Summary
In recent years, highly sensitive assays have been developed that detect HIV-1 drug resistance mutations when present at less than 1% of the viral population. These assays are powerful tools when attempting to determine the clinical implications of these low level resistant virions after the administration of single-dose nevirapine. This report demonstrates that non-drug resistant polymorphisms in the primer binding site for the allele specific PCR (ASPCR) assay impact primer binding resulting in significant discrepancies in the assay's performance. Specifically, the use of a “universal” set of ASPCR primers caused an overestimation of the K103N (ntAAC) mutation at position 103 of reverse transcriptase when primer binding site polymorphisms resided close to the 3′ end of the allele specific primer. Drug resistance was predicted at values ranging from 0.69–7.69% for a sample containing only 1% resistance mutations and 3.35–31.84% for a sample containing 5% mutations. Conversely, the use of polymorphism specific primers detected 1.15–1.36% and 5.20–5.71% resistance for the same 1% and 5% samples. The results demonstrate the need to account for sequence polymorphisms when designing and implementing this highly specific assay.
Keywords: HIV, drug resistance, minor variant, allele specific PCR, nevirapine, subtype C
1. Introduction
Commercial genotyping assays are limited to the detection of HIV-1 drug resistance when resistant viruses comprise at least 20% of the viruses in an infected individual (Brun-Vezinet et al., 2004; Church et al., 2006; Grant et al., 2003; Palmer et al., 2005). There is widespread interest in understanding the clinical relevance of minor variant drug resistance, that is, when drug resistant viruses are present at less than 20%. This has become increasingly important in light of the growing concern regarding the development of nevirapine resistance in mothers and babies following the use of single-dose nevirapine for the prevention of mother-to-child transmission (PMTCT) of HIV in developing countries (Eshleman et al., 2001; Jackson et al., 2000). The exact nature of the clinical importance of minor variants remains unclear, but data suggests these populations of drug resistant viruses contribute to virologic failure (Jourdain et al., 2004; Lecossier et al., 2005), and may allow for the prediction of virologic failure at an earlier stage. As a result, several new assays have been developed to detect and quantify minor variant resistance (Bergroth et al., 2005; Flys et al., 2005; Nissley et al., 2005; Palmer et al., 2005; Shi et al., 2004), and the application of such techniques to HIV-1 subtype C samples has already provided insight into the persistence of drug resistance following single-dose nevirapine (Loubser et al., 2006; Palmer et al., 2006a).
Allele-specific polymerase chain reaction (ASPCR), one of the most powerful of these new techniques, can detect and quantify drug resistant viruses when present at less than 1% (Halvas et al., 2006; Paredes et al., 2007). ASPCR consists of two quantitative PCR (qPCR) reactions differing only in the primer that overlaps the drug resistance mutation: one reaction contains an “allele-specific” primer matching the drug resistance mutation and the other contains an “allele-specific” primer matching the wild type sequence. It has previously been reported that the presence of nucleotide polymorphisms in the allele-specific primer binding site, causing primer-template mismatch, reduces the accuracy of ASPCR, specifically resulting in the underestimation of the percentage of drug resistant viruses (Palmer, 2006b). Given the high degree of background genetic variation that is characteristic of HIV, this has a significant impact when applying ASPCR on a large scale basis to clinical samples for the purpose of assessing the clinical relevance of low levels of drug resistance.
In an effort to understand the underlying cause of this discrepancy, an in vitro analysis was performed to assess the impact of naturally occurring, non-resistance, nucleotide polymorphisms in the ASPCR binding site on the accuracy of the assay. This reports shows that primer site mismatches caused by naturally occurring nucleotide polymorphisms can result in dramatic overestimates of the percentage of drug resistant viruses present in the sample at the 103 position of reverse transcriptase. However, the use of polymorphism specific primers and corresponding standard curves eliminates the problem, allowing accurate quantitation of minor variants, and thus should be applied when ASPCR is used in highly polymorphic regions of HIV.
2. Materials And Methods
2.1. Clinical Samples
Plasma samples from women in Botswana who received single-dose nevirapine for PMTCT were obtained one to 19 months after drug exposure and prior to the initiation of highly active antiretroviral therapy (HAART). The samples were genotyped for nevirapine drug resistance by ViroSeq according to manufacturer protocol (Applied Biosystems, Foster City, CA). In 18 samples, no drug resistance was detected by this conventional sequencing method. Of these 18 sequences, 17 were HIV-1 subtype C and one was HIV-1 subtype A. The genotypes of these 18 sequences were reproduced in plasmids for in vitro testing of the ASPCR method.
