Abstract
The k-space readout of propeller-type sequences may be accelerated by the use of parallel imaging (PI). For PROPELLER, the main benefits are reduced blurring due to T2 decay and SAR reduction, while for EPI-based propeller acquisitions such as Turbo-PROP and SAP-EPI, the faster k-space traversal alleviates geometric distortions. In this work, the feasibility of calculating a 2D GRAPPA kernel on only the undersampled propeller blades themselves is explored, using the matching orthogonal undersampled blade. It is shown that the GRAPPA kernel varies slowly across blades, therefore an angularly continuous 2D GRAPPA kernel is proposed, in which the angular variation of the weights is parameterized. This new angularly continuous kernel formulation greatly increases the numerical stability of the GRAPPA weight estimation, allowing the generation of fully sampled diagnostic quality images using only the undersampled propeller data.
Keywords: parallel imaging, GRAPPA, self-calibration, propeller, SAP-EPI
Introduction
With a parallel imaging (PI) reconstruction technique such as GRAPPA (1), k-space lines can be synthesized from neighboring acquired lines using the fact that the spatially localized coil sensitivities make the data locally dependent on each other in k-space, allowing for a reduction in the amount of acquired data. Besides a general reduction in acquisition time, for EPI-based propeller sequences like SAP-EPI(2), GRAPPA further reduces both TE and geometric distortions, the latter in proportion to the reduction factor. For example, at the 288×288 target resolution with 64×288 SAP-EPI blades and R=2, the geometric distortions become about five times lower compared to a conventional ss-EPI (3). For FSE-based propeller trajectories on the other hand, parallel imaging allows for a direct reduction in SAR, scan time, less T2 related blurring, and better phase navigation - the latter due to wider blades.
For regularly undersampled Cartesian acquisitions, the distances as well as the orientation between the missing datum and its local neighborhood of acquired data (defined by the 2D GRAPPA kernel (4,5)) are constant throughout k-space. Therefore, a single GRAPPA kernel (or ‘set of weights’) can be used to fill in the missing data in the entire k-space. For propeller trajectories (Fig. 1a), the above applies to a single blade since the blade itself is Cartesian. However, the coil geometry as seen from each blade space varies with blade angle. Therefore, GRAPPA weights will also vary across blades. Traditionally, GRAPPA acquisitions acquire a fully sampled strip, which is typically located around the center of k-space while the rest of k-space is R times undersampled along the phase encoding direction. This is not particularly suitable for propeller trajectories for the following reasons: For conventional propeller trajectories whereby the readout direction is oriented along the long-axis of the blade (PROPELLER(6), Turbo-PROP(7), propeller-EPI(8), Fig. 1a, left panel), the limited number of phase encoding lines (typically 16-32 lines) would make the accelerated portion of each blade very limited, significantly reducing the net acceleration factor. Moreover, for any EPI based sequence (including long-axis (“LAP”) (8) and short-axis (SAP) (2) readout propeller-EPI), a dual density k-space sampling scheme introduces another challenge originating from the different sensitivities to off-resonance distortions for the fully sampled and accelerated portions of k-space (9).
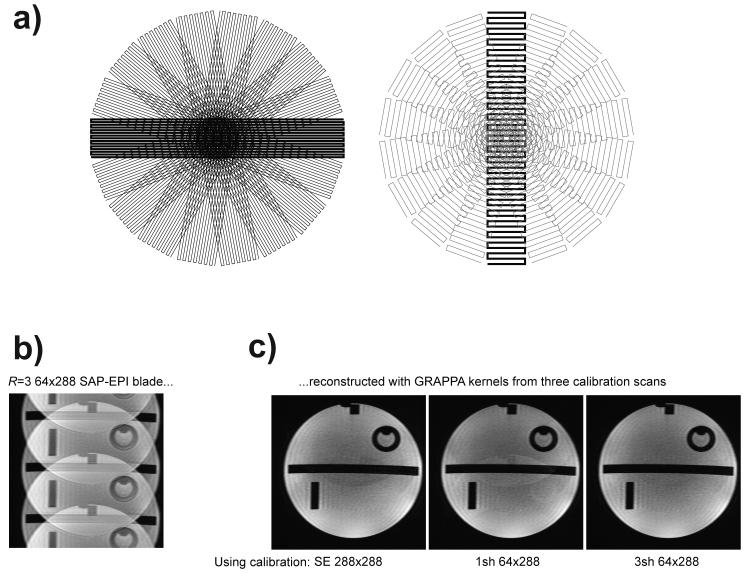
Figure 1.
a) Principle sketch of standard PROPELLER (left) and SAP-EPI (right) k-space trajectories. b) An image of an R=3 times undersampled SAP-EPI propeller blade on a phantom used as data for GRAPPA reconstructions using GRAPPA kernels calculated from three different calibration datasets; Conventional SE, single-shot EPI and R-shot EPI. Resulting images are shown in c).
Given the above, the use of external R=3 GRAPPA calibration scans and acquiring each blade at full R-fold acceleration would be desirable in a propeller acquisition. In the context of propeller trajectories, GRAPPA has been reported with PROPELLER (10), LAP-EPI (11) and SAP-EPI (3). For an R=3 accelerated SAP-EPI blade like in Fig. 1b, the best image quality is obtained if the calibration scan has the same off-resonance level as the accelerated scan. This means that three interleaves per blade should ideally be used for GRAPPA calibration as shown in Fig. 1c, right. For comparison, using GRAPPA kernels derived from either a high SNR SE calibration scan (free of geometric distortions) or a three times higher distorted single-shot EPI calibration scan leads to more residual aliasing (Fig. 1c, left, middle panels). The downside of an R-shot scan on the other hand — if needed for an N-blade propeller scan — is the lengthy calibration time, which becomes R times longer than the GRAPPA accelerated scan itself. Furthermore, any motion that occurs between the shots in the calibration scan will cause ghosting in the calibration data, which will propagate into subsequent GRAPPA reconstructions.
