Abstract
The Alzheimer's Diseases Neuroimaging Initiative project has brought together geographically distributed investigators, each collecting data on the progression of Alzheimer's disease. The quantity and diversity of the imaging, clinical, cognitive, biochemical, and genetic data acquired and generated throughout the study necessitated sophisticated informatics systems to organize, manage, and disseminate data and results. We describe, here, a successful and comprehensive system that provides powerful mechanisms for processing, integrating, and disseminating these data not only to support the research needs of the investigators who make up the Alzheimer's Diseases Neuroimaging Initiative cores, but also to provide widespread data access to the greater scientific community for the study of Alzheimer's Disease.
Keywords: ADNI, Informatics, Alzheimer's disease, Data management
1. Introduction and rationale
Organizing, annotating, and distributing biomedical data in useable and structured ways have become essential elements to many scientific efforts. Driven by advances in clinical, genetic, and imaging electronic data collection, data are being collected at an accelerating rate. The need to store and exchange data in meaningful ways in support of data analysis, hypothesis testing, and future analysis is considerable. However, there are many challenges to achieving value-added data sharing that become more acute when data are collected variably, documentation is insufficient, some postprocessing and/or data analysis has already occurred, or the coordination among multiple sites is less than optimum. If analyzed data are to be distributed, the problem is worse because knowledge of which subject and version of data were used for a particular analysis is critically important to the interpretation and reuse of derived results. Nevertheless, the statistical and operational advantages of distributed science in the form of multisite consortia have continued to push this approach in many population-based investigations.
The biomedical science community has seen increased numbers of multisite consortia, each with varying degrees of success, driven in part by advances in infrastructure technologies, the demand for multiscale data in the investigation of fundamental disease processes, the need for cooperation across disciplines to integrate and interpret the data, and the movement of science in general toward freely available information. Numerous imaging examples can be found in the ubiquitous application of neuroimaging to the study of brain structure and function in health and disease [1–4]. A simple internet search finds several well known cooperative efforts [5] to construct and populate large brain databases [6, 7] and genetics [8]. Inherent in these multilaboratory projects are sociologic, legal, and even ethical concerns that must be resolved satisfactorily before they can work effectively or be widely accepted by the scientific community.
1.1. Sharing
The 2003 National Science Foundation Blue-Ribbon Advisory Panel on Cyberinfrastructure envisioned “[A]n environment in which raw data and recent results are easily shared, not just within a research group or institution but also between scientific disciplines and locations” [9]. However, databases frequently still suffer from some reluctance on the part of a community that harbors doubts about their trustworthiness, belief that there are too many difficulties associated with sharing, and concern about how others will use their data. In the last 10 years, considerable attention has been given to neuroimaging databases from the Organization of Human Brain Mapping (Governing Council of the Organization for Human Brain Mapping, 2001), who expressed concern about the quality of brain imaging data being deposited into such archives, how such data might be reused, and the potential for their being represented in new publication. The question of data ownership, in particular, was a primary concern in initial attempts to archive data. Concerns about the sharing of the primary data still exist and may have, for some, prohibited their availability [10]. With rapid advances being made in neuroimaging technology, data acquisition, and computer networks, the successful organization and management of neuroimaging data has become more important than ever before [11, 12]. A recent data ownership controversy (Abbott, 2008) aggravated further the still tenuous nature of data ownership, re-use, research ethical standards, and the pivotal role that peer-reviewed journals play in this process [13]. The implications of disagreements concerning appropriate data re-use also affects the users of data archives and how researchers might draw from them to publish results independently.
Despite all these potentially serious impediments, we are in the midst of a fundamental paradigm shift. The Alzheimer's Diseases Neuroimaging Initiative (ADNI) exemplifies a remarkably successful, open, shared, usable, and efficient database.
2. ADNI informatics core
The ADNI project has brought together geographically distributed investigators with diverse scientific capabilities to join forces in studying biomarkers that signify the progression of Alzheimer's disease (AD). The quantity of imaging, clinical, cognitive, biochemical, and genetic data acquired and generated throughout the study has required powerful mechanisms for processing, integrating and disseminating these data not only to support the research needs of the investigators who make up the ADNI cores, but also to provide widespread data access to the greater scientific community who benefit from having access to these valuable data. At the junction of this collaborative endeavor, the Laboratory of Neuro Imaging has provided an infrastructure to facilitate data integration, access, and sharing across a diverse and growing community.