2.2. Site-Directed Mutagenesis
The HIV-1 subtype C (HIV-1C) infectious molecular clone pMJ4 (Ndung'u et al., 2001) provided the reverse transcriptase (RT) that served as the backbone for subsequent mutagenesis; the Apa I site located in the pBlueScript vector of pMJ4 was deleted by partial Apa I digest followed by Klenow fill-in. This allowed the 1.6 kb fragment encompassing RT to be cut out by Apa I and Hpa I digest. It was cloned into a pCR2.1 (Invitrogen, Carlsbad, CA) vector modified to include a Hpa I resctriction site. The resulting HIV-1C RT subclone, pCLB11, served as the template for QuikChange II (Stratagene, La Jolla, CA) site-directed PCR mutagenesis to recreate the HIV-1C nucleotide polymorphisms of interest. Briefly, the DNA template is denatured and forward/reverse mutagenic primers containing the desired mutation are annealed to the template followed by extension with PfuUltra DNA polymerase. The parental strand is digested with Dpn I endonuclease, specific for methylated DNA.
Using this technique, 32 plasmids were generated corresponding to the 8 unique sequences seen in the Botswana samples. These sequences corresponded to the changes in the ASPCR primer binding region for position 103 of reverse transcriptase. A plasmid was generated for each of the 8 sequences with an A, C, T, and G at the third position of amino acid 103 of reverse transcriptase.
2.3. Allele-Specific PCR (ASPCR)
ASPCR is a nested PCR assay combining a standard first-round PCR using universal primers and a quantitative, second round PCR with allele-specific primers. The first round primers were RT-18 (5′-GGA AAC CAA AAA TGA T AG GGG GAA TTG GAG G-3′) and NE1 (5′-CCT ACT AAC TTC TGT ATG TCA TTG ACA GTC CAG CT-3′). In a 50 μL reaction, 38.5 μL of water, 5 μL of 10X reaction buffer, 2 uL of dNTPs, 1 μL of each primer NE1 and RT-18 (10 μM concentration each) and 0.5 μL of FastStart High Fidelity Enzyme Blend (Roche Applied Science, Indianapolis, IN) were mixed with 10 ng of each plasmid. The cycling parameters for the PCR were 94° C for 5 min followed by 35 cycles of 94° C for 20 s, 55° C for 20 s, and 72° C for 2 min, with a final extension at 72° C for 7 minutes. The first round PCR generated a 957 bp amplicon comprising nucleotides 2359 to 3316 (HXB2 numbering) which was gel purified by electrophoresis on 1% agarose followed by QiaQuick gel extraction per manufacturer protocol (Qiagen, Valencia, CA).
The first-round PCR amplicon (107 copies as measured by spectrophotometry) served as the template for the second-round, quantitative PCR step. The forward primer was constant for all reactions (2090F-5′-AAG TGG AGA AAA TTA GTA GAT TTC AGG GA-3′). Different sets of reverse primers were created. The universal primer set is exactly the same set of primers used in the protocol devised for subtype C (Palmer et al., 2006a), with the addition of a specific primer for the ntAAG. Allele-specific reverse primers were also constructed to match the varying template sequence in the primer binding site. The reverse primers always included a single nucleotide mismatch at the penultimate 3′ position to enhance reaction specificity. The final 3′ position of the allele-specific primer falls on the third position of the RT aa103 codon, the variation of which confers the lysine to asparagine (K103N) drug resistance mutation. Primers were constructed containing each of the four nucleotides at the final position. For example, the group I reverse primers, designed to match the “universal primers” described previously, consisted of the following:
AAA-103K: 2218R 5′-CCC ACA TCT AGT ACT GTC ACT GAT TCT-3′
AAC-103N: 2218R 5′-CCC ACA TCT AGT ACT GTC ACT GAT TGG-3′
AAT-103N: 2218R 5′-CCC ACA TCT AGT ACT GTC ACT GAT TCA-3′
AAG-103K: 2218R 5′-CCC ACA TCT AGT ACT GTC ACT GAT TGC-3′.
The second round reaction was performed on the Applied Biosystems Sequence Detector System 7500 in a 25 μL reaction. The reaction mix consisted of 12.5 μL of Power Sybr Green PCR Master Mix (Applied Biosystems) and 1.25 uL of both forward and reverse primers (concentration of 6 μM each) mixed with 10 uL of template (PCR amplicon). The cycling conditions had an initial denaturation step of 95° C for 10 minutes followed by 40 cycles of 95° C for 15s, and 60° C for 1 minute. PCR products were subjected to thermal denaturation.
Independent standard curves were generated for each template as follows: a series of five 10-fold dilutions of first round amplicon (108 to 104 copies) were quantified in triplicate in two independent second-round assays and the cumulative data (six points at each dilution) were used to generate a regression line. The slopes of the regression lines for all 32 of the independent standard curves generated in this way ranged from -3.52 to -3.82. The standard curve was calibrated to each subsequent run by including in each run the 106 standard in triplicate and using the data for that sample to adjust the y-intercept of the standard curve.