In the interest of reduced scan time and a more efficient use of propeller imaging, we propose in this paper to eliminate the acquisition of GRAPPA calibration data altogether and retrieve the missing calibration information from undersampled blades themselves (self-calibration). While this paper presents results for SAP-EPI trajectories (Fig. 1a, right), the same sparsity pattern and arguments hold for all propeller trajectories. Potential sequence specific artifact issues aside, the bulk of the results in this paper are therefore also applicable to PROPELLER, Turbo-PROP, and long-axis propeller (“LAP”) EPI acquisitions.
The basic element in the new GRAPPA estimation procedure proposed in this work is a pair of two orthogonal, R-times undersampled blades (Fig. 2a). Since the blades are undersampled, the 2D GRAPPA kernel of one of the blades needs to be estimated by using target points from the other blade. We will show that this is feasible and can result in robust GRAPPA reconstructions, provided that enough training data locations exist for the weight-estimation and that the blades are properly aligned with each other in k-space. However, with thinner blades and/or increased reduction factors, this method can become numerically unstable, producing unacceptable reconstructions. We hypothesize that GRAPPA weights may vary smoothly with blade angle, potentially allowing for a reduction in the total number of independent GRAPPA kernel weights to be estimated for all blades. A detailed account for how to reduce the number of unknowns in relation to the available training data will be presented.
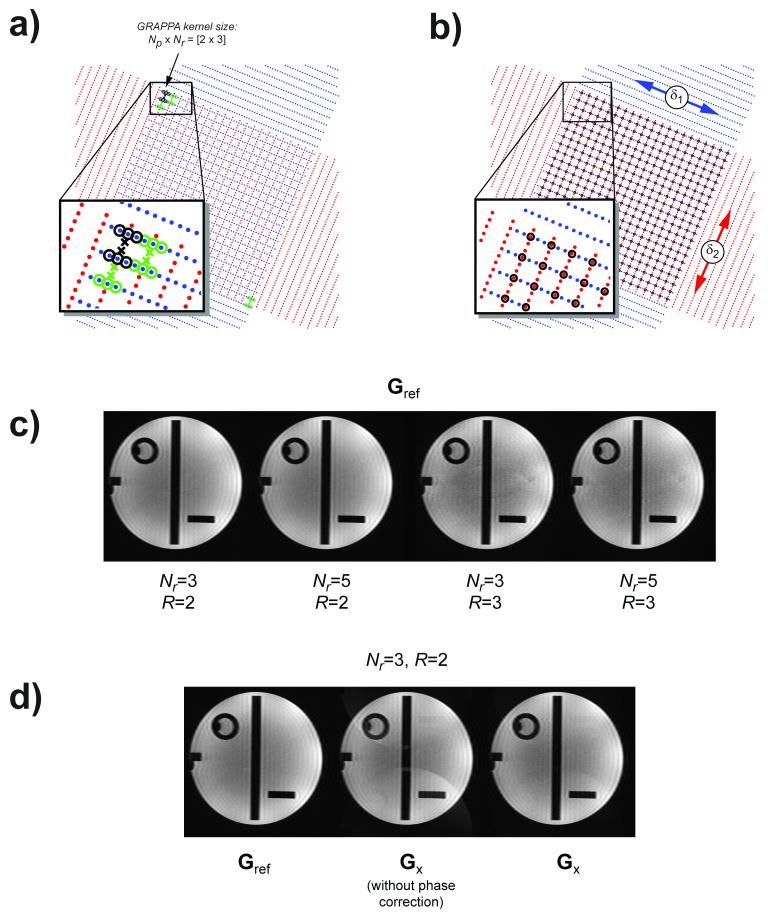
Figure 2.
a) Illustration of how GRAPPA estimation can be done with two R=3 times undersampled orthogonal blades, using the source points (o) from one blade and the target points (×) from the other (black markers in inset). Green markers in insets denote the neighboring training locations, indicating that there is R2 less training locations compared to the case for a fully sampled dataset. b) Black circles denote common k-space locations for the two blades, which should ideally contain the same data. Relative blade alignment is performed by Fourier shifting blades by Ω1 and Ω2 in its fully sampled direction until the overlapping locations (black circles) have minimal difference. c) GRAPPA reconstructions using a 2D kernel with Nr=3 and 5 for R=2 and 3, to show that the smaller kernel width can be used with no or only minor loss in image quality depending on the acceleration factor. d) GRAPPA reconstructions using the fully sampled data (left) and two orthogonal undersampled blades, without (middle) and with (right) the use of the relative blade alignment method in b).
Theory
GRAPPA estimation on two orthogonal undersampled blades
In Fig. 2a, a small GRAPPA kernel of size Nr×Np = 2×3 is shown, with the source lines (o) taken from one blade and the target lines (×), taken from the other. As seen in Fig. 2a (blue markers), the number of k-space locations that can be used to make the linear system involving Np×Nr×Ncoils unknown GRAPPA weights overdetermined is much smaller than if a fully sampled blade would have been used. This is due to two reasons. Firstly, only the square area defined by the overlapping region of the two blades may be used for the calibration. Secondly, the GRAPPA kernel needs to “jump” on the intersections arising from the crossing of the acquired lines, leaving only 1/R2 (e.g. 1/9th for 3-fold acceleration) of the locations compared to a fully sampled blade. Therefore, it is helpful to keep the size of the GRAPPA kernel small in order to reduce the number of unknowns. We will refer to a GRAPPA kernel estimated from fully sampled blades as Gref (golden standard, see Fig. 2c) and our proposed GRAPPA estimation on undersampled data as Gx (where ‘x’ symbolizes the intersection of two blades).
Blade alignment
In addition to the issue of fewer training locations, the k-space peaks of the two orthogonal blades may be misaligned due to phase shifts in the image domain, causing inconsistencies between the target lines and the surrounding source lines included in the GRAPPA kernel estimation. The standard procedure for echo-centering the blades in k-space for propeller acquisitions is to use an image domain phase correction technique (6). Alternatively, one can remove the image phase completely for each blade prior to Fourier transformation back to k-space before gridding the blades together to get the final image (2). However, both approaches only work on fully sampled data, which is why we propose another solution.
