Abstract
Proteins aggregate in response to various stresses including changes in solvent conditions. Addition of alcohols has been recently shown to induce aggregation of disease-related as well as non-disease-related proteins. Here we probed the biophysical mechanisms underlying alcohol-induced protein aggregation, in particular the role of partial protein unfolding in aggregation. We have studied aggregation mechanisms due to benzyl alcohol which is used in numerous biochemical and biotechnological applications. We chose cytochrome c as a model protein, for the reason that various optical and structural probes are available to monitor its global and partial unfolding reactions. Benzyl alcohol induced the aggregation of cytochrome c in isothermal conditions and decreased the temperature at which the protein aggregates. However, benzyl alcohol did not perturb the overall native conformation of cytochrome c. Instead, it caused partial unfolding of a local protein region around the methionine residue at position 80. Site-specific optical probes, two-dimensional NMR titrations, and hydrogen exchange all support this conclusion. The protein aggregation temperature varied linearly with the melting temperature of the Met80 region. Stabilizing the Met80 region by heme iron reduction drastically decreased protein aggregation, which confirmed that the local unfolding of this region causes protein aggregation. These results indicate that a possible mechanism by which alcohols induce protein aggregation is through partial rather than complete unfolding of native proteins.
Keywords: stability, foldon, structure, cytochrome c, benzyl alcohol
Introduction
Protein aggregates are involved in the pathogenesis of various diseases including neurodegenerative diseases 1,2 and sickle cell anemia 3,4. Protein aggregates in therapeutic formulations trigger adverse effects in patients such as immunogenicity to the protein 5-7. Proteins aggregate in response to several stresses including changes in solvent conditions. Recently, addition of alcohols has been shown to aggregate proteins. For example, trifluoroethanol (TFE) induces aggregation of superoxide dismutase 8, ethanol induces insulin aggregation 9, whereas benzyl alcohol (BA) induces the aggregation of human interleukin-1 receptor antagonist 10. However, the mechanisms by which alcohols induce protein aggregation are not known. In this paper, we probe the role of protein unfolding in alcohol-induced protein aggregation.
To study alcohol effects, we chose benzyl alcohol (BA) because of its wide-range of biochemical and biotechnological applications compared to any other alcohol. BA has been used as an antimicrobial agent in protein formulations 11, as an anesthetic 12, as a membrane fluidizer 13, as a heat shock protein inducer 14, as a fragrance component and preservative in cosmetic applications such as hair shampoos, as a food additive, and in other applications 15. However, BA has been shown to induce aggregation of three proteins: interferon-γ 16, interleukin-1 receptor antagonist 10, and human granulocyte colony stimulating factor 17 even at the extremely low levels of 1% v/v. So far, these studies have not provided any mechanistic insight into structural changes that induce protein aggregation, which is critical for the rational development of approaches to inhibit protein aggregation. Earlier studies indicate that BA does not affect global protein stability or structure 10,18,19, raising the question of whether protein stability plays any role in BA-induced protein aggregation. In addition to global unfolding, partial protein unfolding has been proposed to play an important role in general protein aggregation 20,21. We examined here whether alcohol-induced aggregation is caused by unfolding of local protein regions, previously referred to as foldons 22 rather than global unfolding of the entire protein.
For studying the role of partial protein unfolding in protein aggregation, we chose cytochrome c (Cyt c; Fig. 1A) as a model protein, for the reason that various optical and structural probes are available to monitor its global unfolding and especially its distinct partial unfolding reactions 23-25. Cyt c is predominantly an α-helical protein (Fig. 1A), and hence circular dichroism (CD) at 222 nm is a good probe for measuring its global stability. Tryptophan fluorescence is a second probe for Cyt c global stability. Cyt c contains a single tryptophan at position 59 that is not fluorescent in the native protein due to Förster resonance energy transfer to the heme group (Förster distance R0 = 34 Å 26), but becomes highly fluorescent upon protein unfolding. An absorption band at 695 nm due to charge transfer from the Met80-S to the ferric iron provides a probe for unfolding of the local protein region around Met80. In addition, NMR and hydrogen exchange (HX) provide structural probes that can measure changes in protein structure and stability at an individual amino acid level. These various probes have been earlier used to characterize the partial unfolding of Cyt c under various solution conditions 22,27-31. Here we examine the role of such partial unfolding in protein aggregation.
