Abstract
Over 40% of colon cancers have a mutation in K-RAS or N-RAS, GTPases that operate as central hubs for multiple key signaling pathways within the cell. Utilizing an isogenic panel of colon carcinoma cells with K-RAS or N-RAS variations, we observed differences in TNFα-induced apoptosis. When the dynamics of phosphorylated ERK (pERK) response to TNFα were examined, K-RAS mutant cells showed lower activation while N-RAS mutant cells exhibited prolonged duration. These divergent trends were partially explained by differential induction of two ERK-modulatory circuits: negative feedback mediated by DUSP5 and positive feedback by autocrine TGFα. Moreover, in the various RAS-mutant colon carcinoma lines, the TGFα autocrine loop differentially elicited a further downstream chemokine (CXCL1/CXCL8) autocrine loop, with the two loops having opposite impacts on apoptosis. While the apoptotic responses of the RAS-mutant panel to TNFα treatment showed significant dependence on the respective pERK dynamics, successful prediction across the various cell lines required contextual information concerning additional pathways including IKK and p38. A quantitative computational model based on weighted linear combinations of these pathway activities successfully predicted not only the spectrum of cell death responses but also the corresponding chemokine production responses. Our findings indicate that diverse RAS mutations yield differential cell behavioral responses to inflammatory cytokine exposure by means of: [a] differential effects on ERK activity via multiple feedback circuit mechanisms; and [b] differential effects on other key signaling pathways contextually modulating ERK-related dependence.
Keywords: RAS, apoptosis, chemokines, autocrine, mathematical models
Introduction
Upon activation by receptor tyrosine kinases, the RAS family of GTPases (K-RAS4A, K-RAS4B, H-RAS, and N-RAS) signal to multiple downstream effector pathways. Single amino acid mutations at codons 12, 13, or 61 place RAS in a chronically active (GTP-bound) state and are oncogenic (1). Mutations in both K-RAS and N-RAS are found in colon cancer; however, K-RAS mutations are found in nearly 50% of tumors whereas N-RAS mutations are found in approximately 5% (2, 3). Whether the disparate mutation frequencies reflect underlying biological or functional differences is unknown; however, determining differences between the oncogenic forms of K-RAS and N-RAS could improve our ability to target therapies to these sub-groups of colon cancer patients.
K-RAS and N-RAS are greater than 90% homologous and appear to share many of the same downstream effectors, including RAF and PI3K (4). However, it is unclear how the different RAS proteins compete for the same effectors and impact cellular decisions. Several sources of evidence suggest that K-RAS and N-RAS have distinct physiological functions. Loss of K-Ras is embryonically lethal in mice, while N-Ras knockout mice are viable with defects in immune response (1). Mouse models of K-RasG12D and N-RasG12D expressed in the colonic epithelium show distinct phenotypes, with K-RasG12D stimulating hyperproliferation and N-RasG12D conferring resistance to apoptosis (5). Oncogenic K-RAS promotes butyrate-induced apoptosis in human colon carcinoma cells (6) while N-RAS provides anti-apoptotic signals in mouse embryonic fibroblasts (7), indicating that apoptosis is a key cellular process that the RAS proteins differentially regulate.
The effects of RAS proteins and their oncogenic forms in response to inflammation and apoptotic stimuli are of particular interest for colon cancer. Chronic inflammation has been shown to induce DNA damage and colon tumors in mice (8). Similarly, patients with long-term inflammatory bowel disease (IBD) have an increased risk of developing colon cancer (9). While nearly half of colon carcinomas express oncogenic K-RAS, IBD is not associated with K-RAS mutations (10), and links between IBD and N-RAS mutations have not been investigated. Although inflammation is multi-faceted, mice that overproduce the cytokine tumor necrosis factor-alpha (TNFα) develop an IBD-like phenotype (11) and monoclonal-TNFα antibodies have shown some benefit as a therapy for IBD (12), indicating one cytokine of interest.
In this work we examine the response of an isogenic panel of colon carcinoma cell lines with wildtype RAS, mutant K-RAS, mutant N-RAS, or reduced levels of N-RAS to determine how the different RAS configurations impact the apoptotic response to TNFα. We observed consequent differences in pERK dynamics and identified changes in negative feedback mediated by DUSP5 and positive feedback by autocrine TGFα among the RAS variants. Additionally, we identified TGFα-induced chemokine autocrine loops that provide pro-survival input to the cells. To further refine our understanding of the influence of RAS to the TNFα response, we obtained quantitative dynamic measurements of phosphoprotein signals across multiple pathways and elucidated key combinations of these signals capable of predicting differential apoptosis and chemokine production behavior for the different lines. Our findings indicate that diverse RAS mutations yield differential cell behavioral responses to inflammatory cytokine exposure by means of: [a] differential effects on ERK activity via multiple feedback circuit mechanisms; and [b] differential effects on other key signaling pathways contextually modulating ERK-related dependence.
Materials and Methods
Cell Lines and Treatments
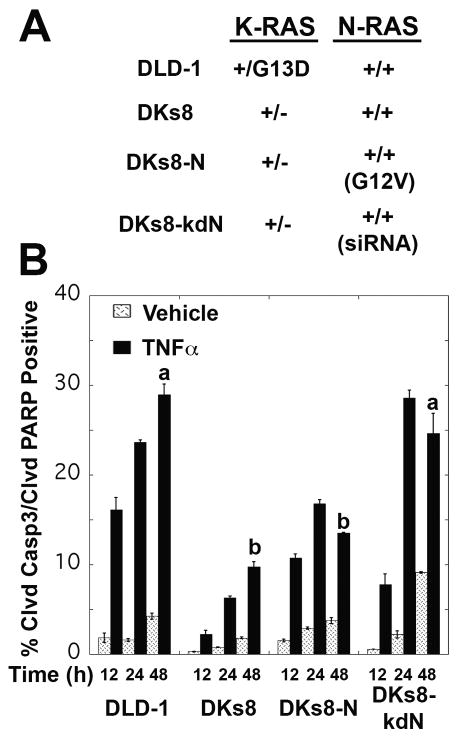
DLD-1, a colon carcinoma cell line with a single copy K-RASG13D mutation, and their isogenic partner, DKs8, where K-RASG12D was removed by homologous recombination, have been previously described (13) (Fig. 1A). DKs8-N, which overexpress mutant N-RASG12V were generated by infection of DKs8 with MSCV retrovirus (14). DKs8-kdN, which are wild-type with respect to K-RAS and have reduced levels of wild-type N-RAS, were generated via lentiviral shRNA using the pSICOR retrovirus and DKs8 (15).