Our approach is outlined in Fig. 2b, with the common nominal k-space points for the two blades marked with black circles. If the two blades are aligned, the difference between the values from the two blades at each of these locations should be minimal. To achieve this, each blade may be shifted along the fully sampled readout dimension of each blade by an amount Ω1 and Ω 2, respectively, without violating the Nyquist sampling criterion. We may drive this linear registration of the two blades (B1 and B2) by minimizing the sum of the absolute differences of the log-intensities of the blades over all intersecting points according to:
| [1] |
The log-intensity is essential here to make all intersecting k-space points contribute significantly to the cost function O(Ω1,Ω2). Furthermore, to find the parameter vector Ω = [Ω1,Ω2], it was found necessary to temporarily take the magnitude of the k-space prior to the minimization procedure. For this low-dimensionality minimization problem the simplex method (Nelder-Mead) algorithm was used (12).
The angularly continuous GRAPPA kernel
The number of training locations for the GRAPPA kernel (Fig. 2a) will decrease by the square of both the reduction factor R and the blade width, which leads to a greatly increased uncertainty in the weight estimation due to the lack of training data. At some point, the number of training locations would be less than the number of weights in the kernel, leading to an underdetermined GRAPPA estimation (inversion) problem. Furthermore, when thinner blades are used, more blades are needed to fill k-space, yielding to smaller blade angle differences and, thus, to smaller changes in apparent coil geometry between blades. Using the same GRAPPA kernel for a certain range of azimuthal angles (‘wedges’) in k-space for GRAPPA estimation has been done before for both spiral and radial data, with reasonable results (13,14). Therefore, we hypothesize that it is possible to construct an angularly continuous GRAPPA kernel, Gxc, (index ‘c’ for continuous), where each of the Nr×Np×Ncoils complex GRAPPA weights are parameterized as a smooth complex function over the entire blade sweep. The smooth functions are modeled by some basis set, defined in a matrix C, consisting of Norder columns and Nb rows. Each of the Norder columns of C represents one term in the basis set and the GRAPPA estimation problem is reformulated such that the optimal coefficients for these terms (‘hyper-weights’) are estimated rather than the GRAPPA weights directly. Provided that the number of terms in the basis set, Norder, can be kept smaller than the number of blades involved without increasing the GRAPPA model fit error, this would make the entire problem more overdetermined, potentially leading to a more numerically stable solution.
The relation between the source lines (shuffled into A), the “missing” (target) lines (shuffled into Y) and the GRAPPA kernel, G, which is a matrix of size [Np×Nr×Ncoils] ×[Ncoils×(R-1)] is given by
| [2] |
where the multiple columns of Y and G come from the standard GRAPPA estimation, whereby independent weights are used for each target coil and each of the R-1 target lines. To simplify the following derivation, we can formulate a continuous kernel for a particular target line i, and target coil j. For blade 1, we get
| [3] |
where “†” denotes the Moore-Penrose inverse of A1, and where x1 (of size [Np×Nr×Ncoils]×1) contains the GRAPPA weights for target line i and target coil j. Also note that A1 only needs to be inverted once for all Ncoils×(R-1) GRAPPA weight sets forming the complete GRAPPA kernel. Now, still assuming independent GRAPPA weights across blades, we can extend Eq. [3] to include all Nb blades (again for only one target line i and target coil j) by the creation of a block diagonal matrix, Abig:
| [4] |
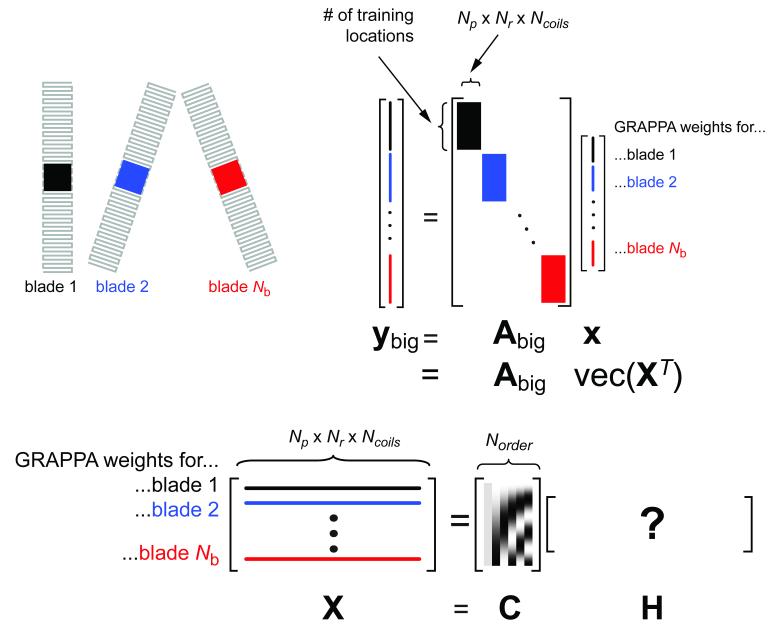
where X is a matrix of size [Np×Nr×Ncoils]×Nb and the “vec” operator unravels the matrix X into a single column, [x1,..., xNb]T. Equation [4] does not provide any computational benefits itself, but to reduce the total number of unknowns in X and to make the GRAPPA kernel a continuous function over the blade angles, X may be constrained by a basis set C of size Nb×Norder across the columns of X, that is X = (CH)T (for a more graphical and intuitive description, please refer to Fig. 3). In this work we have arbitrarily used an orthogonal cosine basis set discretely defined as (15)
| [5] |
where the first column is a constant value modeling the average value of X per column (Fig. 3). Given the basis set C, the task is no longer to estimate the GRAPPA weights in X directly, but rather the elements or ‘hyper-weights’ in H. Using the following algebraic rule: vec(XY) = vec(X), the elements of H can be calculated from Abig and ybig directly via
| [6] |
where “⊗” denotes the Kronecker product (16). For the entire GRAPPA reconstruction, we have (R-1) ×Ncoils different ybig column vectors, which can be stacked together into another matrix, Ybig. The final expression for the continuous GRAPPA kernel, now for all target lines and target coils, becomes:
| [7] |
Figure 3.