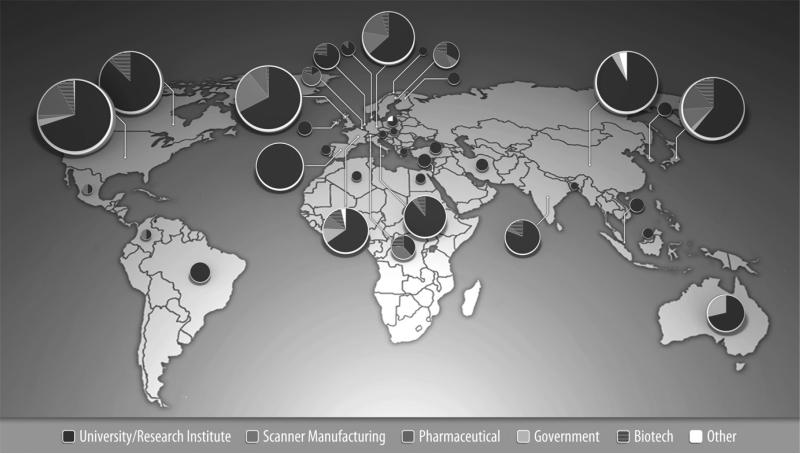
ADNI is made up of the eight cores responsible for conducting the study as well as the extended ADNI family of external investigators who have requested and been authorized to use ADNI data. The various information systems used by the cores result in an intricate flow of data into, out of, and between information systems, institutions, and among individuals. Ultimately the data flow into the ADNI data repository at LONI where they are made available to the community. Well-curated scientific data repositories allow data to be accessed by researchers across the globe and to be preserved over time [9]. To date, more than 1,300 investigators have been granted access to ADNI data resulting in extensive download activity that exceeds 800,000 downloads of imaging, clinical, biomarker, and genetic data. ADNI investigators come from 35 countries across various sectors, as shown in Fig. 1.
Fig. 1.
Global distribution of ADNI investigators by sector.
The ADNI Informatics Core has made significant progress in meeting our aim to provide a user-friendly, web-based environment for storing, searching, and sharing data acquired and generated by the ADNI community. In the process, our LONI Data Archive (LDA) has grown to meet the evolving needs of the ADNI community and we continue making strides toward a more interactive environment for data discovery and visualization. The automated systems we have developed include components for de-identification and secure archiving of imaging data from the 57 ADNI sites, managing the image workflow whereby raw images transition from quarantine status to general availability and then proceed through preprocessing and postprocessing stages; integrating nonimaging data from other cores to enrich the search capabilities, managing data access and data sharing activities for the 1,0001 investigators using these data, and providing a central repository for disseminating data and related information to the ADNI community.
Parallel efforts by the Australian Imaging Biomarkers and Lifestyle Flagship Study of Ageing (AIBL) have resulted in a subset of AIBL data being placed in the LDA where it has been made available to the scientific community. The AIBL data were acquired using the same magnetic resonance imaging and positron emission tomography (PET) imaging protocols, making them compatible for cross-study collaboration [14]. Investigators may apply for data access from the ADNI and AIBL studies, either individually or in combination, and may search across and obtain data from both projects simultaneously using a common LDA search interface.
2.1. Image data workflow
In short, the acquisition sites collect data from participants and enter or upload data into the clinical and imaging databases; the imaging cores perform quality control and preprocessing of the MR and PET images; the ADNI image analysts perform postprocessing and analysis of the preprocessed images and related data; the biochemical samples are processed and the results compiled; and investigators download and analyze data as best fits their individual research needs.
2.1.1. Raw image data
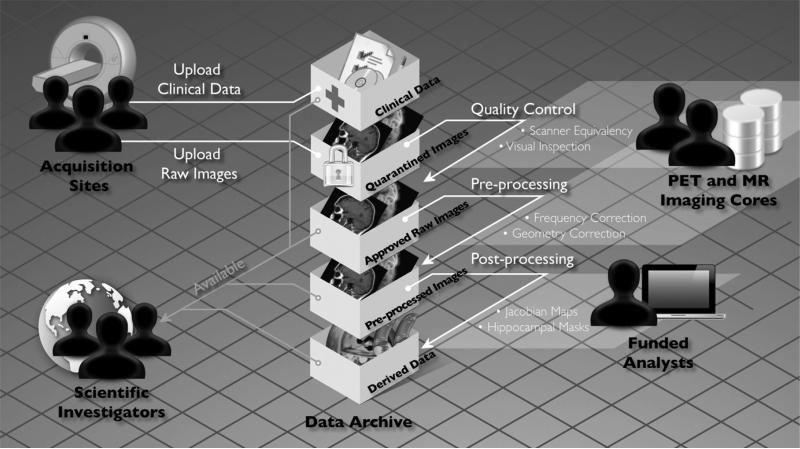
In keeping with the objectives of the ADNI project to make data available to the scientific community, without embargo, while meeting the needs of the core investigators, the image data workflow shown in Fig. 2 was adopted. Initially, each acquisition site uploads image data to the repository through the LDA a web-based application that incorporates a number of data validation and data de-identification operations, including validation of the subject identifier, validation of the dataset as human or phantom, validation of the file format, image file de-identification, encrypted data transmission, database population, secure storage of the image files, and metadata and tracking of data accesses. The image archiving portion of the system is both robust and extremely easy to use with the bulk of new users requiring little, if any, training. Key system components supporting the process of archiving raw data are as follows:
The subject identifier is validated against a set of acceptable, site-specific IDs.