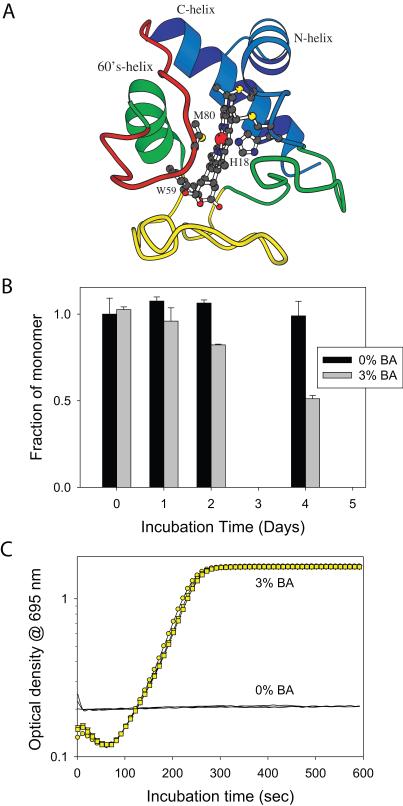
Fig. 1.
BA induces Cyt c aggregation. (A) Structure of Cyt c. Cyt c contains three α-helices and three Ω-loops. Individual cooperatively unfolding regions (foldons) detected in the oxidized Cyt c, at neutral pH and room temperature are colored in terms of their increasing stability: Red, Yellow, Green, and Blue. The structure also shows the residues Met80 and His18, the two axial ligands of the heme, and the single tryptophan residue at position 59. (B) Fraction of soluble monomer remaining in solution after incubation at 37°C with shaking, as determined by the SEC gel filtration. The black and grey bars represent the values without and with 3% v/v BA. (C) Aggregation kinetics of Cyt c at 60°C in the absence (Black curve) and presence of 3% v/v BA (Yellow symbols).
Although Cyt c has been a well-studied model protein for five decades to understand mechanisms of protein folding and stability, its aggregation mechanisms have not been examined. Our results show that BA induces Cyt c aggregation. Increasing BA concentration accelerates aggregation kinetics and decreases the temperature at which Cyt c aggregates. However, BA did not significantly alter Cyt c’s global structure. Instead, it caused partial unfolding of a local protein region around Met80. Selective stabilization of this region decreased the aggregation. These results demonstrate that partial unfolding, rather than global unfolding, of Cyt c results in BA-induced aggregation. This study, for the first time, provides a structural insight into the role of unfolding of local protein regions in alcohol-induced protein aggregation. This work opens a new direction in understanding the effects of alcohols on partially unfolded states (foldons) and their role in protein aggregation. The newly emerging foldon dimension of protein molecules seems to control many other protein behaviors including protein folding and protein function 30.
Materials and Methods
Materials
Equine Cyt c (type VI) was obtained from Sigma Chemical Co. Prior to experiments, protein was oxidized using potassium ferricyanide to remove any trace amounts of the reduced form, dialyzed extensively against the desired buffer, and filtered through 0.22 μm filter. For experiments with the reduced form, Cyt c was reduced with ascorbate. BA was obtained from Merck (Germany) and deuterated BA was from Isotec (Sigma-Aldrich). Optical grade GdmCl was from MP Biomedicals. All other reagents used in this study were of the highest grade available from Fisher Scientific or Sigma Chemicals.
Size Exclusion Chromatography (SEC)
Cyt c (2 mM in 0.1 M sodium phosphate, 0.15 M sodium chloride, pH 7) was incubated at 37°C on a rotator (Thermo Scientific Labquake Shaker Rotisserie) and samples were taken at desired intervals. Concentration of the monomer was estimated by injecting 5 μl onto a TSKgel 5 μm G3000SWxl column on an Agilent 1100 HPLC. The mobile phase used for this column was 0.1 M sodium phosphate, 0.1 M sodium sulfate, pH 6.7 at a flow rate of 0.7 ml/min. Absorbance at 280 nm was used to estimate the protein concentration. The average of 0% v/v BA triplicates on day 0 was used to normalize the peak area of subsequent sample sets.