Pictorial explanation of how the undersampled blade data is formed for the angularly continuous kernel estimation in Eqs. [4]-[6]. Only three out of the Nb blades are shown for clarity. Note that Abig is a sparse block diagonal matrix and that all GRAPPA weights for all blades are contained in the vector x (“what we want to know”). By reshaping x into a matrix X with rows being different blades, we can fit a model or basis set C to this matrix with coefficients contained in H. The product CH forms the GRAPPA weights, constrained to vary slowly across blades (rows).
Note that Gxc,hyper is shown here in its compact form, where elements in the matrix are the hyper-weights. The ‘unpacked’ GRAPPA weights themselves are easily obtained by multiplication with Q
| [8] |
In summary, all the source and target points (for a given slice location) from all blades are put into Abig and Ybig, respectively, and only Eq. [8] is needed to estimate the GRAPPA weights for all blades, target lines and target coils simultaneously. Thus the number of unknowns to be estimated has been reduced by a factor of Nblades / Norder.
Materials & Methods
Phantom and human brain data were acquired on a healthy volunteer using a SAP-EPI sequence on a 1.5T GE Excite whole body unit (GE Healthcare, Waukesha, WI). All study protocols were conducted within the guidelines from the Institutional Review Board and consent was obtained from the participant. RF excitation was applied with the body coil, and signal reception was performed with an eight-channel head array (Invivo Corporation, Pewaukee, WI). Parameter settings for the phantom experiment included four 5 mm thick axial slices with TE/TR = 17/2000 ms, FOV = 22×22 cm. Sixteen propeller blades of size 64×288 (freq.×phase) were acquired, yielding a target resolution of 288×288. For the phantom data, each blade was acquired with 36 interleaves (with 8 phase encoding lines per excitation) to increase the phase encoding bandwidth along the phase encoding dimension enough to avoid interaction between the GRAPPA kernel estimation and off-resonance effects. From this acquisition, three datasets were created: i) with blade widths of 64 (using every second blade, only just fulfilling Nyquist at the k-space perimeter), ii) 32 and iii) 24 (using all 16 blades). From the fully sampled SAP-EPI datasets, GRAPPA weights were calculated independently on each fully sampled blade to serve as the gold standard (Gref). Then, for all three datasets, only every Rth (= 2 & 3) phase encoding line from each blade was kept from which Gx and Gxc kernels were calculated using the methods described above. Using all sixteen 64×288 blades, the richness of the cosine basis set, Norder, was varied from 1 to 12 to investigate how rapidly the weight-functions need to vary over blade angles to not introduce increased model fit errors.
A healthy volunteer was scanned twice, with the head positioned in slightly different orientations between the two scans. The same acquisition parameters as the phantom experiment were used, except with TE/TR = 72/4000 ms and with 2 shots (instead of 36 used for the phantom data). With the 2-shot acquisition, the geometric distortions and tissue contrast became equivalent to a real R=2 scan. For the Gref method, both shots were used for GRAPPA estimation, whereas for the Gx and Gxc kernels proposed in this work, only the first shot was used for the GRAPPA estimation. The two data sets were also mixed in two ways to effectively generate two types of motion; a) motion between different blade pairs; and b) motion between shots and blade pairs. After mixing, both Gref and Gxc were calculated again. In-plane rigid body motion correction between the blades was performed in the image domain for the volunteer data by minimizing the sum-of-squares differences between the blades using a quasi-Newton search algorithm (17).
After GRAPPA reconstruction and motion correction, each blade was corrected using a triangular windowing filter (6) in order to echo-center the data in k-space prior to gridding. All blades were gridded together on a 2x grid, using a Kaiser-Bessel kernel (width=4, β=9), followed by apodization correction. A GRAPPA kernel size of 2×3 was used throughout, except for data presented in Fig. 2 which also show results from a 2×5 size kernel. The benefits of using two instead of four source lines in the phase encoding direction have been demonstrated earlier (18), thus GRAPPA kernels with more than two source lines have not been investigated.
Results
GRAPPA reconstructions (Gref) of one blade from the 36-shot phantom scan is shown in Fig. 2c for two different kernel widths and for acceleration factors R=2 and 3. This figure suggests that the image reconstruction is not hampered per se by the use of a small 2×3 over a 2×5 GRAPPA kernel — neither in terms of residual aliasing nor noise, defending the use of smaller kernel widths in this work.
Figure 2d shows the same blade reconstructed with GRAPPA weights obtained from the fully sampled data, Gref (left panel), and the Gx method (middle & right panels). The middle and right panels of Fig. 2d show the reconstructed blade without and with the k-space echo centering approach proposed in this work (see Fig. 2b), respectively. Even on this phantom data, free from geometric distortions and motion, aliasing can be observed in the reconstructed blade, due to minor misalignments between the blades. Using the echo centering leads to a better reconstruction (Fig. 2d, right).