2.4. Comparisons of Universal Primers to Polymorphism-Specific Primers
PCR amplicon (107 copies) was mixed to create three different scenarios for each group of binding site polymorphisms- the first amplicon tested had no mutations present, the second was a mixture consisting of 1% of each 103N (AAC and AAT) mutation in the sample, and the third mixture tested 5% of each 103N mutation. Reactions with both the universal primers and polymorphism specific primers were performed concurrently with primers used to detect all four possible nucleotides at the terminal position of 103 of RT. The amplicon mixtures were performed in triplicate in a 96 well plate. Controls for each set of primers were also done in triplicate.
The percentage of each nucleotide at position 103 of RT was calculated by taking the number of cycles to reach fluorescence (Ct) for each reaction and applying this value to the universal primer standard curve as well as to the appropriate, polymorphism specific standard curve. The determination of percentage of specific allele was determined by:
2.5. Extended Nucleotide Consensus Sequences
Extended consensus sequences showing the type and percentage of nucleotides present at each position were constructed for reverse transcriptase for HIV-1A, HIV-1B, HIV-1C, and HIV-1D. In all cases, the final nucleotide alignments were downloaded from the Los Alamos National Laboratories (LANL) HIV Database on August 17, 2007 (www.hiv.lanl.gov). The searches for HIV-1A, HIV-1C, and HIV-1D RT (p51) sequences were restricted to sub-Saharan Africa (search parameter “geographical region” = sub-Saharan Africa). The search for HIV-1B RT(p51) sequences was not restricted by geographical region. The searches resulted in 70, 1098, 412, and 53 individual nucleotide sequences for subtypes A, B, C, and D respectively. Sequences characterized as “problematic” by the LANL-HIV search engine were not included in the final alignment. An extended consensus sequence was also constructed for the 18 clinical HIV samples from Botswana. A PERL script was used to automate the construction of extended nucleotide consensus sequences.
3. Results
3.1. Nucleotide Polymorphism in the Allele-Specific Primer Binding Site for RT Position 103
The RT sequence from 18 clinical samples from Botswana revealed nucleotide polymorphism at 7 positions within the binding site for the allele-specific reverse primer for position 103 (Figure 1). Eight binding site groups were defined according to the observed polymorphisms with the group number reflecting the expected impact of the polymorphisms on quantitation using the previously described “universal primer” on the basis of relative proximity of the polymorphism(s) to the 3′ end of the primer. For example, Group I matches the universal primer exactly (with the exception of the intentional mismatch included in all primers) and thus is expected to be quantified most accurately whereas Group VIII includes two polymorphisms that are relatively close to the 3′ end and is therefore expected to be quantified with the least accuracy by the “universal primer”.
Figure 1. Polymorphisms in the primer binding site for the ASPCR assay at the 103 position of reverse transcriptase.
These polymorphisms reflect the difference seen in 18 HIV-1 clinical samples from Botswana. Group I matches the published primers exactly (the intentional mismatch at the 2nd position of the primer is done to enhance specificity). The asterisk represents the third nucleotide at the 103 position of reverse transcriptase that confers a drug resistance mutation as the amino acid changes from a lysine (AAA) to an asparagine (AAC or AAT).
3.2. RT Position 103 ASPCR with Universal Primers
Results from the testing of the universal primers with its matched template (Group I) show that the method used to detect low frequency of virus has a sensitivity of the reaction ranging from 0.002%-0.11% when 100% wild-type was analyzed with each of the other three allele specific primers at the 103 position. Mixtures of PCR amplicon were used to determine the ability of ASPCR to detect each 103 allele at 1% and 5% concentration (Group I in Table 1). These results demonstrate that the universal primers are highly accurate when applied to matched template.
Table 1.
Results obtained when universal primers used in ASPCR for each of the different groups of binding site polymorphisms (shown in Figure 1). Each group tested had 0% (A), 1% (B), and 5% (C) mutation at position 103 of HIV-1 reverse transcriptase (both AAC and AAT mutations). Group I matches the universal primers except for the intentional mismatch at the (-2) position from the 3′ end of the primer.