Thermal scanning method
Changes in the optical density of Cyt c solutions at 695 nm was monitored as a function of increasing temperature using an Agilent 8453 UV-Visible spectrophotometer. Cyt c (300 μM) in 0.1 M sodium phosphate, 0.15 M NaCl, pH 7 was used for these experiments. Solution temperature was raised at a steady rate of 1°C/minute followed by 1 minute equilibration before collecting the signal, and the signal was integrated for 0.5 seconds. First derivative of the optical density spectra was used for calculating the midpoint aggregation temperature (TmAgg), using the software provided with the instrument. The melting temperature of the 695 nm absorbance (Tm695) at a particular BA concentration was determined as the temperature at which the absorbance is half of the native state value in the absence of BA.
Isothermal incubation experiments
Cyt c solution was incubated at the desired temperature, and the changes in optical density at 695 nm were measured as a function of the incubation time. For these experiments, the cuvette of buffer solution was initially equilibrated at the desired temperature, and Cyt c (300 μM final concentration) was added to the cuvette. The aggregation kinetics was monitored until the signal reached a plateau. At longer incubation times, the aggregates started settling down to the bottom of the cuvette, resulting in decreased optical density. At that point, the experiment was stopped.
Denaturant melts
Guanidinium Chloride (GdmCl) was used as the denaturant. Protein solutions at varying GdmCl concentrations were prepared and equilibrated overnight before measuring changes in optical signals as a function of denaturant concentration. Concentration of the denaturant was determined using refractive index measurements 32. Changes in fluorescence signal were measured using 5 μM Cyt c in 20 mM Tris, 150 mM NaCl, pH 7 with the excitation and emission wavelengths set at 280 nm and 349 nm respectively, using either a PTI or a SPEX Fluorolog-3 fluorometer. For measuring changes in the 695 nm absorbance, 300 μM Cyt c in 0.1 M Tris, 0.15 M NaCl, pH 7 was used. The ΔG values were determined by fitting the changes in optical signals at different denaturant concentrations to a 2-state unfolding model 33.
FT-IR experiments
IR spectra of Cyt c solutions (1 mM in 0.1 M sodium phosphate, 0.15 M NaCl, H2O, pH 7, room temperature) were recorded using a Biomem MB-series FT-IR spectrometer (ABB Biomem Inc. Canada) and CaF2 cell (Biotools, USA). For each sample, 128 scans were acquired in single beam mode with 4 cm−1 resolution. IR absorbance spectra were processed using GRAMS/AI 7.00 software (Thermo Galactic, Thermo Electron Co., USA).
NMR titration experiments
2D gradient COSY experiments were run to monitor changes in Cyt c amide crosspeaks as a function of BA concentration using a Varian Inova 500 MHz NMR instrument. For these experiments, 3 mM Cyt c (0.1 M sodium phosphate, 0.15 M NaCl, pH 7) and deuterated BA (1-3% v/v) were used. NMR spectra were collected with 8000 Hz spectral width and 512 points in each direction. The spectra were processed in magnitude mode using the nmrPipe software (Delaglio) with zero filling to twice their real points, apodization with nonshifted sine multiplication, exponential broadening, and Gaussian transformation. The spectra were base line corrected in both dimensions. Changes in the peak positions (chemical shifts) and peak volumes were calculated using the nmrDraw package. Cyt c residue assignments available in literature 34 were used for this purpose. 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) was used as a standard to reference the chemical shifts as well as to normalize the measured changes in crosspeak volumes with the addition of BA.