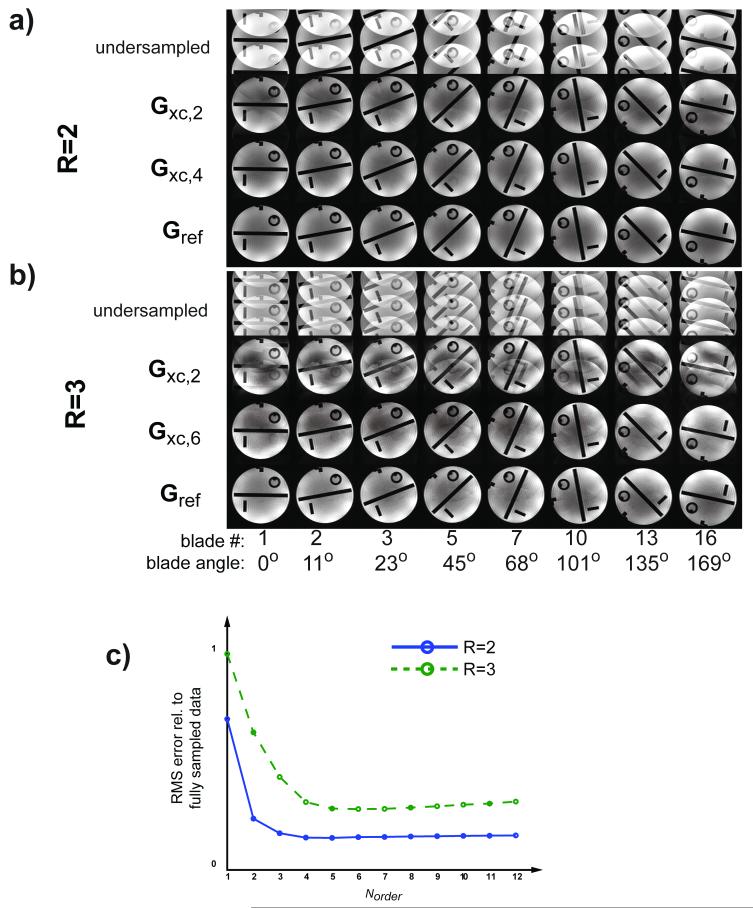
The top row of Figure 4a shows the reconstruction of the R=2 undersampled blades without the use of GRAPPA. The second row of Fig. 4a show the same blades reconstructed with the proposed Gxc method using a very limited basis set of Norder=2 (i.e. C has two columns, see Fig. 3), which largely unalias the object but leaves some residual ghosting in the image. An acceptable image quality can be obtained when using a Gxc,4 (Norder=4) kernel for R=2 (third row), and is quite comparable to the Gref kernel (bottom row) based on fully sampled 2-shot data. Corresponding data for R=3 is given in Fig. 4b, but where the third row show reconstructions using a and Gxc,6 (Norder=6) kernel. Norder= 4 and 6 were the lowest number of terms (i.e.. Norder) deemed acceptable by visual inspection for R=2 and R=3, respectively. This trend is also supported in Fig. 4c, showing the root-mean-square (RMS) error summed over all blade images vs. Norder. Fig. 4c demonstrates the model fit error of the angularly continuous Gxc kernel using a different number of terms, Norder, in the basis set C (for the remainder of the paper, the order of the basis set will be indicated by Gxc,Norder). Certainly, the threshold for Norder can not be generalized and depends on a multitude of parameters, such as the number, size, and location of coil elements, blade angles, reduction factor, et cetera, but from Fig. 4c it becomes evident that Gxc kernels with a modest Norder achieve a sufficiently good reconstruction.
Figure 4.
Eight out of 16 blades reconstructed with a) R=2 and b) R=3. Top row: Undersampled blades reconstructed without GRAPPA. Second row in a) and b) shows Gxc reconstructions using a very limited basis set with Norder=2. Residual aliasing (esp. for R=3) indicates that two basis functions are not enough. Third row in a) Norder=4 (R=2) and b) Norder =6 (R=3) show improved reconstructions thanks to increased basis set of the continuous kernel. c) Sum of the RMS errors over all blades w.r.t. fully sampled blades (no GRAPPA) vs. the richness of the cosine basis set. The RMS values suggest that the Norder values above are indeed optimal for this data and coil configuration.
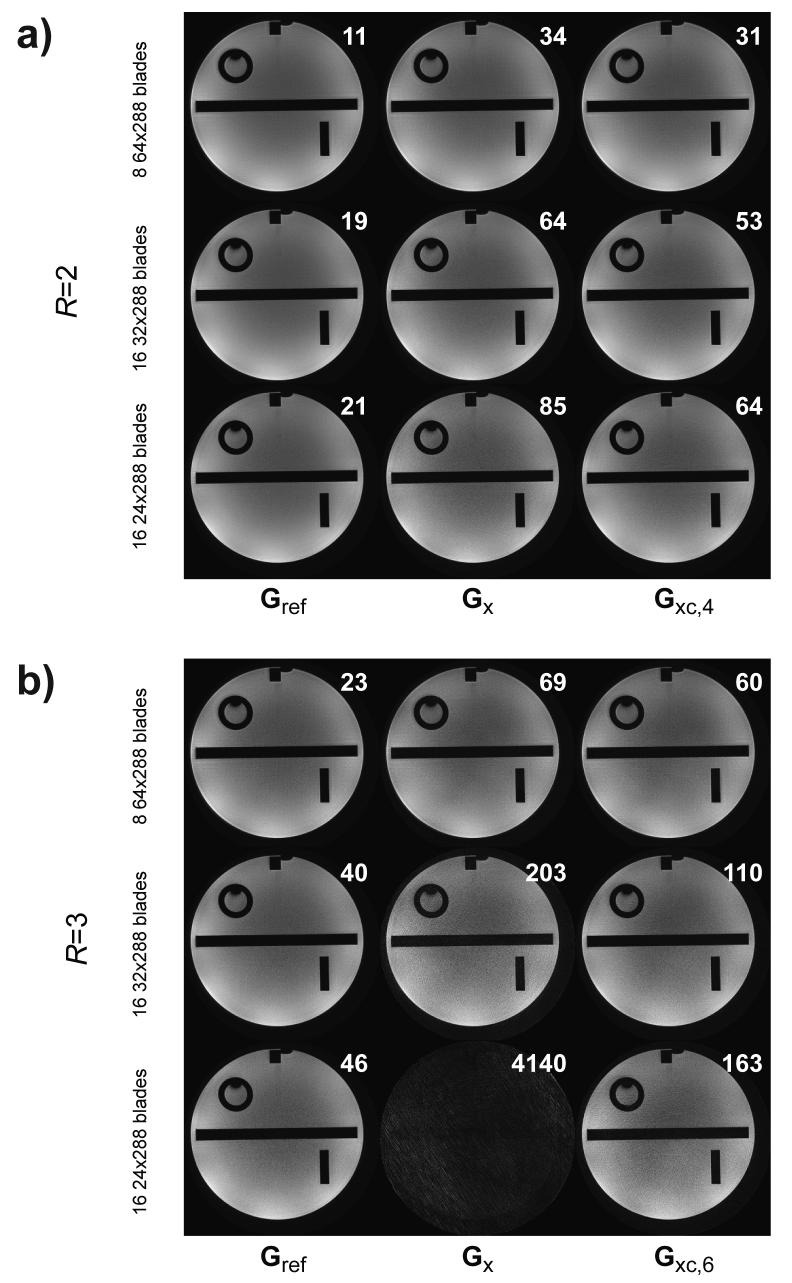
Gridded images from blades that have been reconstructed using GRAPPA variants Gref, Gx, and Gxc are depicted in Figs. 5a (R=2) and 5b (R=3). With wide blades for R=2 (Fig. 5a, top row), the amount of training locations for the two orthogonal blades are enough to make the weight estimation problem well overdetermined, yielding similar apparent image quality for all three methods; although the RMS difference between the GRAPPA reconstructed data and the fully sampled data is three times lower for Gref. The corresponding R=3 reconstruction (Fig. 5b, top tow) shows a minor, but measurable, noise enhancement for Gx compared to Gxc (right) and Gref (left). However, as the blade width is reduced, the number of training locations for the Gx kernel is reduced - leading to an increased noise enhancement and eventually to a complete reconstruction failure as seen for the example with a blade width of 24 and R=3 (Fig. 5b, bottom row). In this case, the number of training locations became exactly equal to the number of parameters in the GRAPPA kernel. Even for a blade width of 32, the increased GRAPPA noise for Gx is evident for a higher acceleration factor R=3 (Fig. 5b). On the contrary, for the angularly continuous GRAPPA kernel, Gxc, the increased number of blades that are required for narrow blades helps to avoid this problem. Compared to the reference method, the Gxc method shows acceptable image reconstructions in all cases, with no residual aliasing and acceptable image noise penalty.