Potentially patient-identifying information is removed or replaced. Raw image data are encoded in the DICOM, ECAT, and HRRT file formats, from different scanner manufacturers and models (e.g., SIEMENS Symphony, GE SIGNA Excite, PHILIPS Intera, etc.). The Java applet de-identification engine is customized for each of the image file formats deemed acceptable by the ADNI imaging cores and any files not of an acceptable format are bypassed. Because the applet is sent to the upload site, all de-identification takes place at the acquisition site and no identifying information is transmitted.
Images are checked to see that they “look” appropriate for the type of upload. Phantom images uploaded under a patient identifier are flagged and removed from the upload set. This check is accomplished using a classifier that has been trained to identify human and phantom images.
Image files are transferred encrypted (HTTPS) to the repository in compliance with patient-privacy regulations.
Metadata elements are extracted from the image files and inserted into the database to support optimal storage and findability. Customized database mappings were constructed for the various image file formats to provide consistency across scanners and image file formats.
Newly received images are placed into quarantine status, and the images are queued for those charged with performing MR and PET quality assessment.
Quality assessment results are imported from an external database and applied to the quarantined images. Images passing quality assessment are made available and images not passing quality control tagged as failing quality control.
Fig. 2.
Clinical and imaging data flows from the acquisition sites into separate clinical and imaging databases. Quality assessments and preprocessed images are generated by the imaging cores and returned to the central archive where image analysts obtain, process, and return to the archive further derived image data and results.
After raw data undergo quality assessment and are released from quarantine, they become immediately available to authorized users.
2.1.2. Processed image data
The imaging cores decided to use preprocessed images as the common, recommended set for analysis. The goals of preprocessing were to produce data standardized across site and scanner and with certain image artifacts corrected [15, 16]. Usability of processed data for further analysis requires an understanding of the data provenance, or information about the origin and subsequent processing applied to a set of data [17, 18]. To provide almost immediate access to preprocessed data in a manner that preserved the relationship between the raw and preprocessed images and that captured processing provenance, we developed an upload mechanism that links image data and provenance metadata. The Extended Markup Language (XML) upload method uses an XML schema, which defines required metadata elements as well as standardized taxonomies. As part of the preprocessed image upload process, unique image identifier(s) of associated images are validated keeping the relationship(s) among raw and processed images unambiguous with a clear lineage. The system supports uploading large batches of preprocessed images in a single session with minimal interaction required by the person performing the upload. A key aspect of this process is agreement on the definitions of provenance metadata descriptors. Using standardized terms to describe processing minimizes variability and aids investigators in gaining an unambiguous interpretation of the data [17].
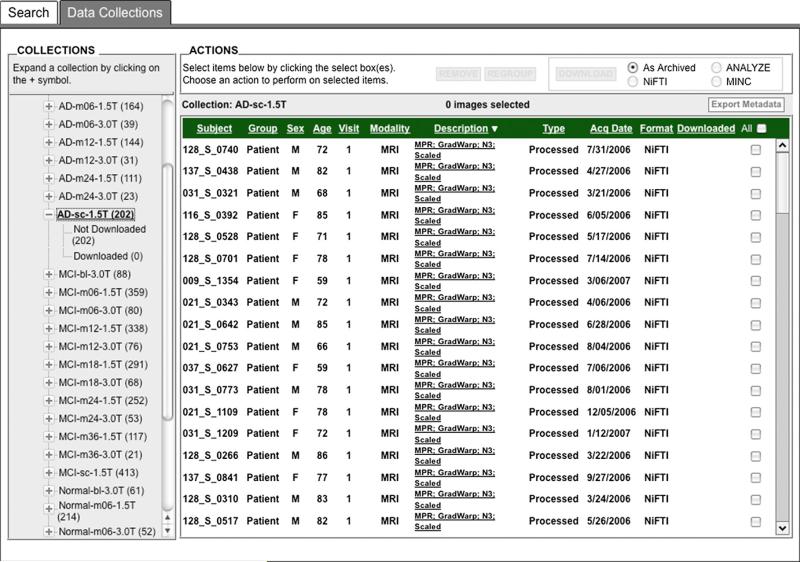
Preprocessed images are uploaded by the quality control sites on a fairly continuous basis and initially, each analyst had to search the data archive to find and then download data uploaded since the investigator's previous session. This was found to be cumbersome, so an automated data collection component was implemented, whereby newly uploaded preprocessed scans are placed into predefined, shared data collections. These shared collections, organized by patient diagnostic group (normal control, mild cognitive impairment, AD) and visit (Baseline, 6 month, etc.), together with a redesigned user interface (Fig. 3) that clearly indicates which images have not previously been downloaded, greatly reduced the time and effort needed to obtain new data. The same process may be used for postprocessed data, allowing analysts to share processing protocols through descriptive information contained in the XML metadata files.