HX
Cyt c (3 mM) whose amides were protons was lyophilized in deionized water. It was dissolved in the appropriate buffer in D2O just before running the HX experiment and filtered through 0.22 μm filter. A series of 2D gradient COSY was recorded back to back using a Varian Inova 600 MHz NMR instrument equipped with a cryoprobe to monitor the HX. NMR spectra were processed using nmrPipe software and the changes in peak volumes due to exchange were measured using nmrDraw software. These were normalized with the intensities of five non-exchangeable crosspeaks (C6H-C7H & C4H-C5H of Trp59, C2H-C3H of Tyr74, CγH - CδH3 of Leu64, and CγH - CδH3 of Leu98). The changes in peak intensities were fitted to a single exponential equation to obtain the exchange rate constants (kex). Using the calibrated HX rate constants in the unfolded state (kchem) calculated using Excel spreadsheets available at http://hx2.med.upenn.edu, the stabilities of the protecting structures were determined using the equation ΔG = −RT ln(kex/kchem) where R is the universal gas constant and T is the absolute temperature 25. For this calculation, we assumed that the HX rates in the protein unfolded state were unaffected by the minimal concentrations of BA (1-3% v/v) used in this study.
Results
BA aggregates Cyt c
We first tested whether BA induces Cyt c aggregation. Using size-exclusion chromatography (SEC) in combination with HPLC, we measured Cyt c monomer concentration as a function of the time the protein was kept shaking at 37 °C in isothermal incubation experiments (Fig. 1B). This temperature was chosen because it is physiologically relevant and also to accelerate the aggregation kinetics compared to room temperature. There was no change in the monomer concentration over a time period of 4 days in the absence of BA whereas the monomer concentration sharply decreased in the presence of 3% v/v BA, indicating that BA induces the aggregation of Cyt c.
Since BA aggregates Cyt c on a time scale of days, we accelerated the aggregation kinetics by increasing the temperature so that the aggregation mechanisms can be probed on a convenient laboratory timescale. We measured protein aggregation kinetics at a fixed temperature (60°C) in terms of the increase in solution turbidity as a function of the incubation time (Fig. 1C). For these measurements, 695 nm wavelength was used such that we can simultaneously measure unfolding of the local protein region around Met80 in addition to protein aggregation. The 695 nm absorbance initially decreased with increasing incubation time. This was followed by a gradual increase in the optical density due to increased solution turbidity because of protein aggregation. At longer incubation times, optical density decreased (data not shown) as protein aggregates start settling down to the bottom of the cuvette. The observation that the decrease in 695 nm absorbance preceded the increase in optical density indicates that the local protein region around Met80 might unfold before protein aggregates, which was confirmed as described below.
BA decreases the temperature at which Cyt c aggregates
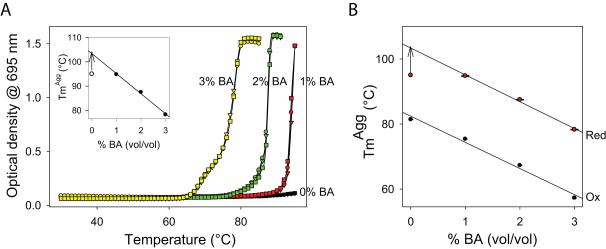
We followed the effect of BA on the temperature at which Cyt c aggregates using a thermal scanning method. The temperature of the solution was increased at a steady rate of 1°C/min, and protein aggregation was estimated by measuring the increase in solution turbidity (Fig. 2A). As before, 695 nm wavelength was used for these measurements to simultaneously measure unfolding of the local protein region around Met80 and protein aggregation. In buffer without BA, the 695 nm absorbance initially decreased as the temperature increased, indicating unfolding of the Met80 region. This was followed by an increase in the optical density at temperatures above 75°C due to increase in solution turbidity because of protein aggregation. When the same thermal transition was monitored using 800 nm wavelength where the protein does not absorb, we observed only the later part of the transition, indicating that the increase in optical density at 695 nm at higher temperatures (Fig. 2A) is because of protein aggregation and not due to increase in the 695 nm absorbance. With the increase in BA concentration, there was a significant decrease in the 695 nm absorbance even at 30°C. Also, 695 nm absorbance melted at lower temperatures in the presence of BA, indicating that BA decreased stability or enhanced unfolding of the local protein region around Met80. The proceeding thermal aggregation followed the same trend, i.e., Cyt c aggregated at lower temperatures in the presence of BA. The midpoint temperature of Cyt c aggregation (TmAgg) decreased from 81°C in 0% v/v BA to 57°C in 3% v/v BA with a linear slope of 8°C/ % v/v BA (Fig. 2B & Table 1). The aggregation temperature decreased linearly with the decrease in the melting temperature of the 695 nm absorbance (Tm695 nm) upon BA addition (Fig. 2C). These results indicate that the unfolding of the local protein region around Met80 by BA promotes protein aggregation, and thus this region acts as a ‘hot-spot’ for triggering protein aggregation.