| A. Plasmids with 100% wt (AAA) at RT position 103. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | Percent | I | II | III | IV | V | VI | VII | VIII |
| 103K AAA | (100%) | 99.85 | 99.39 | 96.06 | 97.44 | 96.13 | 97.45 | 96.22 | 96.01 |
| 103N AAC | (0%) | 0.05 | 0.02 | 1.14 | 0.09 | 0.67 | 0.18 | 0.57 | 0.33 |
| 103N AAT | (0%) | 0.002 | 0.004 | 0.09 | 0.04 | 0.18 | 0.09 | 0.53 | 2.17 |
| 103K AAG | (0%) | 0.11 | 0.58 | 2.71 | 2.43 | 3.03 | 2.28 | 2.68 | 1.49 |
| B. Plasmids with 98% wt (AAA), 1% 103N (AAC) and 1% 103N (AAT) at RT position 103. | |||||||||
| Allele | Percent | I | II | III | IV | V | VI | VII | VIII |
| 103K AAA | (98%) | 97.53 | 98.76 | 95.63 | 94.82 | 91.88 | 91.28 | 92.39 | 89.18 |
| 103N AAC | (1%) | 1.06 | 0.69 | 3.85 | 4.19 | 7.26 | 7.47 | 6.84 | 7.69 |
| 103N AAT | (1%) | 1.28 | 0.45 | 0.10 | 0.38 | 0.26 | 0.28 | 0.23 | 2.26 |
| 103K AAG | (0%) | 0.13 | 0.10 | 0.43 | 0.60 | 0.60 | 0.98 | 0.53 | 0.88 |
| C. Plasmids with 90% wt (AAA), 5% 103N (AAC) and 5% 103N (AAT) at RT position 103. | |||||||||
| Allele | Percent | I | II | III | IV | V | VI | VII | VIII |
| 103K AAA | (90%) | 88.77 | 94.17 | 82.41 | 81.34 | 72.53 | 71.23 | 67.06 | 68.92 |
| 103N AAC | (5%) | 5.00 | 3.35 | 16.71 | 16.61 | 25.86 | 27.05 | 31.84 | 28.71 |
| 103N AAT | (5%) | 6.03 | 2.34 | 0.46 | 1.42 | 0.97 | 0.92 | 0.72 | 1.79 |
| 103K AAG | (0%) | 0.21 | 0.14 | 0.42 | 0.64 | 0.63 | 0.80 | 0.38 | 0.58 |
The results of quantitation of changes at RT position 103 by ASPCR using the “universal primers” on Groups I-VIII are shown in Table 1. For each of the binding site polymorphism groups, the universal primers were used to quantify plasmid-based “samples” consisting of the following three scenarios: 100% 103K (ntAAA); 98% 103K (ntAAA), 1% 103N (ntAAC), and 1% 103N (ntAAT); and 90% 103K (ntAAA), 5% 103N (ntAAC), and 5% 103N (ntAAT). Group II performed well although the percentage of the 103N mutations (ntAAC and ntAAT) were both mildly underestimated. The accuracy of the assay however substantially declined when applied to all other groups demonstrating a consistent over-estimation of the presence of 103N (ntAAC). The magnitude of the inaccuracy was relatively minor for the 100% 103K samples, with the Group VIII showing the greatest overestimation of 103N at 2.17%, but the the discrepancy in results increased as the percentage of 103N variants in the samples increased. The inaccuracy reached a maximum in the 90%/5%/5% samples with a range of cumulative over-estimation of 103N ranging from nearly 17% (Group III) to over 31% (Group VII).
3.3. RT Position 103 ASPCR with Binding Site Polymorphism-Specific Primers
The results of the comparison between the universal primers and the binding site polymorphism specific primers are shown in Table 2. The same mixtures of template were used with the polymorphism-specific primers as performed above with the universal primers.
Table 2.
Results obtained when polymorphism specific primers used in ASPCR for each of the different groups of binding site polymorphisms (shown in Figure 1). Each group tested had 0% (A), 1% (B), and 5% (C) mutation at position 103 of HIV-1 reverse transcriptase (both AAC and AAT mutations.