Figure 5.
Gridded phantom data with blade widths 24, 32 and 64 undersampled by R=2 (a) and R=3 (b). The three columns refer to blade reconstructions using Gref, Gx and Gxc,4 (or Gxc,6 for R=3). The white number denotes the sum of RMS error for all blades involved prior to gridding.
Figure 6 shows four gridded slices from the 2-shot SAP-EPI volunteer scan. Being a 2-shot scan, one has now the same T2 and off-resonance effects as a real R=2 scan. Hence, this figure also encompasses potential effects of distortions, blurring and physiological noise into the GRAPPA kernel estimations. Fig. 6a shows the first shot reconstructed with Gref, calculated from both shots, whereas the Fig. 6b shows the same data unaliased with Gxc,4 solely using information from the first interleave. Despite the moderate off-resonance effects present in this acquisition, the Gxc,4 reconstruction shows no streaks and has - compared to the phantom case earlier - a much lower RMS error elevation relative to the image unaliased with Gref.
Figure 6.
Four axial slices of the volunteer scan using 8 64×288 gridded blades undersampled by R=2 and reconstructed with Gref and Gxc,4. The RMS errors given in the sub-panels show very little noise amplification in vivo between the proposed Gxc,4 and the gold standard (Gref).
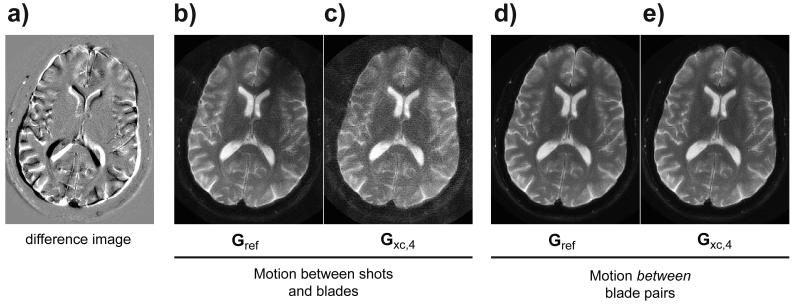
The final figure shows how the reference GRAPPA method as well as the continuous GRAPPA kernel responds to different types of motion. Fig. 7a depicts the difference image obtained by mixing the two fully-sampled datasets acquired with head orientated slightly different between these two acquisitions. These datasets were mixed in such a way that they simulated either motion between shots and blades, or motion between blade pairs. The former is particularly relevant if a full R-shot calibration is performed for each blade (i.e. Gref). As shown in Fig. 7b-c, intra-blade pair motion leads to poor image quality for the continuous kernel, however this is also true for the reference method due to motion also between shots for each blade (shading in frontal part of the brain). However, when motion only occurs between blade pairs, the reconstruction using both methods renders an artifact-free image after motion correction (Fig. 7d,e).
Figure 7.
Controlled motion experiment. In (a), the extent of motion is shown by the difference image between the two acquisitions. With datasets mixed such that motion occurs between shots and intra-blade-pairs, both Gref (b) and Gxc,4 (c) reconstructions lead to inferior weights, leading to shading and streaks despite that the motion correction itself is able to align the blades. On the other hand, if motion occurs between only inter-blade pairs, both methods reconstruct blades accurately and can be realigned prior to gridding like in d-e).
Comparisons of computational speed for the most relevant steps in the reconstruction are shown in Table 1 for the three GRAPPA kernels, Gref, Gx and Gxc,4. As seen in this table, there is practically no speed penalty associated with the newly proposed technique, and the computational bottle-neck is the gridding (despite this part being compiled C-code). This can be considered a significant advantage when comparing this technique to potential alternatives such as conjugate gradient reconstruction or methods that would grid the overlapping area of all blades to generate a fully sampled region of k-space for GRAPPA weight determination.
Table 1.
| values in seconds | Gref | Gx | Gxc,4 |
|---|---|---|---|
| Finding overlap between two undersampled blades | - | 0.1 | 0.1 |
| Peak alignment of blades | - | 2.6 | 2.6 |
| GRAPPA estimation (all blades) | 4.5 | 0.2 | 0.7 |
| GRAPPA data synthesization (all blades) | 0.6 | 0.6 | 0.6 |
| Blade phase correction | 1.5 | 1.5 | 1.5 |
| Gridding (coil-by-coil) | 15.6 | 15.6 | 15.6 |
| Total | 22.2 | 20.6 | 21.1 |
Values are given in seconds per image slice calculated in Matlab2007a on a 64-bit Linux Xeon 3.2GHz server with R=2, 64×288 blade size and a kernel size of 2×3
Discussion & Conclusion
Under ideal circumstances with no scan time restrictions and no motion during the calibration scan, it is clear from Fig. 1b and previous work on Cartesian EPI data (9) that a distortion matched calibration scan produces the best GRAPPA kernel. For SAP-EPI, this would mean an R-shot calibration and for PROPELLER this would correspond to fully sampled blades. For both sequence types, the calibration will take R times longer than the actual scan, which can only be defended for time series data where the calibration phase is either just a small fraction of the entire scan or is made part of the time series.