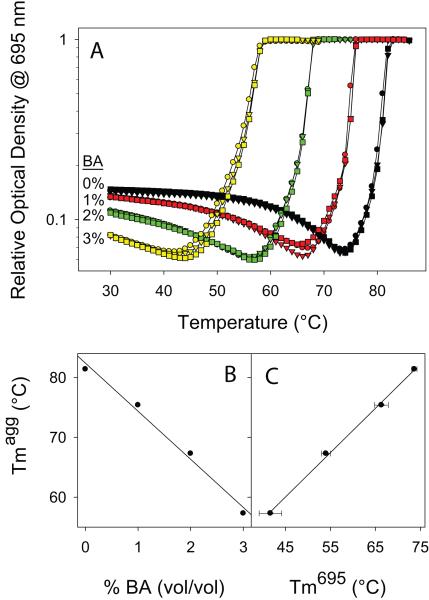
Fig. 2.
BA-induced Cyt c aggregation depends on the unfolding of a local protein region around Met80. (A) Thermal denaturation and aggregation of Cyt c as monitored by the optical density at 695 nm. Three curves were shown for each BA concentration. (B) The midpoint aggregation temperature (TmAgg) decreases linearly at a rate of 8.0 ± 0.1°C/ % v/v BA. Individual TmAgg values are given in Table 1. (C) The TmAgg decreases linearly with the decrease in the melting temperature of the 695 nm absorbance (Tm695), which is an indicator of unfolding of the Met80 region.
Table 1.
Aggregation midpoint temperatures (TmAgg) of oxidized and reduced Cyt c at different BA concentrations. TmAgg was determined from the peak position in the first derivative spectra of the optical density increase with temperature shown in Figs. 2A & 7A. For reduced Cyt c, complete transition was not observed for 0% v/v and 1% v/v BA, and for 3% v/v BA, a minor transition was observed in addition to the major transition. TmAgg for reduced Cyt c at 1% v/v BA was determined assuming that the increase in optical density reaches the same plateau value as that of 2% v/v and 3% v/v BA. The error bar on the TmAgg value was estimated from the three curves shown for each BA concentration.
| Protein | [BA] (v/v) | TmAgg (°C) |
|---|---|---|
| Cyt c Oxidized |
0 % | 81.4 ± 0.1 |
| 1 % | 75.4 ± 0.1 | |
| 2 % | 67.3 ± 0.1 | |
| 3 % | 57.3 ± 0.1 | |
| Cyt c Reduced |
0 % | > 95 |
| 1 % | 94.8 ± 0.2 | |
| 2 % | 87.5 ± 0.1 | |
| 3 % | 69.0 ± 0.4 78.3 ± 0.1 |
BA unfolds the local protein region around Met80
Since 695 nm absorbance was clearly affected by BA even at 30°C (Fig. 2A), we examined changes in the 695 nm absorbance as a function of BA concentration at room temperature (Fig. 3A). Consistent with the results in Fig. 2A, the 695 nm absorbance decreased with increasing BA concentration indicating unfolding of the local protein region around Met80. Under the same conditions, no change in the native state fluorescence was observed indicating that the addition of BA does not increase the Trp59 to heme distance to an extent that results in increased fluorescence, implying that BA does not perturb the overall global protein structure.