Figure 1.
TNFα treatment induced apoptosis in all four RAS-variant cell lines, with the highest levels observed in DLD-1 and DKs8-kdN cells. A, Overview of RAS variant genotypes – full details can be found in Materials and Methods. B, Cells were stained for cleaved caspase-3 and cleaved PARP and analyzed by flow cytometry for double positive (apoptotic) cells. Different letters indicate significant differences between TNFα-treated RAS-variants at 48 hours, p<0.05.
All cell lines were maintained in DMEM plus 10% fetal bovine serum (FBS); DKs8-kdN were also supplemented with 7.5μg/mL puromycin to maintain shRNA selection. For experiments, cells were plated in 10% FBS at 15,000 cells/cm2 (DLD-1, DKs8-N) or 18,000 cells/cm2 (DKs8, DKs8-kdN). After 24 hours, cells were sensitized with 200 units/mL interferon-γ (IFNγ, Roche Applied Science, Indianapolis, IN) in 5% FBS. After 24 hours, cells were treated with either vehicle or 100ng/mL TNFα (Peprotech, Rocky Hill, NJ). In a subset of experiments, cycloheximide or repertaxin were added with TNFα (Sigma, St. Louis, MO; 2.5μg/mL and 0.1μM, respectively) or ab225 was added prior to TNFα (a gift from H.S. Wiley, Pacific Northwest National Laboratory, 10μg/mL).
Lysis and Signaling Measurements
At various times after TNFα stimulation (0, 5, 15, 30, 60, 90, 120, 240, 480, and 720 minutes) cells were lysed using Bio-Plex cell lysis buffer for clarified lysates (Bio-Rad, Hercules, CA) and an SDS-based lysis buffer for whole cell lysates (16). Total protein concentrations were determined using the bicinchoninic acid assay (Pierce, Rockford, IL).
Phosphoproteins (pERK1, pERK2, pIκBα, pJNK, pAKT, and pHSP27) were detected using commercially-available kits for the Luminex system (Bio-Rad). A master positive-reference sample was loaded in each assay for normalization purposes. Cleaved caspase-8 was detected using immunoblots, again with positive reference samples for normalization purposes. For cleaved caspase-8, 50μg of total protein was loaded and probed (Cell Signaling Technology, Danvers, MA #9496). Blots were detected with ECL Advance (GE Healthcare, Piscataway, NJ) and imaged on a Kodak Image Station 1000.
Flow Cytometry
Floating and adherent cells were pooled and analyzed for apoptosis using Annexin V/propidium iodide and cleaved caspase-3/cleaved PARP similar to the previously described methods (16). A minimum of 25,000 cells per condition were analyzed on a BD Biosciences LSRII (part of the Koch Institute Flow Cytometry Core Facility, MIT) and by FlowJo (Tree Star, Inc, Ashland, OR).
ELISAs
Conditioned media was collected and analyzed for TGFα, IL-1ra, IL-1α, and IL-1β by ELISA (R&D Systems, Minneapolis, MN), and screened for 50 cytokines, chemokines, and growth factors with Human Group I and II multiplex assays (Bio-Rad). Follow-up assays for VEGF, CXCL1, CXCL8, and CXCL10 were performed using individual assays (Bio-Rad). For normalization, cells were trypsinized and the live cell fraction was counted using a ViCell XR (Beckman Coulter, Fullerton, CA). TACE levels were determined by ELISA (R&D Systems) from lysates of IFNγ-sensitized cells according to the manufacturer's instructions and normalized to total protein determined by bicinchoninic acid assay.
Quantitative RT-PCR
Expression levels of DUSP5 and GAPDH were determined by quantitative RT-PCR for RNA collected at 0, 30, 90, and 240 minutes after TNFα treatment, Each assay was run with a standard curve of stock cDNA from untreated DKs8 cells and analyzed by the relative standard curve method.
CXCR Immunofluorescence
DLD-1 cells were plated on acid-washed coverslips and IFNγ sensitized for 24 hours, fixed with 4% paraformaldehyde and stained with anti-CXCR1 (1:25, BD Pharmigen, Franklin Lakes, NJ) or anti-CXCR2 (1:5, R&D Systems) overnight at 4°C, then detected with Alexa 488-donkey-anti-mouse IgG (1:400) and phalloidin-rhodamine (1:200, Invitrogen, Carlsbad, CA). Slides were imaged on a DeltaVision microscope (Applied Precision, Tacoma, WA) with a 60× oil-objective (NA=1.4). 0.2μm sections were captured and the stack compressed with the maximum value at each pixel displayed.
Partial Least Squares Regression (PLSR) Modeling and Statistical Analysis
The compiled data set represents approximately 1,800 individual proteomic measurements. Multi-pathway models for this data set were generated using the PLSR algorithm in SimcaP (Umetrics, Kinnelon, NJ - see (17) for details). Signal and response data were unit-variance scaled (16) and models were tested by cross-validation. The independent variable block for the full PLSR model included 64 measurements – six phosphorylated proteins at 10 time points and 4 measures of cleaved caspase-8.
Data are represented as average ± SEM, with three independent measurements for each treatment condition. Comparisons were performed by ANOVA and Tukey-HSD, with significance set to p<0.05.