| A. Plasmids with 100% wt (AAA) at position 103. Results of ASPCR using polymorphism specific primers. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Allele | Percent | I | II | III | IV | V | VI | VII | VIII |
| 103K AAA | (100%) | 99.85 | 99.94 | 99.87 | 99.77 | 99.81 | 99.51 | 99.98 | 99.92 |
| 103N AAC | (0%) | 0.05 | 0.01 | 0.05 | 0.18 | 0.05 | 0.02 | 0.01 | 0.004 |
| 103N AAT | (0%) | 0.002 | 0.002 | 0.02 | 0.011 | 0.04 | 0.001 | 0.001 | 0.005 |
| 103K AAG | (0%) | 0.11 | 0.05 | 0.07 | 0.04 | 0.10 | 0.47 | 0.01 | 0.07 |
| B. Plasmids with 98% wt (AAA), 1% 103N (AAC) and 1% 103N (AAT) at position 103. Results of ASPCR using polymorphism specific primers | |||||||||
| Allele | Percent | I | II | III | IV | V | VI | VII | VIII |
| 103K AAA | (98%) | 97.53 | 97.50 | 97.49 | 97.68 | 97.12 | 97.19 | 97.29 | 97.52 |
| 103N AAC | (1%) | 1.06 | 1.17 | 1.24 | 1.15 | 1.34 | 1.36 | 1.36 | 1.29 |
| 103N AAT | (1%) | 1.28 | 1.27 | 1.16 | 1.16 | 1.38 | 1.41 | 1.30 | 1.18 |
| 103K AAG | (0%) | 0.13 | 0.06 | 0.12 | 0.006 | 0.15 | 0.04 | 0.05 | 0.009 |
| C. Plasmids with 90% wt (AAA), 5% 103N (AAC) and 5% 103N (AAT) at position 103. Results of ASPCR using polymorphism specific primers | |||||||||
| Allele | Percent | I | II | III | IV | V | VI | VII | VIII |
| 103K AAA | (90%) | 88.77 | 89.46 | 88.90 | 89.82 | 88.23 | 88.13 | 88.82 | 89.27 |
| 103N AAC | (5%) | 5.00 | 5.33 | 5.26 | 5.20 | 5.71 | 5.66 | 5.71 | 5.29 |
| 103N AAT | (5%) | 6.03 | 5.11 | 5.58 | 4.97 | 5.75 | 6.15 | 5.34 | 5.42 |
| 103K AAG | (0%) | 0.21 | 0.11 | 0.27 | 0.02 | 0.31 | 0.06 | 0.12 | 0.02 |
When 100% 103K (ntAAA) was introduced into the polymorphism-specific reactions, the accuracy of the results was very good with all of the tests for drug resistant alleles being less than 0.05% with the exception of one group (group IV) being mildly elevated with 0.18% 103N (ntAAC) when only wild-type was entered into the assay. Group VI had an elevated recording of 0.47% 103K (ntAAG) when only 103K (ntAAA) was entered into the reaction.
The scenario with 98% 103K (ntAAA), 1% 103N (ntAAC), and 1% 103N (ntAAT) of each Group (I-VIII) entered into the polymorphism-specific reactions showed excellent results ranging from 1.06% to 1.41% of both 103N alleles (ntAAC and ntAAT). Similarly, the mixture of template with 90% 103K (ntAAA), 5% 103N (ntAAC), and 5% 103N (ntAAT) demonstrated the polymorphism-specific primers performed quite well with results ranging from 4.97% to 6.15% for both mutation alleles when 5% mutation was introduced into the sample.
3.4. Results of Consensus Sequence Generation
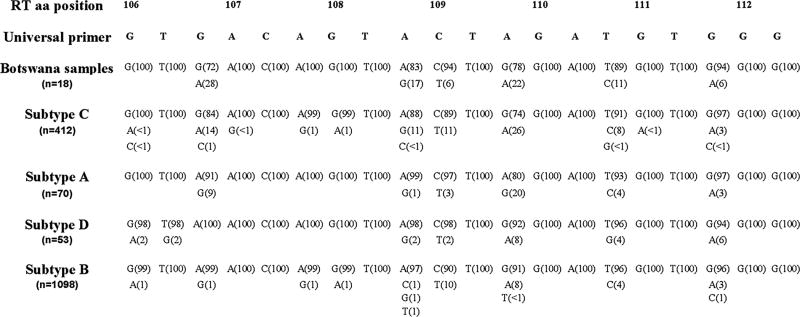
The results from the generation of the consensus sequence are shown in Figure 2. Although the initial work was based on a small sample of 18 Botswana samples, large population scale data supports the findings that polymorphisms are prevalent in the binding site region for the 103 primer across all subtypes evaluated.
Figure 2.
Polymorphisms in the primer binding site region for the ASPCR primers near position 103 of reverse transcriptase. Listed above are the amino acid positions and the universal primer sequence from positions 106-112 of reverse transcriptase, the region of polymorphisms tested in vitro based on the sequences of the 18 Botswana samples. Data from subtype C, A, and D were included as they are the predominant subtypes in Africa. Subtype B included for comparison.
4. Discussion
Several studies have shown that the use of commercial genotyping kits to detect HIV-1 drug resistance lack the ability to detect minor variants, mutations that are less than approximately 20% of the clinical sample, and newer techniques can determine resistance with much greater sensitivity. These new methods that detect fewer than 20% of resistance mutations are important tools in furthering the understanding of mutations that arise with the use of single-dose nevirapine and will ultimately provide valuable insight into the clinical impact of low levels of resistant virions. The results of this in vitro analysis demonstrate that naturally occurring nucleotide polymorphisms in the binding site for the allele-specific primer in the ASPCR technique can have significant impact on the accurate quantitation of drug resistance variants.