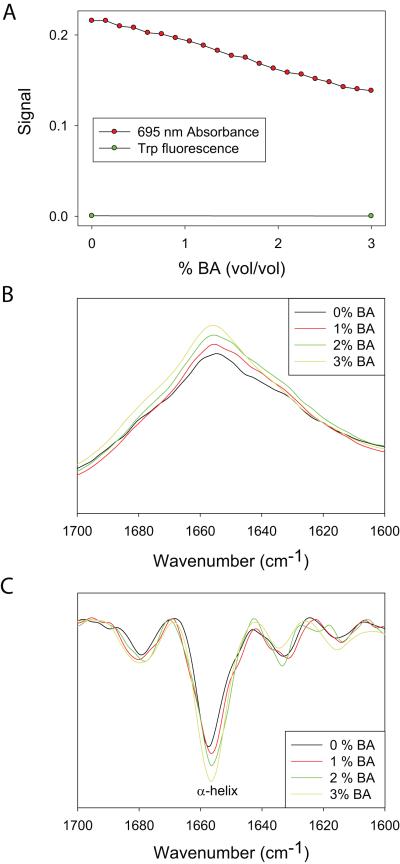
Fig. 3.
Effect of BA on Cyt c global and local structure. (A) Changes in the fluorescence and 695 nm absorbance. With the addition of BA, 695 nm absorbance decreased, whereas there was no change in the native state fluorescence. (B) Changes in the FT-IR spectra and (C) its second derivative. With the addition of BA, α-helix content (1654 cm−1) increases.
We were unable to use CD signal at 222 nm to measure changes in the protein secondary structure with the addition of BA, because BA strongly absorbs in this wavelength region. Alternatively, we monitored the secondary structure change using vibrational bands in the amide I region of the FT-IR spectrum (Fig. 3B&C). Although individual secondary structures absorb over multiple wavelengths, they have characteristic absorption bands in the second derivative absorption spectra 35. The band at 1654 cm−1 is a signature of α-helices. Cyt c, being an α-helical protein, showed an intense band at 1654 cm−1 (Fig. 3C). With the addition of BA, the α-helical content (1654 cm−1) increased significantly.
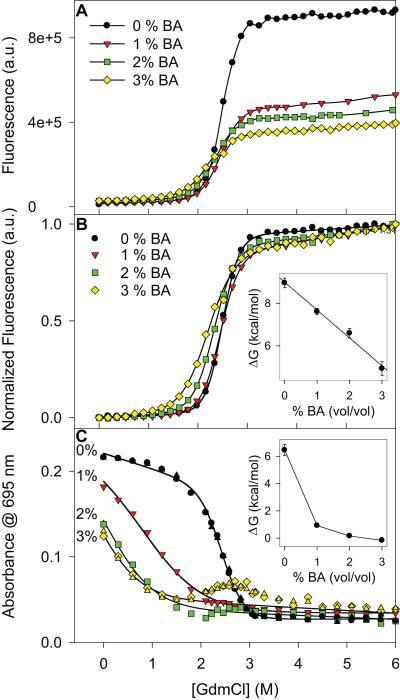
We examined the BA effects on the global and local stability using denaturant melts with GdmCl as the denaturant. As mentioned above, we could not use CD at 222 nm because BA strongly absorbs in this wavelength region. Alternatively, we used tryptophan fluorescence to measure the changes in global stability. The equilibrium denaturant melts did not show significant variation at different BA concentrations (Fig. 4A). Cyt c retained its overall global structure as there was no change in the native fluorescence signal with the addition of BA at zero denaturant concentration. Fluorescence of the unfolded state decreased at higher BA concentrations (Fig. 4A), probably due to quenching by BA or due to the formation of residual structure in the unfolded state upon adding BA leading to tryptophan fluorescence quenching by the heme. The normalized melting curves (Fig. 4B) showed a slight shift in the midpoint of the transition to lower denaturant values and a decrease in the steepness of the melt with increasing BA concentration, indicating a decrease in global stability with the addition of BA. Assuming that only native and unfolded states are present at equilibrium with no intermediates at all denaturant concentrations (2-state folding), the change in global stability was determined by fitting the melting curves to a 2-state model 33. The Gibb’s free energy ΔG varied linearly with the BA concentration with a slope of 1.3 kcal/mol/% v/v BA (Fig. 4B inset). Importantly, since there was no change in the native fluorescence signal after the addition of BA in the absence of denaturant, no global structural change occurred, consistent with the results in Fig. 3A. In contrast to fluorescence, 695 nm absorbance showed a drastic variation with the addition of BA (Fig. 4C). In buffer without denaturant, a clear decrease in 695 nm absorbance was seen with the