Results
TNFα Treatment Induces RAS-Specific Levels of Apoptosis
To examine the impact of RAS mutations on the cellular response to TNFα, we utilized an isogenic panel of colon carcinoma cells (Fig. 1A) that express both TNFR1 and TNFR2 (Supplementary Fig. S1). TNFα treatment resulted in a significant increase in apoptosis as measured by cleaved caspase-3 and cleaved PARP (Fig. 1B, Supplementary Fig. S2, p<0.0001). RAS variations impacted the extent of TNFα-induced apoptosis, with mutations in K-RAS (DLD-1) or reductions in N-RAS (DKs8-kdN) showing the highest levels of apoptosis. These results are consistent with other reports of K-RAS mutations being pro-apoptotic and N-RAS having a protective role (7, 15, 18). Despite the biochemical similarities in K-RAS and N-RAS mutants, the Dks8-N cells do not exhibit this heightened sensitivity to the apoptotic stimuli. Resistance to apoptosis has been described as a necessary step for tumor development (19). Therefore, it may seem counter-intuitive that colon carcinoma cell lines with the common K-RAS mutation are more sensitive to TNFα than cells with wildtype RAS (Fig.1B). However, K-RAS mutations are less common in IBD-associated cancers than in the general population of colon cancers (10).
Negative Regulators of pERK Dynamics Differ Among RAS Variants
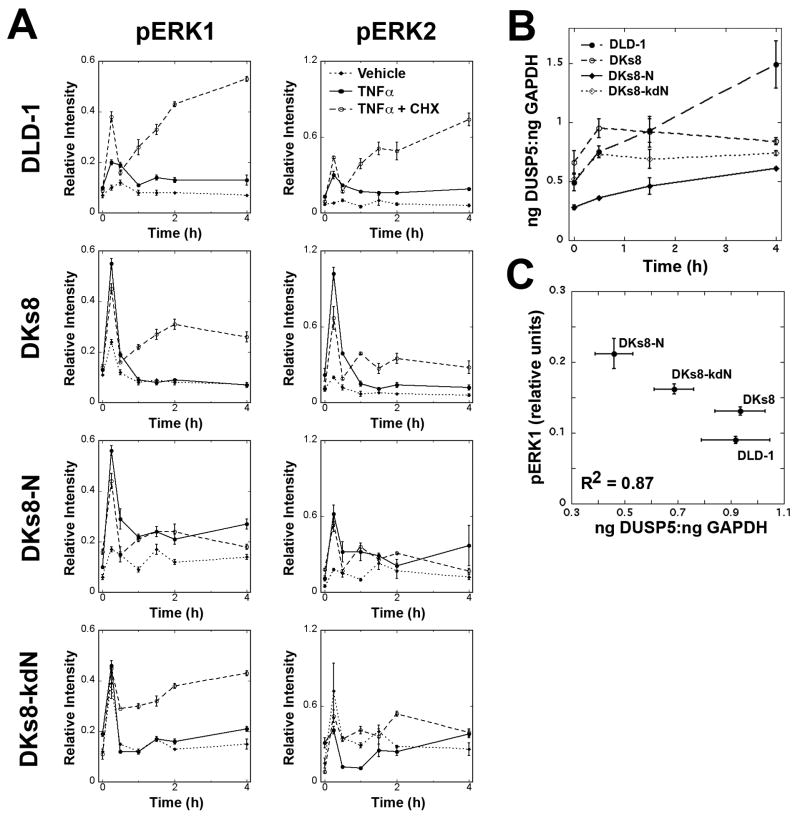
To examine how changes in RAS expression level and mutation status impacted TNFα-induced apoptosis, we first examined the dynamics of pERK1 and pERK2 following treatment (Fig. 2A). These assays were performed using the Luminex platform, which was validated to determine that measurements were reliably quantitative (Supplementary Fig. S3, (20)). RAS variants had different early and late patterns for pERK. DLD-1 cells had much lower levels of pERK in the early peaks (around t=15 minutes) while DKs8-N cells had pERK levels that did not return to baseline by 4 hours.
Figure 2.
pERK levels and phosphatase induction differed between RAS-variants following TNFα treatment. A, Levels of pERK1 and pERK2 were determined by Luminex assay for each cell line following treatment with vehicle, TNFα, or TNFα and cycloheximide. B, Quantitative RT-PCR analysis of DUSP5 in TNFα-treated cells. Expression levels were normalized to GAPDH. C, Relationship between pERK1 levels (at 2 hours) and DUSP5 (at 1.5 hours).
The importance of pathway-specificity in determining the activation of dual-specificity phosphatases (DUSPs) has been recently highlighted (21). Therefore, we reasoned that the RAS variants may impact which DUSPs are turned on in response to TNFα and this could explain the extended signaling observed in the DKs8-N cells. To determine whether the different patterns of pERK required newly synthesized proteins, cells were treated with cycloheximide and TNFα. DLD-1, DKs8, and DKs8-kdN had elevated pERK signals when treated with cycloheximide and TNFα, compared to TNFα alone (Fig. 2A). DKs8-N cells showed little difference with cycloheximide, indicating that an induced phosphatase could differ between the RAS variants. Primers were validated for nine DUSPs that recognize ERK as a substrate ((22), Supplementary Fig. S4). An initial screen identified DUSP5 as a target DUSP that was induced in response to TNFα treatment (Supplementary Table S1). When assayed by quantitative RT-PCR, DUSP5 was induced in all four cell lines (Fig. 2B). The overall level and extent of induction was lower in DKs8-N cells, and a strong inverse correlation was observed between the normalized level of DUSP5 at 1.5 hours and the level of pERK1 at 2 hours (Fig. 2C).