Fig. 3.
The Data Collections interface provides access to shared data collections and is organized to meet the workflow needs of the analysts. The ability to easily select only images not previously downloaded by the user saves time and effort.
2.2. Data integration
A subset of data from the clinical database was integrated into the LDA to support richer queries across the combined set. The selection of the initial set of clinical data elements was based on user surveys in which participants identified the elements they thought would be most useful in supporting their investigations. As a result, a subset of clinical assessment scores, as well the initial diagnostic group of each subject, was integrated into the LDA to be used in searches and incorporated into the metadata files that accompany each downloaded image. Because the clinical data originate in an external database, automated methods for obtaining and integrating the external data were developed that validate and synchronize the data from the two sources and ensure that data from the same subject visit are combined.
2.3. Infrastructure
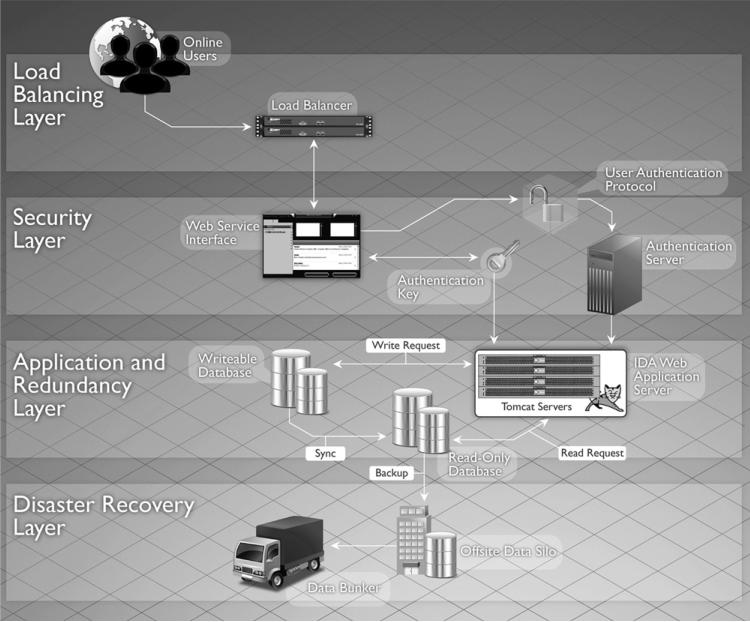
A robust and reliable infrastructure is a necessity for supporting a resource intended to serve a global community. The hardware infrastructure we built provides high performance, security, and reliability at each level. The fault–tolerant network infrastructure has no single points of failure. There are multiple switches, routers, and Internet connections. A firewall appliance protects and segments the network traffic, permitting only authorized ingress and egress. Multiple redundant database, application, and web servers ensure service continuity in the event of a single system failure and also provide improved performance through load balancing of requests across the multiple machines. To augment the network-based security practices and to ensure compliance with privacy requirements, the servers use Secure Sockets Layer (SSL) encryption for all data transfers. Post-transfer redundancy checking on the files is performed to guarantee the integrity of the data.
Communication with the LDA is managed by a set of redundant load balancers that divides client requests among groups of web servers for optimized resource use. One group of web servers is dedicated to receiving image files from contributors, and the other group sends data to authorized downloaders. Each web server communicates with redundant database servers that are organized in a master–slave configuration. The image files stored in the LDA reside on a multi-node Isilon storage cluster (Isilon Systems Inc., Seattle, WA). The storage system uses a block-based point-in-time snapshot feature that automatically and securely creates an internal copy of all files at the time of creation or modification. In the event of data loss or corruption in the archive, we can readily recover copies of files stored in the snapshot library without resorting to external backups.