addition of BA (similar to Figs. 2A & 3A), indicating decreased stability or enhanced unfolding of the local region around Met80. With increasing BA concentration, the 695 nm absorbance melted at lower denaturant concentrations, again indicating decreased stability of the local protein region around Met80. Assuming that the 695 nm absorbance has the same value in the native state in the absence and presence of BA, fitting the melting curves to a 2-state equation resulted in a sharp decrease in ΔG, even with just 1% v/v BA concentration (Fig. 4C inset). It is interesting to note that at higher denaturant concentrations in the presence of 2% or 3% v/v BA, 695 nm absorbance seems to reappear at 2 M GdmCl concentration and disappear at concentrations above 3.5 M (Fig. 4C). This artifact is because of the increased solution turbidity (measured by light scattering) in overnight equilibrated protein samples at these particular BA and denaturant concentrations.
Fig. 4.
Effect of BA on Cyt c global and local stability. (A) Changes in the fluorescence signal as a function of GdmCl concentration at different BA concentrations. (B) Normalized curves shown in panel A. Inset shows the ΔG values obtained by fitting these curves to a Santoro-Bolen 2-state equation. Global ΔG varies linearly with a slope of 1.3 ± 0.1 kcal/mol/% v/v BA. (C) Changes in the 695 nm absorbance with GdmCl concentration at different BA concentrations. Two sets of data points were shown for 0% and 3% v/v BA concentrations. Inset shows the variation in ΔG of the Met80 region obtained by fitting to a 2-state equation, assuming that the 695 nm absorbance has the same value in the native state at all BA concentrations.
BA increases dynamics of the local protein region around Met80
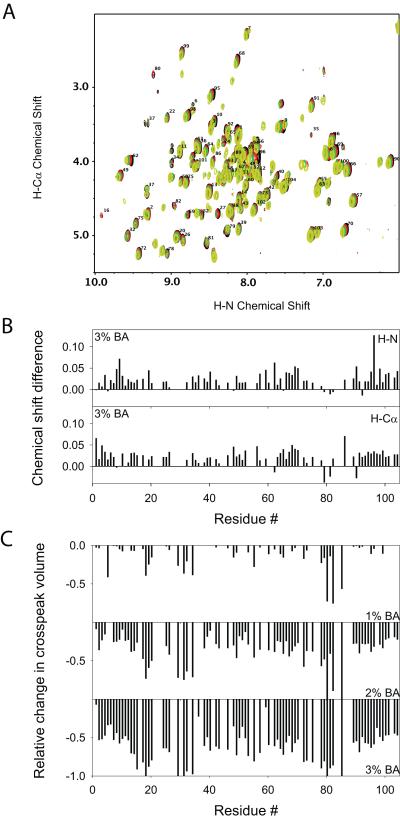
We monitored changes in the global protein structure using 2D NMR COSY spectra recorded as a function of increasing BA concentration (Fig. 5A). No significant changes were observed in the positions of individual amide crosspeaks, indicating that BA did not perturb the overall global protein structure. The chemical shifts of mainchain amide protons and Cα protons of all residues were within 0.1 ppm (Fig. 5B). Although there was no change in the native state chemical shifts, some new weak peaks appeared, indicating an altered conformation in the presence of BA. When we mapped the locations of the residues for which we can identify the appearance of new peaks based on the disappearance of the adjacent old peak, we did not see any clustering of residues in the protein 3D structure, indicating that no specific binding pockets exist for BA. In relation, isothermal titration calorimetry experiments showed that the enthalpy changes observed as a function of the molar ratio of BA to Cyt c were minimal (below 0.07 kcal/mol) (data not shown) similar to that observed for other proteins 10,19, indicating that BA does not have any strong binding sites on Cyt c and the structural perturbation is small.