Differences in TGFα Autocrine Loop Among RAS Variants
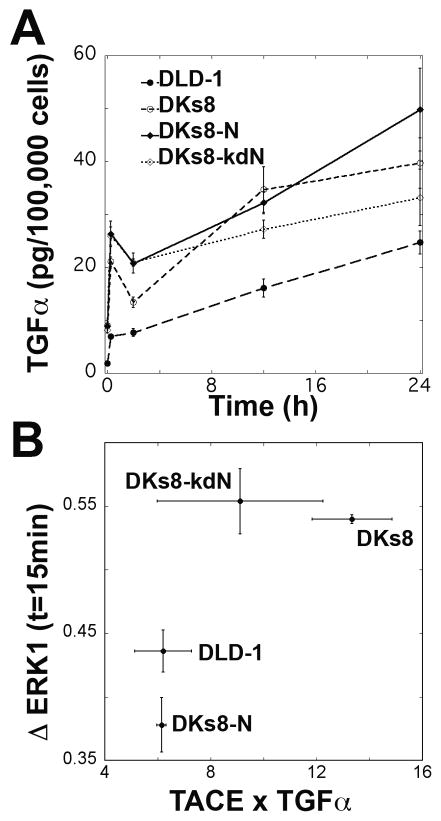
TNFα treatment has been previously shown to induce a TGFα autocrine cascade in the B-RAF mutant HT-29 colon carcinoma cells (23). To determine if this cascade was present in our RAS-variant panel, cells were treated with TNFα and ab225, a monoclonal antibody that blocks EGFR and prevents uptake of released TGFα. Significant increases in the levels of TGFα in the cell culture media were seen within 15 minutes of TNFα treatment (Fig. 3A, p=0.0001). Over time, TGFα accumulated in the media at a rate between 0.6 and 1.3 pg/100,000 cells/hour; the long-term rates were lower than the initial bursts (9.8 – 43.8 pg/100,000 cells/hour). The very rapid initial increase in TGFα production suggests a non-transcriptional mechanism such as transactivation (24). Cells treated with TNFα and ab255 had significantly reduced levels of pERK1 at 15 minutes (Fig. 3B), indicating that this autocrine loop is responsible for much of the early peak (Fig. 2A). Different levels of pro-TGFα, TACE/ADAM17 (tumor necrosis factor-converting enzyme/a disintegrin and metalloprotease) -- the metalloprotease implicated in the ecto-domain cleavage of pro-TGFα (25), and/or EGFR could conceivably explain the difference in early ERK behavior. Levels of EGFR were not significantly different between RAS variants, but DKs8 and DKs8-kdN cells exhibited significantly higher levels of TACE and DKs8-dkN cells demonstrated substantially lower levels of pro-TGFα (Supplementary Fig. S5) indicating both these factors could be influential. Indeed, the product of pro-TGFα and TACE levels at the time of TNFα treatment correlated monotonically with short-term TGFα-induced increases in pERK1 (Fig. 3B). Interestingly, the relative magnitudes of this TGFα autocrine loop-mediated pERK signaling for the different RAS variants does not appear to immediately correspond to their respective TNFα-induced apoptosis responses (Fig. 1B). This observation motivated us to consider further consequences of the TGFα autocrine loop as well as other pathway effects.
Figure 3.
TGFα autocrine cascade is induced by TNFα. A, TGFα levels were assayed in conditioned media from TNFα and ab225 treated cells. Growth factor levels were normalized to concurrent cell counts. B, Relationship between the potential TGFα release (quantified by the intensity of pro-TGFα by Western blot times the amount of TACE by ELISA) and the increase in pERK1 at 15 minutes between cells treated with TNFα and ab225 or TNFα alone.
TNFα-Induced Production of Multiple Chemokines and Growth Factors
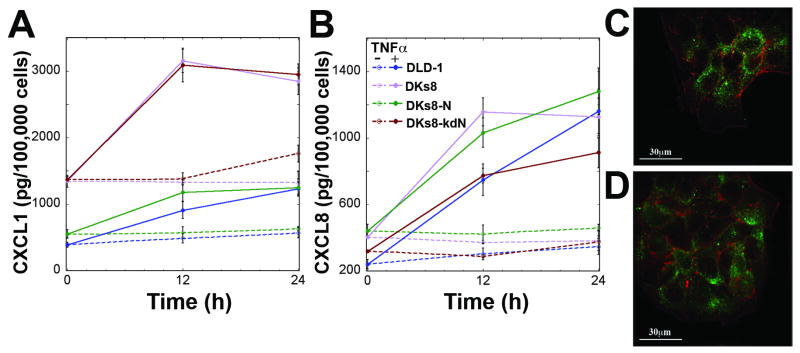
The TNFα-induced TGFα autocrine loop has been shown to induce additional autocrine loops, including an IL-1α/IL-1ra cascade (in HT-29 cells (23)) and an IL-1α/IL-1β/IL-1ra cascade (primary rat hepatocytes (26)). The RAS-variant cell lines did not secrete detectable levels of IL-1α, IL-1β, or IL-1ra (data not shown). A screen of media from TNFα-treated cells for 50 cytokines, chemokines, and growth factors revealed detectable levels of CCL2, CCL7, CXCL1, CXCL8, CXCL9, CXCL10, CXCL12, ICAM1, MIF and VEGF. Of these ten positive results, CXCL1, CXCL8, CXCL10, and VEGF showed significant increases with TNFα treatment (Fig. 4, Supplementary Fig. S6A and S7A). VEGF and CXCL8 (IL-8) levels were similar between all four RAS-variants while CXCL1 (GROα) and CXCL10 (IP-10) were substantially lower in K-RAS and N-RAS mutant cells both before and after TNFα treatment. To determine whether secretion of these proteins could lead to additional autocrine cascades, cells were examined at the time of TNFα treatment for the appropriate receptors by immunofluorescence or PCR. None of the RAS-variant cells expressed VEGF-R2 (Supplementary Fig. S7B). DLD-1 cells expressed CXCR1 (for CXCL8), CXCR2 (for CXCL1 and CXCL8), and CXCR3 (for CXCL10), indicating possible autocrine loops (Fig. 4C,D and Supplementary Fig. S6B).
Figure 4.