While the clinical implication of minor variant drug resistance is unclear, a recent study from Botswana has demonstrated that exposure to nevirapine during delivery impacts negatively on maternal outcome with HAART if the medications are started within 6 months postpartum (Lockman et al., 2007). The fading of drug resistance mutations after nevirapine exposure may be responsible for the successful outcomes seen when patients delay initiation of NNRTI-containing HAART.
The studies from developing countries that use single-dose nevirapine are quite heterogeneous with respect to the techniques used and the frequency of nevirapine resistance mutations detected in different HIV-1 subtypes (Flys et al., 2005; Johnson et al., 2005; Lalonde et al., 2007; Loubser et al., 2006; Palmer et al., 2006a). All of the studies demonstrate that the commercial techniques are suboptimal in their ability to detect minor variants, but no consensus emerges from the data regarding the degree of resistance and the duration of persistence of minor variant resistance after nevirapine exposure. It is possible that some of the heterogeneity in results could be attributed to the different assays used, and an overestimation of resistance may occur in some cases, as demonstrated in this in vitro study. Techniques such as the oligonucleotide ligation assay (OLA) or assays dependent on hybridization probes may be subject to similar problems as the ASPCR assay in that the polymorphism in the region being studied may lead to differential binding of oligonucleotides or probes impacting accurate quantitation as seen with the ASPCR primers. The ability of the new genotyping methods to accurately detect and quantify resistance needs to be closely evaluated so that results using the various methods involving different HIV-1 subtypes can be accurately compared.
The discrepancies in the results seen in this study follow a predictable pattern when a universal ASPCR primer set for a specific subtype and polymorphism specific primers are used. The data demonstrate that with polymorphisms in the template close to 3′ end of the allele specific primer, the accuracy of the results is significant impacted. This was clearly demonstrated when looking for the K103N mutation (AAA→AAC) at position 103 of reverse transcriptase, the most commonly seen mutation in clinical samples from Botswana in women who received nevirapine for PMTCT (Shapiro et al., 2006).
The mechanism responsible for the different results when the different sets of primers are used on the same template likely reflects the natural difference in bond strength that occurs between nucleotides. As a result of the three hydrogen bonds between the G-C nucleotides compared to the two hydrogen bonds between the A-T nucleotides, the G-C pair is more stable. The impact of this increased bond strength is critical when it occurs at the 3′ end of the allele specific primer. In the case of the K103N mutation (ntAAC), the mutation specific primer that ends in a guanine (G) and binds to the cytosine (C) will bind more tightly than the thymine (T) in the wild-type primer as it binds to the A (adenine). In the setting of other polymorphisms in the primer binding site, the terminal G-C binding would act as an anchor for the mutation primer with relatively increased (ie. not as significant of a decrease) binding to the template compared to the wild-type allele specific primer and its A-T bond. Polymorphisms close to the 3′ end of the primer and multiple polymorphisms in the binding site create a scenario that makes the terminal bond more significant in the allele specific reactions. In essence, the crossing threshold (CT) of the wild-type reaction (ntAAA) is shifted further to the right (amplifies later) compared to the CT of the mutation primer (ntAAC). When applied to the corresponding standard curve, this change will be calculated as less total wild-type template present in a sample than truly exists, thereby falsely underreporting wild-type and overcalling the 103N (ntAAC) mutation.
Each group analyzed in our data showed vast improvements in the accuracy of the assay when polymorphism specific primers were used, with the exception of Group II, which had a single polymorphism at a site distant from the 3′ end of the primers. Additionally, the ordering of the groups generally followed the anticipated increasing error rate between the two methods. Overall, the proximity of the polymorphisms to the 3′ end and the number of polymorphisms demonstrate clear situations where the ASPCR method falters unless primer binding site polymorphism specific primers are used.
Our technique differs from the protocol developed by others using universal primers specific to samples of HIV-1 subtype C (Palmer et al., 2006a). In this previous approach, a common reverse primer was used that did not overlie the site of interest and this allows for the quantitation of a total number of copies of amplicon in each sample. The three primers (wild-type and both 103N mutation primers) were each used on the samples and the results were calculated based on a percentage of each amplicon over the total copy number obtained by the reverse primer. No attempt was made to quantify the infrequent ntAAG (103K) polymorphism in their analysis. It was decided to eliminate the common primer from the calculation and substitute a fourth primer containing the ntAAG polymorphism at position 103 as this would enable us to detect the four possible nucleotides at the position of interest without the need to calculate total copy number by a separate reaction, and be able to detect AAG when it is actually present in our samples rather than inferring it. The calculated number of copies from all four primers served as the total copy number and calculated the % wild-type or mutant based on number of copies that each represented of this total.