Backup systems are designed to ensure data integrity and to protect data in the event of catastrophic failure. Incremental backups are performed nightly, with full backups stored on tape every week and sent off site. Full backups follow the industry's standard grandfather-father-son rotation scheme. To augment the data snapshot functionality, we perform nightly incremental and monthly full backups of the entire data repository. This automated backup is stored on tape in our secondary data center located in another campus building to protect against data loss in the case of a catastrophic event in our primary data center where the storage subsystem is housed. Additionally, we provide a tertiary level of protection against data loss by performing completely independent weekly tape backups of the entire collection which are deposited to an offsite vaulting service (Iron Mountain Inc., Boston, MA). This multipronged approach to data protection minimizes the risk of loss and ensures that a pristine copy of the data archive is always available (Fig. 4).
Fig. 4.
Redundant hardware and multiple backup systems ensure data are secure and accessible.
2.4. Performance
ADNI policy requires participating sites to upload new data within 24 hours of acquisition. To prevent a large number of downloaders from competing with resources needed by uploaders, the application servers are divided by upload/ download functionality. To prevent a single downloader from dominating a web server with multiple requests, the activity of each downloader is monitored and his/her download rate is throttled accordingly. Additionally, users are discouraged from downloading the same image files multiple times through the use of dialogs that interrupt and confirm the download process. These measures help to ensure ADNI data, and resources are equitably shared while maximizing the efficiency of the upload processes.
2.5. Data access and security
Access to ADNI data is restricted to those who are site participants and those who have applied for access and received approval from the Data Sharing and Publication Committee (DPC). Different levels of user access control the system features available to an individual. Those at the acquisition sites are able to upload data for subjects from their site but are not able to access data from other sites, whereas the imaging core leaders may upload, download, edit, or delete data. All data uploads, changes, and deletions are logged.
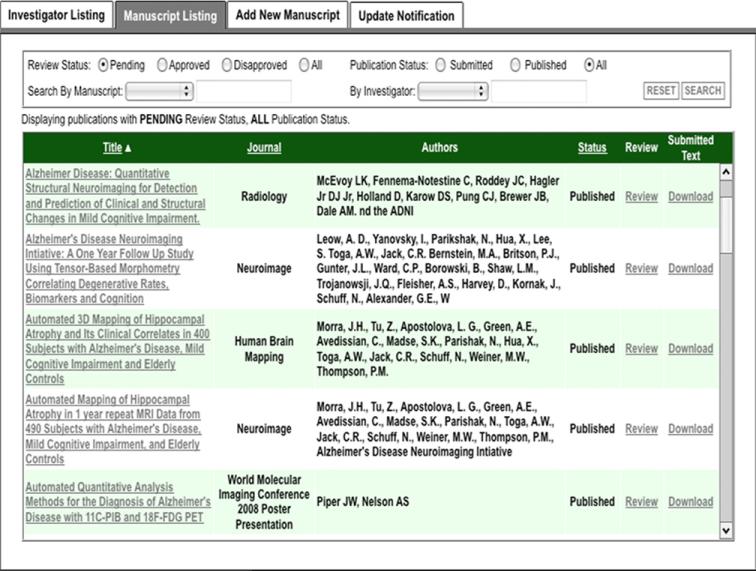
The ADNI DPC oversees access by external investigators. An online application and review feature is integrated into the LDA so that applicant information and committee decisions are recorded in the database and the e-mail communications acknowledging application receipt, approval, or disapproval are automatically generated. Approved ADNI data users are required to submit annual progress reports, and the online system provides mechanisms for this function along with related tasks, such as adding team members to an approved application and receiving manuscripts for DPC review. All data accesses are logged and numbers of uploaded and downloaded image data are available to project managers through interactive project summary features.
More than 100,000 image datasets (more than five million files) and related clinical imaging, biomarker, and genetic datasets are available to approved investigators. More than 800,000 downloads of raw, pre-, and postprocessed scans have been provided to authorized investigators. Downloads of the clinical, biomarker, image analysis results and genetic data have been downloaded more than 4,300 times.