RAS-variant cells express CXCR1 and CXCR2 and produce CXCL1 and CXCL8 in response to TNFα treatment. A,B, CXCL1 and CXCL8 were quantified by Luminex assay and normalized to concurrent cell counts. C, CXCR1 and D, CXCR2 were observed in DLD-1 cells by immunofluorescence. Green = CXCR1,2; Red = phalloidin for actin filaments.
TGFα and Chemokine Autocrine Loops are Linked and Contrapositively Impact Apoptosis
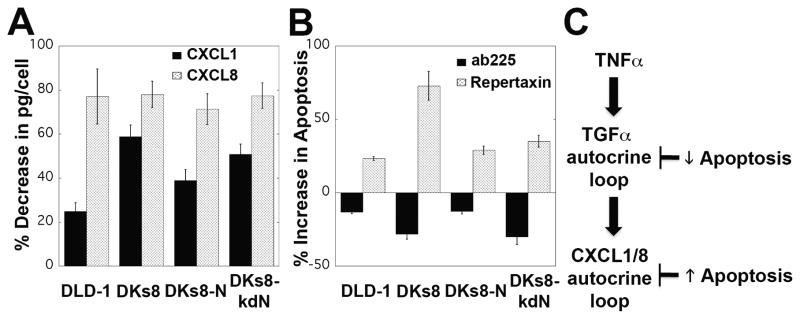
To determine if the increase in CXCL1, CXCL8, and CXCL10 following TNFα treatment was mediated by the TGFα autocrine cascade, the RAS-variant cells were treated with TNFα and ab225. Levels of all three chemokines were substantially reduced with ab225 co-treatment, indicating that the chemokine response to TNFα was a consequence of the induced TGFα autocrine loop (Fig. 5A and data not shown). The impact of these autocrine loops on apoptosis was examined by co-treatment with TNFα and ab225 or repertaxin (a non-competitive allosteric inhibitor of CXCR1/2 (27)). Blockade of the TGFα loop with ab225 decreased apoptosis in all four RAS-variants, while repertaxin treatment increased apoptosis (Fig. 5B). Exogenous treatment with CXCL10 to supplement TNFα and ab225 co-treated cells had no effect on apoptosis at 24 hours, suggesting that CXCL10 does not directly impact the apoptotic decision (Supplementary Fig. S6C). The molecular logic of the TNFα/TGFα/CXCL1/8 autocrine cascades (Fig. 5C) suggests that the TGFα autocrine loop has multiple downstream effects, including the described pERK effects and induction of chemokines. Despite the induction of a pro-survival chemokine loop, the net effect of TGFα was pro-apoptotic. To further examine these complex influences and help gain further insight into the effect of the RAS mutations on the interpretation of these loops, we measured phosphoprotein signals across multiple pathways alongside that of ERK.
Figure 5.
The TGFα and chemokine autocrine loops are linked. A, CXCL1 and CXCL8 levels decreased when cells were treated with ab225 and TNFα for 12 hours compared to TNFα alone. B, Co-treatment of TNFα with ab225 for 24 hours decreased apoptosis in all RAS variants, while co-treatment with repertaxin increased apoptosis. C, The molecular logic of the TGFα and CXCL1/8 autocrine loops following TNFα treatment.
Multi-Pathway Models Can Predict Apoptosis and Chemokine Levels
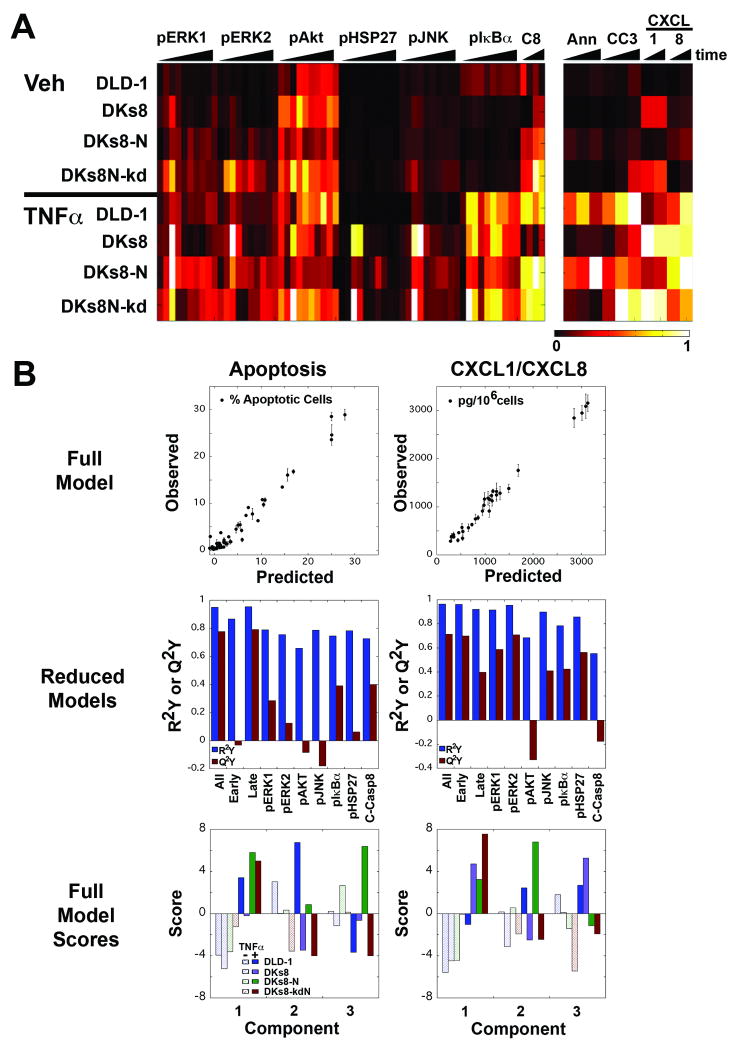
Lysates from vehicle and TNFα treated RAS-variant cells were analyzed for additional signaling molecules that are downstream of the TNF receptor or RAS (pIκBα, pHSP27, pAKT, pJNK, and cleaved caspase-8). The resulting data set (Fig. 6A) demonstrates substantial differences in pIκBα, pHSP27, pJNK, and cleaved caspase-8 with TNFα treatment, while pAKT signaling does not appear to be dependent on TNFα. DKs8 cells have strong peaks in pJNK and pHSP27 at early times, while DKs8-N have higher signals compared to the other RAS-variants at later times, similar to the pERK data (Fig. 2A and 6A). Interestingly, although they have similar levels of apoptosis (Fig. 1B), DLD-1 and DKs8-kdN cells have different signaling patterns for multiple molecules (Fig. 6A).