Hence, for propeller trajectories there is a great need to reduce the calibration time, and in this work we have shown that it is possible to eliminate the need to acquire extra calibration data altogether. Although the amount of training data per blade becomes substantially reduced without additional calibration information, it has been shown here that the use of small kernels with the angularly continuous GRAPPA representation can produce acceptable reconstructions. This is particularly important for the narrower blades often used in the RF-refocused PROPELLER sequence. We have shown that even a 2×3 GRAPPA kernel produce images of comparable image quality compared to the more common 2×5 sized kernel, and that even a very limited basis set with 4-6 terms is enough to model angular the angularly continuous GRAPPA kernel. Increasing the number of terms does not reduce the GRAPPA model fit error to the data used in this work (Fig. 4c). While the cosine basis set is a general purpose orthogonal set, it is subject to future work to prove that this basis set is the most optimal choice for all coil geometries or even the coil geometry used in this paper. The appearance of GRAPPA reconstruction artifacts for a blade is similar to that of conventional Cartesian imaging (Fig. 4), while in the gridded image (Fig. 5) these artifacts primarily manifest as increased noise.
Using this technique for time series data, such as fMRI, perfusion MRI or multiple ‘b=0’ images for DTI, would be quite beneficial as the amount of training locations goes up linearly with the number of time frames, which will further suppress noise in the Gxc reconstruction. While inter-volume motion can occur within a time series, the use of the continuous GRAPPA kernel is expected to be robust since the training locations are built up from pairs of orthogonal blades consecutively over time.
As the GRAPPA estimation is performed on two orthogonal blade pairs it is both important that their k-space peaks are overlapping and that the blades indeed are orthogonal. An even number of blades must therefore be used, with blade angles chosen such that every blade has an orthogonal counter part. The conventional image phase correction that is performed prior to gridding in propeller imaging addresses the k-space echo centering well for fully sampled blades, but we have found this does not work for undersampled data prior to GRAPPA estimation. Therefore, we have here presented (Fig. 2b) a new method that is able to echo center two undersampled orthogonal blades relative to each other without violating the Nyquist criterion, by moving each blade only along its fully sampled direction until their difference is minimized (Eq. [1]). However, this procedure only performs linear shifts in k-space. In the event of large non-linear image phase differences between the two blade pairs, inconsistencies between the blade pairs could occur, leading to suboptimal GRAPPA weights.
Another issue is rigid body motion. As long as no motion occurs for the data used for GRAPPA calibration, standard rigid body in-plane motion correction of the GRAPPA reconstructed propeller blades is trivial. In contrast to Cartesian imaging, robustness to motion is one of the well known strengths of propeller-type acquisitions, since densely overlapped blades (although slightly rotated after motion correction) do not introduce ghosting in the final image. However, we have also shown that motion between shots for Gref as well as intra-blade-pair motion for Gxc leads to inferior GRAPPA reconstruction. Nevertheless, for the latter method proposed here, there are two good solutions that we intend to implement to overcome this: Firstly, we may use a dual-blade SAP-EPI acquisition (19) where two orthogonal blades are read out following a single excitation. In this case, the intra-blade pair motion is eliminated and hence the estimation of Gxc will be independent of the inter-blade pair motion unless it is extremely large. Secondly, the angularly continuous GRAPPA representation also has another advantage in the context of motion. By observing the residual error associated with the GRAPPA kernel estimation, it is possible to identify the blade pairs which have been affected by motion, since — from our experience from Cartesian GRAPPA — motion and residual error in GRAPPA are very strongly correlated (unpublished data). After identification of motion hampered blade pairs, the hyper-weights of the Gxc kernel can be re-estimated without these blades by using a modified Q matrix in Eq. [7] obtained from a C matrix with the associate rows taken out. Then, the ‘unpacked’ GRAPPA weights for all (including the omitted) blades are obtained by using the nominal Q matrix.
An alternative approach to the one presented here is to use iterative Conjugate Gradient reconstruction methods that have recently been very popular for many non-Cartesian scenarios, not least in the context of parallel imaging and motion (20-22). The advantage with iterative reconstructions is that it is possible to drive the reconstruction to quite an artifact free image - provided that the model incorporates the sources of artifacts well and that the proper amount of regularization is used. Most likely, this can lead to lower noise and less residual aliasing than presented here using the same data. This has recently in part been shown as an attractive solution using a pre-calculated GRAPPA kernel and data consistency constraints (23). Yet, it comes with the price of lengthy reconstruction times (22), in some cases hours - which ought to be seen in context with the relatively short propeller-acquisition times of a few minutes or less. Substantial reduction of this computation time for time series data has recently been presented for SENSE reconstructions using the k-SPA method (21).
In an early stage of this work, we also tried to use ideas from (24) to generate training data for each blade using samples from all other blades, solely for the GRAPPA estimation step. In our experience, this did not improve the GRAPPA kernel for any dataset we have tested so far - neither in terms of noise or aliasing properties, despite the increased number of training locations generated after the gridding process.
Finally, with ideas from Ref. (25), which used a discrete set of GRAPPA weights for different line distances in 1D non-Cartesian imaging, it may be possible to expand the angularly continuous kernel representation presented here to an angular and radially continuous kernel, whose weights not only as a function of azimuthal angle but also radial distance. This could be useful for radial and spiral and other non-Cartesian trajectories.
In conclusion, it has been demonstrated that it is possible to reconstruct undersampled propeller blades without relying on the acquisition of additional PI calibration data. With the use of an angularly continuous 2D GRAPPA kernel (for 1D acceleration), tailored specifically for propeller trajectories, the numerical stability and accuracy of the GRAPPA kernel was shown to be very similar to GRAPPA kernels derived from fully sampled data. This method has here been shown to work adequately for SAP-EPI, but is applicable to propeller trajectories in general.