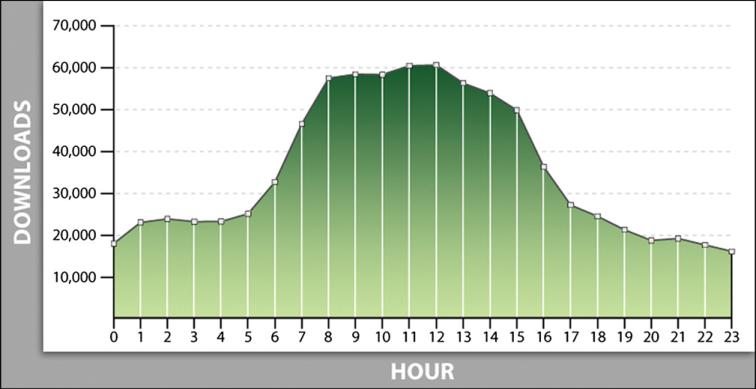
Data download activity has increased each year since the data became available, increasing from 154,200 image data-sets downloaded in 2007 to almost 290,000 image datasets downloaded in 2009. With users from across the globe accessing the archive, activity occurs around the clock (Fig. 5).
Fig. 5.
The number of image downloads by hour of the day shows maximum activity occurring during U.S. working hours but still a significant amount of activity at other times in line with the numbers of investigators inside and outside the U.S.
2.6. Data management
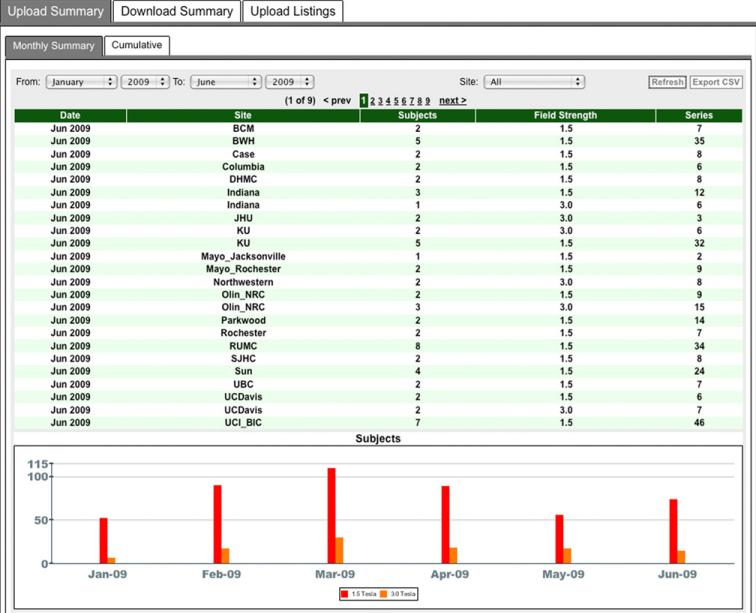
With responsibilities for data and access oversight and administration spread across multiple institutions, we built a set of components to help those involved manage portions of the study. These include a set of data user management tools for reviewing data use applications, managing manuscript submissions, and sending notifications to investigators whose annual ADNI update is due (Fig. 6), and also a set of project summary tools that support interactive views of upload and download activities by site, user, time period, and provide exports of the same (Fig. 7). Other information, documents, and resources geared toward apprising investigators about the status of the study and data available in the archive are provided through the website.
Fig. 6.
Project summary components include a tabular listing (above) and a graphical representation (below). Specific sites and time ranges control the information displayed and tabular data may be exported for use, such as in reports and in further analysis.
Fig. 7.
User management components support the work of the DPC, the body charged with reviewing, approving, and tracking ADNI data usage and related publications.
3. Discussion
The informatics core of ADNI provides a mechanism to distribute and share data, results, and information not only between and among the participants, but also to the scientific community at large. This informatics model enables a far more extensive array of analytic strategies and approaches to interpreting these data. It could be argued that these dissemination aspects of ADNI are among the most important to its success. The databases we developed have contributed to the field of AD research as well as others. In fact, several other neurological and psychiatric disease studies are already underway or in the planning stages that emulate systems developed in the Informatics Core of ADNI.
The specific informatics characteristics necessary to achieve this level of success are several. A spectrum of database models has been proposed [19] that range from simple FTP sites to fully curated efforts containing data from published studies. The databasing, querying, examination, and processing of datasets from multiple subjects necessitate efficient and intuitive interfaces, responsive answers to searches, coupled analyses workflows, and comprehensive descriptions and provenance with a view toward promoting independent re-analysis and study replication.