Figure 6.
A multi-pathway model can accurately predict apoptosis and chemokine production. A, Heat map of the four RAS-variant cell lines treated with vehicle (top) or TNFα (bottom). Luminex assays were used to measure pERK1, pERK2, pAKT, pHSP27, pJNK, and pIκBα. Quantitative Western blots were used to measure cleaved caspase-8 (C8). Apoptosis was measured by flow cytometry for Ann (Annexin+/propidium iodide-) and CC3 (cleaved caspase-3+/cleaved PARP+). CXCL1 and CXCL8 were quantified by Luminex assay and normalized to cell counts. Each box represents the average of three independent measurements at one time, normalized across all times and cells for that measurement. B, PLSR models for apoptosis (left) and chemokine levels (right) were constructed using all (top, bottom) or subsets (middle) of signals.
To analyze this multi-pathway data set, we utilized partial least squares regression (PLSR), which has been described (28) and applied elsewhere (17, 29). In PLSR, the X matrix (here the signaling data set) is regressed against the Y matrix (here, either apoptosis or chemokine levels). PLSR reduces the dimensionality of the data matrix to fewer variables by emphasizing the independent measurements that strongly co-vary with the dependent outcomes – in essence, PLSR attempts to develop a model in which similar signaling “signatures” are associated with similar functional responses. PLSR models are constructed in an iterative process by calculating principal components - linear combinations of variables in the original independent and dependent blocks. The first principal component captures the strongest variation in the original data matrix, while succeeding principal components capture remaining variation. The number of principal components that results in the minimum error signifies the model with the maximum useful information captured without extending to include variation from experimental noise.
An important element of PLSR modeling is how the data is pre-processed (16). We constructed models using the raw data from each assay, data normalized to concurrently-run master lysates, data normalized to the values for each signal at the zero timepoint of that cell line, and data normalized to the maximum signal across all conditions and times. Only data normalized to the concurrently-run master lysates was successful in building predictive models (Q2Y > 0.7) for both chemokines and apoptosis outcomes (Fig. 6B, top and data not shown). Previous studies in our lab have utilized signal-derived metrics, such as time derivatives and area-under-the-curve measures as part of the X matrix (30). Our results indicated that these metrics did not substantially change model fit (R2Y) or predictiveness (Q2Y), likely since they primarily represent linear combinations of the X variables (data not shown).
To assess the importance of the various signals in the PLSR models, we evaluated reduced models, which used various subsets of the independent variables to fit the response data (Fig. 6B, middle). Example subsets include early times (0-60 minutes), late times (after 60 minutes), and individual signal measurements. While R2Y is not strongly affected in the resulting three component models, Q2Y varied widely. For apoptosis, the best predicting signals were pIκBα and cleaved caspase-8, while pERK1, pERK2, and pHSP27 were stronger predictors for CXCL1 and CXCL8. Importantly, models built without these dominant signals were still nearly as predictive as the full models (Q2Y of 0.577 for apoptosis and 0.587 for chemokines). Early time signals were more predictive for chemokine levels while late time signals were more predictive for apoptosis.
The models based on the full signal data sets were analyzed in more detail to determine how the RAS-variants and treatments are captured in the terms of the model (Fig. 6B, bottom and Supplementary Tables S2-S8). Loadings describe how strongly each signal projects along that individual principal component, while scores describe how strongly each treamtent condition projects (31). For both models, the scores for the treatment conditions indicate that the first principal component describes the impact of TNFα treatment, while the second and third principal components capture the RAS variations. The two models have different scores patterns for these later components, with DLD-1 cells projecting positively in both models' second component, while DKs8-N projects positively in the third component for apoptosis, but the second component for chemokines. This corresponds to the difference in DLD-1 and DKs8-N with respect to apoptosis (Fig. 1B) but not chemokine levels (Fig. 4A,B). Examination of the top 20 variable importance of projection values (Supplementary Table S2) indicated that pIkBα and pERK1 dominate the apoptosis model, while the important signals are more broadly distributed for chemokine production.
High loadings in the first principal components of each model were skewed to pIκBα and pERK1 (a result of the TNFα-TGFα autocrine cascade, Fig. 3A), consistent with the interpretation of the first component as a ‘TNFα treatment’ axis (Supplementary Table S3, S6). The second principal component of the apoptosis model included large loadings for many of the zero timepoint values, consistent with the differences seen in baselines for DLD-1 (Supplementary Table S4, Fig. 6A). The third principal component for the apoptosis model includes strong negative loadings for pAKT measures, which are lower in the DKs8-N cells (Supplementary Table S5). The second and third components of the chemokine model included strong positive and negative loadings for several signaling molecules at various times, potentially highlighting pan-RAS mutant effects (Supplementary Table S7-S8).
Discussion
Our results demonstrate that cells with oncogenic K-RAS, oncogenic N-RAS, or no RAS mutations differentially affect multiple pathways to impact cell fate. These perturbations to the cell network influence the ERK pathway by convoluting positive and negative feedback circuits, as well as additional pathways that together direct cell behavior. Importantly, despite the clear differences in ERK, prediction of the RAS impact on apoptosis and chemokine levels requires incorporation of both ERK and additional pathways which provide a context for the ERK variations.