The use of three specific primers and one common primer (for calculation of the total copies) rather than four specific primers would not negate the effect of the binding site polymorphisms on the results seen. The ntAAC at position 103, when polymorphisms exist in the binding site, would still result in more robust amplification by comparison to the ntAAA or ntAAT. Whether the total copies is calculated separately (using a common primer) or by summing the four different nucleotide amplifications, the effect is the same in that the numerator will be falsely increased in the case of ntAAC due to the strength of the terminal G-C anchor. The resulting calculation predicts a falsely elevated percentage of minor variants.
The in vitro design of this study allows for close scrutiny of the technique prior to its application to clinical samples. The 8 different groups of binding-site polymorphisms in this analysis reflect those seen in a limited sample set from Botswana. Importantly, the polymorphisms seen are reflective of the heterogeneous nature of this region of reverse transcriptase. The consensus sequences shown in this report only include the areas of polymorphism tested in our experiments. The HIV-1 subtypes found in Africa all have changes in this region that could impact the ability to correctly apply ASPCR. Further heterogeneity exists in the region of amino acid 104 in all subtypes and at position 105 in subtypes B and C (data not shown) that were not seen in our samples. These variations in the genome would be expected to cause greater discrepancy in amplification of the ASPCR primers as they reside close to the 3′ end of the primers, and would likely overcall minor variant drug resistance at position 103.
The limitations of this technique as it applies to clinical samples remains the fact that sequencing of the region of interest must occur prior to applying the ASPCR technique in order to determine which primer pair set must be used in the analysis. The bulk sequencing technique used to determine the primers will not detect minor polymorphisms in the binding site, and this will possibly mitigate the exact determination of the percentage of minor variants. However, the majority of the variants in the binding site will be detected and by using the appropriate primers and corresponding standard curves, it will allow for closer approximation of the true drug resistant population.
The clinical significance of minor drug resistant variants remains unknown but the determination of a threshold of drug resistance below which good clinical outcomes can be expected is a potential use of this research particularly as it relates to women who received single-dose nevirapine for PMTCT. The use of highly sensitive techniques must be accompanied by careful examination of the impact of polymorphisms on theses assays. This in vitro examination of the ASPCR assay confirms the ability of ASPCR technique to detect minor drug resistance variants, but specific matching primer sets should be applied to clinical samples to ensure template specificity as minor changes in the area of drug resistance mutations can impact the results dramatically.
Acknowledgments
Supported by NIAID K08 AI067014 (C.F.R.). The authors would like to thank Daniel Kuritzkes, MD, for his assistance with the ASPCR assay and for his comments regarding the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Christopher F. Rowley, Email: crowley1@bidmc.harvard.edu.
Christian L. Boutwell, Email: christian_boutwell@student.hms.harvard.edu.
Shahin Lockman, Email: slockman@hsph.harvard.edu.
M. Essex, Email: messex@hsph.harvard.edu.
References
- Bergroth T, Sonnerborg A, Yun Z. Discrimination of lamivudine resistant minor HIV-1 variants by selective real-time PCR. J Virol Methods. 2005;127:100–107. doi: 10.1016/j.jviromet.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Brun-Vezinet F, Costagliola D, Khaled MA, Calvez V, Clavel F, Clotet B, Haubrich R, Kempf D, King M, Kuritzkes D, Lanier R, Miller M, Miller V, Phillips A, Pillay D, Schapiro J, Scott J, Shafer R, Zazzi M, Zolopa A, DeGruttola V. Clinically validated genotype analysis: guiding principles and statistical concerns. Antivir Ther. 2004;9:465–478. [PubMed] [Google Scholar]
- Church JD, Jones D, Flys T, Hoover D, Marlowe N, Chen S, Shi C, Eshleman JR, Guay LA, Jackson JB, Kumwenda N, Taha TE, Eshleman SH. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn. 2006;8:430–432. doi: 10.2353/jmoldx.2006.050148. quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshleman SH, Krogstad P, Jackson JB, Wang YG, Lee S, Wei LJ, Cunningham S, Wantman M, Wiznia A, Johnson G, Nachman S, Palumbo P. Analysis of human immunodeficiency virus type 1 drug resistance in children receiving nucleoside analogue reverse-transcriptase inhibitors plus nevirapine, nelfinavir, or ritonavir (Pediatric AIDS Clinical Trials Group 377) J Infect Dis. 2001;183:1732–1738. doi: 10.1086/320728. [DOI] [PubMed] [Google Scholar]
