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Mascola JR, Sun Y, Gorgone DA, Buzby AP, Xu L, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra S, Seaman MS, Xu L, Barouch DH, Lord CI, Lifton MA, et al. Replication-defective adenovirus serotype 5 vectors elicit durable cellular and humoral immune responses in nonhuman primates. J Virol. 2005;79:6516–6522. doi: 10.1128/JVI.79.10.6516-6522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deml L, Bojak A, Steck S, Graf M, Wild J, Schirmbeck R, et al. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol. 2001;75:10991–11001. doi: 10.1128/JVI.75.22.10991-11001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André S, Seed B, Eberle J, Schraut W, Bültmann A., and, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Campbell M, Nasioulas G, Felber BK., and, Pavlakis GN. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao LA, Wu L, Khan AS, Satishchandran A, Draghia-Akli R., and, Weiner DB. Intradermal/subcutaneous immunization by electroporation improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine. 2008;26:440–448. doi: 10.1016/j.vaccine.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Otten G, Schaefer M, Doe B, Liu H, Srivastava I, zur Megede J, et al. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine. 2004;22:2489–2493. doi: 10.1016/j.vaccine.2003.11.073. [DOI] [PubMed] [Google Scholar]
- Otten GR, Schaefer M, Doe B, Liu H, Megede JZ, Donnelly J, et al. Potent immunogenicity of an HIV-1 gag-pol fusion DNA vaccine delivered by in vivo electroporation. Vaccine. 2006;24:4503–4509. doi: 10.1016/j.vaccine.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Luckay A, Sidhu MK, Kjeken R, Megati S, Chong SY, Roopchand V, et al. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J Virol. 2007;81:5257–5269. doi: 10.1128/JVI.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati M, Bergamaschi C, Valentin A, Kulkarni V, Jalah R, Alicea C, et al. DNA vaccination in rhesus macaques induces potent immune responses and decreases acute and chronic viremia after SIVmac251 challenge. Proc Natl Acad Sci USA. 2009;106:15831–15836. doi: 10.1073/pnas.0902628106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Hokey DA, Morrow MP, Corbitt N, Harris K, Harris D, et al. Novel SIVmac DNA vaccines encoding Env, Pol and Gag consensus proteins elicit strong cellular immune responses in cynomolgus macaques. Vaccine. 2009;27:3260–3266. doi: 10.1016/j.vaccine.2009.01.065. [DOI] [PubMed] [Google Scholar]
- Sumida SM, Truitt DM, Lemckert AA, Vogels R, Custers JH, Addo MM, et al. Neutralizing antibodies to adenovirus serotype 5 vaccine vectors are directed primarily against the adenovirus hexon protein. J Immunol. 2005;174:7179–7185. doi: 10.4049/jimmunol.174.11.7179. [DOI] [PubMed] [Google Scholar]
- Liu J, Ewald BA, Lynch DM, Denholtz M, Abbink P, Lemckert AA, et al. Magnitude and phenotype of cellular immune responses elicited by recombinant adenovirus vectors and heterologous prime-boost regimens in rhesus monkeys. J Virol. 2008;82:4844–4852. doi: 10.1128/JVI.02616-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, O'Brien KL, Lynch DM, Simmons NL, La Porte A, Riggs AM, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannanganat S, Kapogiannis BG, Ibegbu C, Chennareddi L, Goepfert P, Robinson HL, et al. Human immunodeficiency virus type 1 controllers but not noncontrollers maintain CD4 T cells coexpressing three cytokines. J Virol. 2007;81:12071–12076. doi: 10.1128/JVI.01261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boaz MJ, Waters A, Murad S, Easterbrook PJ., and, Vyakarnam A. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T cell responses is associated with nonprogression in HIV-1 infection. J Immunol. 2002;169:6376–6385. doi: 10.4049/jimmunol.169.11.6376. [DOI] [PubMed] [Google Scholar]
- Emu B, Sinclair E, Favre D, Moretto WJ, Hsue P, Hoh R, et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005;79:14169–14178. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- Cox KS, Clair JH, Prokop MT, Sykes KJ, Dubey SA, Shiver JW, et al. DNA gag/adenovirus type 5 (Ad5) gag and Ad5 gag/Ad5 gag vaccines induce distinct T-cell response profiles. J Virol. 2008;82:8161–8171. doi: 10.1128/JVI.00620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985;183:1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- Korber B, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J.et al. (1998Human Retroviruses and AIDS Los Alamos National Laboratory: Los Alamos, NM [Google Scholar]
- Larder BA, Purifoy DJ, Powell KL., and, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- Larder BA, Kemp SD., and, Purifoy DJ. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc Natl Acad Sci USA. 1989;86:4803–4807. doi: 10.1073/pnas.86.13.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JF, 2nd, Hostomska Z, Hostomsky Z, Jordan SR., and, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Schatz O, Cromme FV, Grüninger-Leitch F., and, Le Grice SF. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- Mizrahi V, Usdin MT, Harington A., and, Dudding LR. Site-directed mutagenesis of the conserved Asp-443 and Asp-498 carboxy-terminal residues of HIV-1 reverse transcriptase. Nucleic Acids Res. 1990;18:5359–5363. doi: 10.1093/nar/18.18.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt AD, Shiue L., and, Varmus HE. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- Wiskerchen M., and, Muesing MA. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youil R, Toner TJ, Su Q, Casimiro D, Shiver JW, Chen L, et al. Comparative analysis of the effects of packaging signal, transgene orientation, promoters, polyadenylation signals, and E3 region on growth properties of first-generation adenoviruses. Hum Gene Ther. 2003;14:1017–1034. doi: 10.1089/104303403766682278. [DOI] [PubMed] [Google Scholar]
- Chapman BS, Thayer RM, Vincent KA., and, Haigwood NL. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC., and, Rottman FM. The 3'-flanking sequence of the bovine growth hormone gene contains novel elements required for efficient and accurate polyadenylation. J Biol Chem. 1992;267:16330–16334. [PubMed] [Google Scholar]