Similar to previous reports in HT-29 cells (23) and mammary epithelial cells (24), TNFα treatment led to transactivation of the EGFR by TGFα in the RAS-variant cells. TACE has been implicated as the enzyme responsible for cleaving TGFα from the cell surface (25). Interestingly, activation of ERK has been linked to phosphorylation of TACE, which results in trafficking of TACE to the cell surface (32). Despite their constitutively active K-RAS allele, DLD-1 cells exhibit lower levels of basal pERK, which is mitigated by DUSP6 (5). The reduced levels of basal pERK combined with the lower levels of TACE (Supplementary Fig. S5C) likely explain the reduced TGFα release and subsequent lower activation of pERK in DLD-1.
Differences in pERK at later times among the RAS-variants appear to be mediated by a transcriptionally-induced protein, DUSP5. Induction of the negative-feedback DUSP genes was recently shown to be pathway-specific process (21). DUSP5 is a nuclear-localized phosphatase with ERK-specificity (22) that is induced by growth factors and stress (33). In the DKs8-N cells, normalized DUSP5 levels are lower, and induction is delayed, correlating with the extended duration of pERK in these cells with oncogenic N-RAS (Fig. 2C). DUSP6, a cytoplasmic ERK-specific phosphatase, has been previously shown to impact basal pERK in K-RAS mutant cells (5) and is induced during cellular transformation by oncogenic RAS (34). To our knowledge, this is the first report of RAS-dependent differential activation of DUSP5.
The TNFα-induced TGFα autocrine loop was previously shown to initiate a pro-death IL-1 loop in HT-29 colon carcinoma cells (23). In the RAS mutant cell lines, we found no evidence for the IL-1 loop, and instead, our results suggest that TGFα initiated a pro-death loop as well as a pro-survival CXCL1 and/or CXCL8 cascade (Fig. 5C). CXCL1 has been reported to be elevated in colon cancer (35) and associated with greater proliferation and invasiveness in colon carcinoma cells (36), while CXCL8 constitutes a pro-proliferative autocrine loop in HCT-116 (37). Recent reports have begun to highlight unexpected autocrine roles for chemokines, including CXCR2/p53-dependent senescence (38), which were not observed in this panel of p53-mutant cell lines (13). It will be important in future studies to examine the relative importance of the paracrine and autocrine effects of chemokine production on tumor development.
To interpret the broader effects of the RAS variations on the cellular signaling network, and how these changes are integrated into decisions, we utilized a large phosphoproteomic data set and PLSR (Fig. 6). PLSR has been previously used to provide evidence for induced autocrine cascades, demonstrate common effector processing for cell specific responses, and predict production of interleukins (23, 39, 40). In these studies, we demonstrate that the same compendium of signals can predict diverse outcomes (apoptosis and CXCL1/8). The two models have different components (Supplemental Tables S2-S8), indicating that parts of the signaling network are more responsible for one outcome versus another. We also demonstrate that models built without the ‘dominant’ signals are still predictive. This observation is important as it [a] suggests that the data and model allow us to observe how a change in one signal is propagated throughout a network and [b] indicates that even by only collecting information about a few molecules, we can still capture important network behavior. The separate PLSR models show time-dependence, suggesting that the ‘early’ signals result in the production of chemokines for the CXCL1/8 autocrine loop. The ‘late’ signals, which may represent the effects of these chemokine loops, then determine the apoptotic decision.
In conclusion, we have demonstrated multiple differences that result from changes in the form of mutant RAS expressed by cells. While pERK signals in response to TNFα are clearly different through changes to both positive and negative feedback circuits, only with the inclusion of additional pathway context can we predict the differences seen in apoptosis between the RAS variants. Combined, our data suggests that multi-pathway models can interpret the influence of the oncogenic RAS proteins by including both direct effects (pERK) and contextual effects such as how the TGFα-chemokine autocrine cascade impact other signaling pathways.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Michael Yaffe, Dr. Tyler Jacks, and members of the Lauffenburger lab for helpful discussions. We would also like to acknowledge Glenn Paradis of the Koch Institute Flow Cytometry Core Facility for technical assistance. This work was funded by National Institutes of Health Grants U54-CA112967 and P50-GM68762, and by the American Cancer Society (PF-08-026-01-CCG to P.K.K).
Financial Support: This work was funded by National Institutes of Health Grants U54-CA112967 and P50-GM68762, and by the American Cancer Society (PF-08-026-01-CCG to P.K.K).