Successful informatics solutions must build a trust with the communities they seek to serve and provide dependable services and open policies to researchers. Several factors contribute to a database's utility, including whether it actually contains viable data, and these are accompanied by a detailed description of their acquisition (e.g., meta-data); whether the database is well-organized and the user interface is easy to navigate; whether the data are derived versions of raw data or the raw data itself; the manner in which the database addresses the sociological and bureaucratic issues that can be associated with data sharing; whether it has a policy in place to ensure that requesting authors give proper attribution to the original collectors of the data; and the efficiency of secure data transactions. These systems must provide flexible methods for data description and relationships among various meta-data characteristics [12]. Moreover, those that have been specifically designed to serve a large and diverse audience with a variety of needs and that possess the qualities described earlier, represent the types of databases that can have the greatest benefit to scientists looking to study the disease, assess new methods, examine previously published data, or with interests in exploring novel ideas using the data [19].
A list of lessons learned by the Informatics Core of ADNI follows many of the same principles identified by Arzberger et al. [20] and Beaulieu [21]. These are as follows: (1) the data archive—information should be open and unrestricted; (2) The database should be transparent in terms of activity and content; (3) The individuals and institutions responsible for what should be clear. The exact formal responsibilities among the stakeholders should be stated; (4) Technical and semantic interoperability between the database and other online resources (data and analyses) should be possible; (5) Clear curation systems governing quality control, data validation, authentication, and authorization must be in place; (6) The systems must be operationally efficient and flexible; (7) There should be clear policies of respect for intellectual property and other ethical and legal requirements; (8) The needs to be management accountability and authority; (9) A solid technological architecture and expertise are essential; (10) There should be systems for reliable user support. Additional issues involve Health Insurance Portability and Accountability Act (HIPAA) compliance [22], concern over incidental findings [23], anonymization of facial features [24, 25], and skull stripping [26]. The most trusted and hence successful informatics solutions tend to be those that meet these principles and have archival processes that are sufficiently mature and demonstrate solid foundation and trust. At least until 2009, a valid analogy could be that informatics systems and databases are to the science community as financial systems and banks are (were) to the financial community.
4. Future direction
Scientific research is increasingly data intensive and collaborative, yet the infrastructure needed for supporting data intensive research is still maturing. An effective database should strive to blur the boundary between the data and the analyses, providing data exploration, statistics, and graphical presentation as embedded toolsets for interrogating data. Interactive, iterative interrogation and visualization of scientific data lead to enhanced analysis and understanding and lowers the usability threshold [27]. Next steps for the informatics core will involve development of new tools and services for data discovery, integration, and visualization. Components for discovering ADNI data must include contextual information that allows data to be understood, reused, and the results reproduced. Integrating a broader spectrum of ADNI data and providing tools for interrogating and visualizing those data will enable investigators to more easily and interactively investigate broader scientific questions. We plan to integrate additional clinical and biochemical data and analysis results into the database and build tools for interactively interrogating and visualizing the data in tabular and graphical forms.
Acknowledgments
Data used in preparing this article were obtained from the Alzheimer's Disease Neuroimaging Initiative database (www.loni.ucla.edu/ADNI). Consequently, many ADNI investigators contributed to the design and implementation of ADNI, or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators is available at www.loni.ucla.edu/ADNI/Collaboration/ADNI_Citation.shtml. This work was primarily funded by the ADNI (Principal Investigator: Michael Weiner; NIH grant number U01 AG024904). ADNI is funded by the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and the Foundation for the National Institutes of Health, through generous contributions from the following companies and organizations: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging (ISOA), with participation from the U.S. Food and Drug Administration. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. This study was also supported by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 RR021813, entitled Center for Computational Biology (CCB). Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics. Additional support was provided by grants P41 RR013642 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH).
We thank Scott Neu and the Data Archive team for their contributions.