- Flys T, Nissley DV, Claasen CW, Jones D, Shi C, Guay LA, Musoke P, Mmiro F, Strathern JN, Jackson JB, Eshleman JR, Eshleman SH. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005;192:24–29. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- Grant RM, Kuritzkes DR, Johnson VA, Mellors JW, Sullivan JL, Swanstrom R, D'Aquila RT, Van Gorder M, Holodniy M, Lloyd RM, Jr, Reid C, Morgan GF, Winslow DL. Accuracy of the TRUGENE HIV-1 genotyping kit. J Clin Microbiol. 2003;41:1586–1593. doi: 10.1128/JCM.41.4.1586-1593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvas EK, Aldrovandi GM, Balfe P, Beck IA, Boltz VF, Coffin JM, Frenkel LM, Hazelwood JD, Johnson VA, Kearney M, Kovacs A, Kuritzkes DR, Metzner KJ, Nissley DV, Nowicki M, Palmer S, Ziermann R, Zhao RY, Jennings CL, Bremer J, Brambilla D, Mellors JW. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J Clin Microbiol. 2006;44:2612–2614. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JB, Becker-Pergola G, Guay LA, Musoke P, Mracna M, Fowler MG, Mofenson LM, Mirochnick M, Mmiro F, Eshleman SH. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS. 2000;14:F111–F115. doi: 10.1097/00002030-200007280-00001. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Li JF, Morris L, Martinson N, Gray G, McIntyre J, Heneine W. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- Jourdain G, Ngo-Giang-Huong N, Le Coeur S, Bowonwatanuwong C, Kantipong P, Leechanachai P, Ariyadej S, Leenasirimakul P, Hammer S, Lallemant M. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med. 2004;351:229–240. doi: 10.1056/NEJMoa041305. [DOI] [PubMed] [Google Scholar]
- Lalonde MS, Troyer RM, Syed AR, Bulime S, Demers K, Bajunirwe F, Arts EJ. Sensitive oligonucleotide ligation assay for low-level detection of nevirapine resistance mutations in human immunodeficiency virus type 1 quasispecies. J Clin Microbiol. 2007;45:2604–2615. doi: 10.1128/JCM.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Shulman NS, Morand-Joubert L, Shafer RW, Joly V, Zolopa AR, Clavel F, Hance AJ. Detection of minority populations of HIV-1 expressing the K103N resistance mutation in patients failing nevirapine. J Acquir Immune Defic Syndr. 2005;38:37–42. doi: 10.1097/00126334-200501010-00007. [DOI] [PubMed] [Google Scholar]
- Lockman S, Shapiro RL, Smeaton LM, Wester C, Thior I, Stevens L, Chand F, Makhema J, Moffat C, Asmelash A, Ndase P, Arimi P, van Widenfelt E, Mazhani L, Novitsky V, Lagakos S, Essex M. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- Loubser S, Balfe P, Sherman G, Hammer S, Kuhn L, Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS. 2006;20:995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndung'u T, Renjifo B, Essex M. Construction and analysis of an infectious human Immunodeficiency virus type 1 subtype C molecular clone. J Virol. 2001;75:4964–4972. doi: 10.1128/JVI.75.11.4964-4972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissley DV, Halvas EK, Hoppman NL, Garfinkel DJ, Mellors JW, Strathern JN. Sensitive phenotypic detection of minor drug-resistant human immunodeficiency virus type 1 reverse transcriptase variants. J Clin Microbiol. 2005;43:5696–5704. doi: 10.1128/JCM.43.11.5696-5704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Boltz V, Martinson N, Maldarelli F, Gray G, McIntyre J, Mellors J, Morris L, Coffin J. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci USA. 2006a;103:7094–7099. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Boltz V, Maldarelli F, McIntyre J, Morris L, Hopley M, Mayers D, Robinson P, Mellors J, Coffin J. Optimization of allele-specific PCR for drug-resistant HIV variants using patient-specific consensus sequences for primer design. Proceedings of the 13th Conference on Retroviruses and Opportunistic Infections; February 5–8, 2006; Denver, CO. 2006b. Abstract 628. [Google Scholar]
- Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes R, Marconi V, Campbell T, Kuritzkes D. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J Virol Methods. 2007;146:136–146. doi: 10.1016/j.jviromet.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RL, Thior I, Gilbert PB, Lockman S, Wester C, Smeaton LM, Stevens L, Heymann SJ, Ndung'u T, Gaseitsiwe S, Novitsky V, Makhema J, Lagakos S, Essex M. Maternal single-dose nevirapine versus placebo as part of an antiretroviral strategy to prevent mother-to-child HIV transmission in Botswana. AIDS. 2006;20:1281–1288. doi: 10.1097/01.aids.0000232236.26630.35. [DOI] [PubMed] [Google Scholar]
- Shi C, Eshleman SH, Jones D, Fukushima N, Hua L, Parker AR, Yeo CJ, Hruban RH, Goggins MG, Eshleman JR. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]