References
- 1.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–7. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 4.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 5.Haigis KM, Kendall KR, Wang Y, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008 doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klampfer L, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. Oncogenic Ras promotes butyrate-induced apoptosis through inhibition of gelsolin expression. J Biol Chem. 2004;279:36680–8. doi: 10.1074/jbc.M405197200. [DOI] [PubMed] [Google Scholar]
- 7.Wolfman JC, Palmby T, Der CJ, Wolfman A. Cellular N-Ras promotes cell survival by downregulation of Jun N-terminal protein kinase and p38. Mol Cell Biol. 2002;22:1589–606. doi: 10.1128/mcb.22.5.1589-1606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meira LB, Bugni JM, Green SL, et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J Clin Invest. 2008;118:2516–25. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaden JA, Mayberry JF. Colorectal cancer complicating ulcerative colitis: a review. Am J Gastroenterol. 2000;95:2710–9. doi: 10.1111/j.1572-0241.2000.02297.x. [DOI] [PubMed] [Google Scholar]
- 10.Umetani N, Sasaki S, Watanabe T, et al. Genetic alterations in ulcerative colitis-associated neoplasia focusing on APC, K-ras gene and microsatellite instability. Jpn J Cancer Res. 1999;90:1081–7. doi: 10.1111/j.1349-7006.1999.tb00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–98. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 12.Stokkers PC, Hommes DW. New cytokine therapeutics for inflammatory bowel disease. Cytokine. 2004;28:167–73. doi: 10.1016/j.cyto.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–8. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 14.Keller JW, Franklin JL, Graves-Deal R, Friedman DB, Whitwell CW, Coffey RJ. Oncogenic KRAS provides a uniquely powerful and variable oncogenic contribution among RAS family members in the colonic epithelium. J Cell Physiol. 2007;210:740–9. doi: 10.1002/jcp.20898. [DOI] [PubMed] [Google Scholar]
- 15.Keller JW, Haigis KM, Franklin JL, Whitehead RH, Jacks T, Coffey RJ. Oncogenic K-RAS subverts the antiapoptotic role of N-RAS and alters modulation of the N-RAS: gelsolin complex. Oncogene. 2007;26:3051–9. doi: 10.1038/sj.onc.1210103. [DOI] [PubMed] [Google Scholar]
- 16.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–90. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Kumar N, Wolf-Yadlin A, White FM, Lauffenburger DA. Modeling HER2 Effects on Cell Behavior from Mass Spectrometry Phosphotyrosine Data. PLoS Comput Biol. 2007;3:e4. doi: 10.1371/journal.pcbi.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfman JC, Wolfman A. Endogenous c-N-Ras provides a steady-state anti-apoptotic signal. J Biol Chem. 2000;275:19315–23. doi: 10.1074/jbc.M000250200. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 20.Joslin EJ, Opresko LK, Wells A, Wiley HS, Lauffenburger DA. EGF-receptor-mediated mammary epithelial cell migration is driven by sustained ERK signaling from autocrine stimulation. J Cell Sci. 2007;120:3688–99. doi: 10.1242/jcs.010488. [DOI] [PubMed] [Google Scholar]
- 21.Amit I, Citri A, Shay T, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–12. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 22.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–13. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 23.Janes KA, Gaudet S, Albeck JG, Nielsen UB, Lauffenburger DA, Sorger PK. The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell. 2006;124:1225–39. doi: 10.1016/j.cell.2006.01.041. [DOI] [PubMed] [Google Scholar]
- 24.Rodland KD, Bollinger N, Ippolito D, et al. Multiple mechanisms are responsible for transactivation of the epidermal growth factor receptor in mammary epithelial cells. J Biol Chem. 2008;283:31477–87. doi: 10.1074/jbc.M800456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merchant NB, Voskresensky I, Rogers CM, et al. TACE/ADAM-17: a component of the epidermal growth factor receptor axis and a promising therapeutic target in colorectal cancer. Clin Cancer Res. 2008;14:1182–91. doi: 10.1158/1078-0432.CCR-07-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosgrove BD, Cheng C, Pritchard JR, Stolz DB, Lauffenburger DA, Griffith LG. An inducible autocrine cascade regulates rat hepatocyte proliferation and apoptosis responses to tumor necrosis factor-alpha. Hepatology. 2008;48:276–88. doi: 10.1002/hep.22335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertini R, Allegretti M, Bizzarri C, et al. Noncompetitive allosteric inhibitors of the inflammatory chemokine receptors CXCR1 and CXCR2: prevention of reperfusion injury. Proc Natl Acad Sci U S A. 2004;101:11791–6. doi: 10.1073/pnas.0402090101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geladi P, Kowalski BR. Partial least-squares regression: a tutorial. Anal Chim Acta. 1986;185:1–17. [Google Scholar]
- 29.Janes KA, Kelly JR, Gaudet S, Albeck JG, Sorger PK, Lauffenburger DA. Cue-signal-response analysis of TNF-induced apoptosis by partial least squares regression of dynamic multivariate data. J Comput Biol. 2004;11:544–61. doi: 10.1089/cmb.2004.11.544. [DOI] [PubMed] [Google Scholar]
- 30.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–53. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 31.Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–8. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 32.Soond SM, Everson B, Riches DW, Murphy G. ERK-mediated phosphorylation of Thr735 in TNFalpha-converting enzyme and its potential role in TACE protein trafficking. J Cell Sci. 2005;118:2371–80. doi: 10.1242/jcs.02357. [DOI] [PubMed] [Google Scholar]
- 33.Ishibashi T, Bottaro DP, Michieli P, Kelley CA, Aaronson SA. A novel dual specificity phosphatase induced by serum stimulation and heat shock. J Biol Chem. 1994;269:29897–902. [PubMed] [Google Scholar]
- 34.Bluthgen N, Legewie S, Kielbasa SM, et al. A systems biological approach suggests that transcriptional feedback regulation by dual-specificity phosphatase 6 shapes extracellular signal-related kinase activity in RAS-transformed fibroblasts. FEBS J. 2009;276:1024–35. doi: 10.1111/j.1742-4658.2008.06846.x. [DOI] [PubMed] [Google Scholar]
- 35.Wen Y, Giardina SF, Hamming D, et al. GROalpha is highly expressed in adenocarcinoma of the colon and down-regulates fibulin-1. Clin Cancer Res. 2006;12:5951–9. doi: 10.1158/1078-0432.CCR-06-0736. [DOI] [PubMed] [Google Scholar]
- 36.Li A, Varney ML, Singh RK. Constitutive expression of growth regulated oncogene (gro) in human colon carcinoma cells with different metastatic potential and its role in regulating their metastatic phenotype. Clin Exp Metastasis. 2004;21:571–9. doi: 10.1007/s10585-004-5458-3. [DOI] [PubMed] [Google Scholar]
- 37.Brew R, Erikson JS, West DC, Kinsella AR, Slavin J, Christmas SE. Interleukin-8 as an autocrine growth factor for human colon carcinoma cells in vitro. Cytokine. 2000;12:78–85. doi: 10.1006/cyto.1999.0518. [DOI] [PubMed] [Google Scholar]
- 38.Acosta JC, O'Loghlen A, Banito A, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–18. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 39.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–8. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 40.Kemp ML, Wille L, Lewis CL, Nicholson LB, Lauffenburger DA. Quantitative network signal combinations downstream of TCR activation can predict IL-2 production response. J Immunol. 2007;178:4984–92. doi: 10.4049/jimmunol.178.8.4984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.