References
- 1.Toga AW, Thompson PM. Genetics of brain structure and intelligence. Annu Rev Neurosci. 2005;28:1–23. doi: 10.1146/annurev.neuro.28.061604.135655. [DOI] [PubMed] [Google Scholar]
- 2.Toga AW, Mazziotta JC. Brain Mapping: The Methods. 2nd ed. Academic Press; San Diego, CA: 2002. [Google Scholar]
- 3.Mazziotta JC, Toga AW, Frackowiak RSJ. Brain Mapping: The Disorders. Academic Press; San Diego, CA: 2000. [Google Scholar]
- 4.Toga AW, Mazziotta JC. Brain Mapping: The Systems. Academic Press; San Diego, CA: 2000. [Google Scholar]
- 5.Olson GM, Zimmerman A, Bos N. Introduction. In: Olson GM, Zimmerman A, Bos N, editors. Scientific Collaboration on the Internet. MIT Press; Cambridge, MA: 2008. pp. 1–12. [Google Scholar]
- 6.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, et al. A four-dimensional probabilistic atlas of the human brain. J Am Med Inform Assoc. 2001;8:401–30. doi: 10.1136/jamia.2001.0080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–8. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- 9.Atkins DE, Droegemeier KK, Feldman SI, Garcia-Molina H, Klein ML, Messerschmitt DG, et al. Revolutionizing Science and Engineering Through Cyberinfrastructure: Report of the National Science Foundation Blue-Ribbon Advisory Panel on Cyberinfrastructure. National Science Foundation; Arlington, VA: 2003. [Google Scholar]
- 10.Koslow SH. Opinion: sharing primary data: a threat or asset to discovery? Nat Rev Neurosci. 2002;3:311–3. doi: 10.1038/nrn787. [DOI] [PubMed] [Google Scholar]
- 11.Poliakov AV. Unobtrusive integration of data management with fMRI analysis. Neuroinformatics. 2007;5:3–10. doi: 10.1385/ni:5:1:3. [DOI] [PubMed] [Google Scholar]
- 12.Hasson U, Skipper JI, Wilde MJ, Nusbaum HC, Small SL. Improving the analysis, storage and sharing of neuroimaging data using relational databases and distributed computing. Neuroimage. 2008;39:693–706. doi: 10.1016/j.neuroimage.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox PT, Bullmore E, Bandettinig PA, Lancaster JL. Protecting peer review: correspondence chronology and ethical analysis regarding Loghothetis vs. Shmuel and Leopold. Hum Brain Mapp. 2009;30:347–54. doi: 10.1002/hbm.20682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher J. Alzheimer's researchers open the doors to data sharing. Lancet Neurol. 2007;6:480–1. doi: 10.1016/S1474-4422(07)70118-7. [DOI] [PubMed] [Google Scholar]
- 15.Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–91. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi A, Koeppe RA, Fessler JA. Reducing between scanner differences in multi-center PET studies. Neuroimage. 2009;46:154–9. doi: 10.1016/j.neuroimage.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmhan YL, Plale B, Gannon D. A survey of data provenance in e-science. SIGMOD Rec. 2005;34:31–6. [Google Scholar]
- 18.Mackenzie-Graham AJ, Van Horn JD, Woods RP, Crawford KL, Toga AW. Provenance in neuroimaging. Neuroimage. 2008;42:178–95. doi: 10.1016/j.neuroimage.2008.04.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Horn JD, Grethe JS, Kostelec P, Woodward JB, Aslam JA, Rus D, Rockmore D, Gazzaniga MS. The functional Magnetic Resonance Imaging Data Center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Philos Trans R Soc Lond B Biol Sci. 2001;356:1323–39. doi: 10.1098/rstb.2001.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arzberger P, Schroeder P, Beaulieu A, Bowker G, Casey K, Laaksonen L, Moorman D, Uhlir P, Wouters P. Sceicne and government. An international framework to promote access to data. Science. 2004;303:1777–8. doi: 10.1126/science.1095958. [DOI] [PubMed] [Google Scholar]
- 21.Beaulieu A. Voxels in the brain: neuroscience, informatics and changing notions of objectivity. Soc Stud Sci. 2001;31:635–80. doi: 10.1177/030631201031005001. [DOI] [PubMed] [Google Scholar]
- 22.Kulynych J. Legal and ethical issues in neuroimaging research: human subjects protection, medical privacy, and the public communication of research results. Brain Cogn. 2002;50:345–57. doi: 10.1016/s0278-2626(02)00518-3. [DOI] [PubMed] [Google Scholar]
- 23.Illes J, De Vries R, Cho MK, Schraedley-Desmond P. ELSI priorities for brain imaging. Am J Bioeth. 2006;6:W24–31. doi: 10.1080/15265160500506274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bischoff-Grethe A, Ozyurt IB, Busa E, Quinn BT, Fennema-Notestine C, Clark CP, et al. A technique for the deidentification of structural brain MR images. Hum Brain Mapp. 2007;28:892–903. doi: 10.1002/hbm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neu SC, Toga AW. Automatic localization of anatomical point landmarks for brain image processing algorithms. Neuroinformatics. 2008;6:135–48. doi: 10.1007/s12021-008-9018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang AH, Valentino DJ, Toga AW. Skull-stripping magnetic resonance brain images using a model-based level set. Neuroimage. 2006;32:79–92. doi: 10.1016/j.neuroimage.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 27.National Science Foundation (US) and Cyberinfrastructure Council . Cyberinfrastructure vision for 21st century discovery. National Science Foundation; Arlington, VA: 2007. [Google Scholar]