Acknowledgements
This work was supported in part by the Swedish Research Council (K2007-53P-20322-01-4), NIH (1R01EB002711), the Center of Advanced MR Technology at Stanford (P41RR09784), the Lucas Foundation, and the Oak Foundation.
References
- 1.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 2.Skare S, Newbould RD, Clayton DB, Bammer R. Propeller EPI in the other direction. Magn Reson Med. 2006;55:1298–1307. doi: 10.1002/mrm.20890. [DOI] [PubMed] [Google Scholar]
- 3.Skare S, Newbould RD, Clayton DB, Bammer R. Diffusion imaging using MinD-SAP-EPI. Proceedings of the 14th Annual Meeting of ISMRM; Seattle, WA, USA. 2006.p. 857. [Google Scholar]
- 4.Qu P, Shen GX, Wang C, Wu B, Yuan J. Tailored utilization of acquired k-space points for GRAPPA reconstruction. J Magn Reson. 2005;174:60–67. doi: 10.1016/j.jmr.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Wang J, Detre JA. Improved data reconstruction method for GRAPPA. Magn Reson Med. 2005;54:738–742. doi: 10.1002/mrm.20601. [DOI] [PubMed] [Google Scholar]
- 6.Pipe JG. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn Reson Med. 1999;42:963–969. doi: 10.1002/(sici)1522-2594(199911)42:5<963::aid-mrm17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Pipe JG, Zwart N. Turboprop: improved PROPELLER imaging. Magn Reson Med. 2006;55:380–385. doi: 10.1002/mrm.20768. [DOI] [PubMed] [Google Scholar]
- 8.Chuang TC, Huang TY, Lin FH, Wang FN, Chung HW, Chen CY, Kwong K. Propeller EPI with SENSE parallel imaging using a circularly symmetric phase array RF coil. Proceedings of the 12th Annual Meeting of ISMRM; Kyoto, Japan. 2004; p. 535. [DOI] [PubMed] [Google Scholar]
- 9.Skare S, Newbould RD, Clayton DB, Albers GW, Nagle S, Bammer R. Clinical multishot DW-EPI through parallel imaging with considerations of susceptibility, motion, and noise. Magn Reson Med. 2007;57:881–890. doi: 10.1002/mrm.21176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beatty PJ, Brau AC. ISMRM workshop on Non-Cartesian MRI. Sedona, AZ, USA: 2007. Parallel Imaging PROPELLER with Across-Blade Autocalibration. [Google Scholar]
- 11.Chuang TC, Huang TY, Lin FH, Wang FN, Ko CW, Kwong K, Chung HW. PROPELLER EPI with Parallel Imaging on High Resolution DTI at 3T. Proceedings of the 13th Annual Meeting of ISMRM; Miami Beach, FL, USA. 2005.p. 9. [Google Scholar]
- 12.Nelder JA, Mead R. Computer Journal. 1965;7:308–313. [Google Scholar]
- 13.Griswold MA, Heidemann RM, Jakob PM. Direct Parallel Imaging Reconstruction of Radially Sampled Data Using GRAPPA with Relative Shifts. Proceedings of the 11th Annual Meeting of ISMRM; Toronto, Ontario, Canada. 2003.p. 2349. [Google Scholar]
- 14.Heberlein K, Hu X. Auto-calibrated parallel spiral imaging. Magn Reson Med. 2006;55:619–625. doi: 10.1002/mrm.20811. [DOI] [PubMed] [Google Scholar]
- 15.Friston K, Ashburner J, Andersson J, Holmes A, Poline J-B, Ciuciu P,TN, Brett M, Glauche V, Gitelman D. Statistical Parametric Mapping. The Wellcome Departement of Imaging Neuroscience, Institute of Neurology, UCL; London: 2005. [Google Scholar]
- 16.Jain AK. Fundamentals of Digital Image Processing. Prentice Hall; 1989. [Google Scholar]
- 17.Davidon WC. Variable Metric Method for Minimization. Siam J Optimization. 1991;1:1–17. [Google Scholar]
- 18.Nana R, Zhao T, Hu X. Automatic Kernel Selection For Optimal GRAPPA Reconstruction. Proceedings of the 15th Annual Meeting of ISMRM; Berlin, Germany. 2007.p. 747. [Google Scholar]
- 19.Skare S, Newbould R, Holdsworth SJ, Nordell A, Bammer R. ISMRM workshop on Non-Cartesian MRI. Sedona, AZ: 2007. GRAPPA accelerated DW SAP-EPI using dual blade readouts. [Google Scholar]
- 20.Bammer R, Aksoy M, Liu C. Augmented generalized SENSE reconstruction to correct for rigid body motion. Magn Reson Med. 2007;57:90–102. doi: 10.1002/mrm.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, Bammer R, Moseley ME. Parallel imaging reconstruction for arbitrary trajectories using k-space sparse matrices (kSPA) Magn Reson Med. 2007;58:1171–1181. doi: 10.1002/mrm.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh EN, McKenzie CA, Ohliger MA, Sodickson DK. Parallel magnetic resonance imaging with adaptive radius in k-space (PARS): constrained image reconstruction using k-space locality in radiofrequency coil encoded data. Magn Reson Med. 2005;53:1383–1392. doi: 10.1002/mrm.20490. [DOI] [PubMed] [Google Scholar]
- 23.Lustig M, Pauly JM. Iterative GRAPPA: a General solution for the GRAPPA Reconstruction from Arbitrary k-pace Sampling. Proceedings of the 15th Annual Meeting of ISMRM; Berlin, Germany. 2007.p. 333. [Google Scholar]
- 24.Hu P, Meyer CH. BOSCO: Parallel Image Reconstruction Based on Successive Convolution Operations. Proceedings of the 14th Annual Meeting of ISMRM; Seattle, WA, USA. 2006.p. 10. [Google Scholar]
- 25.Heidemann RM, Griswold MA, Seiberlich N, Nittka M, Kannengiesser SA, Kiefer B, Jakob PM. Fast method for 1D non-cartesian parallel imaging using GRAPPA. Magn Reson Med. 2007;57:1037–1046. doi: 10.1002/mrm.21227. [DOI] [PubMed] [Google Scholar]