Fig. 5.
Effect of BA on Cyt c solution structure and dynamics. (A) Changes in the 2D NMR COSY fingerprint region with the addition of BA. Black, Red, Green, and Yellow represent the spectra at 0%, 1%, 2% and 3% v/v BA respectively. (B) Changes in chemical shifts of protein mainchain protons at 3% BA. H-N and H-Cα represent the protons attached to the amide and α-carbon atoms respectively. (C) Relative changes in H-N to H-Cα crosspeak volumes as a function of BA concentration when compared to those in the absence of BA. 0 represents no change in volume whereas -1 represents 100% decrease in the peak volume (absence of the peak). With the increase in BA concentration, no changes in native chemical shifts were observed indicating no global structural change, whereas significant changes were observed in the crosspeak volumes indicating increased dynamics.
Although chemical shifts did not show significant variation, native state crosspeak volumes decreased with increasing BA concentration (Fig. 5C). At 1% v/v BA, maximum volume changes were observed for residues Met80, Phe82, and Ile85. Decreased crosspeak volumes with no change in chemical shifts indicate that the native state is in exchange with a partially unfolded state in the presence of BA (slow to intermediate NMR exchange regime). The increased dynamics of the Met80 region appeared to be propagated to the other side of the heme group. In Cyt c, Met80 is ligated to the heme on one side whereas His18 is ligated on the other side of the heme. Some significant crosspeak volume changes were seen at residues around His18 and around Asn31. With increased BA concentration, the protein regions that showed large changes in crosspeak volumes expanded. For example, at 3% v/v BA concentration, all residues close to Met80 became more dynamic so that no crosspeaks were seen for many residues in this region, whereas at 1% v/v BA, only the three residues Met80, Phe82 and Ile85 showed significant volume changes compared to other residues in the Met80 region. However, since there was no change in the native state chemical shifts (Figs. 5A & B) or in the native state fluorescence signal (Fig. 3A) even at 3% v/v BA, the protein retained its overall global structure, except that it became more dynamic with the addition of BA.
BA effects on partial unfolding and foldon substructure of Cyt c
Since BA increased the α-helix content (Figs. 3B & C) and at the same time unfolded the protein region around Met80 (Figs. 1C, 2A, 3A & 4C), we probed how BA affects the stabilities of various local protein regions using HX methods. If the induced α-helix is stable over the native structure, the stabilities of those regions where the α-helix is formed should increase. Similarly, the stability of those regions that unfold with BA should decrease. Such study on alcohol effects on the stability of local protein regions or foldon substructure was not performed earlier on any protein; most earlier studies were on monitoring the alcohol effects on global protein structure and stability.
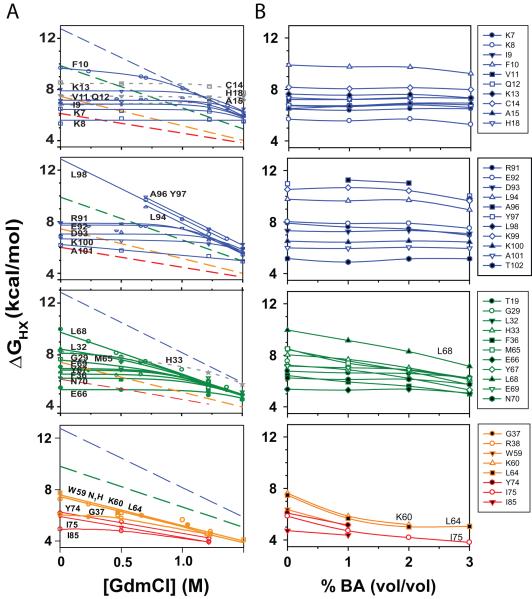
HX in combination with 2D NMR measures the local stability of structures around individual amino acids 25. For a proton to be exchanged with the solvent, it needs to be exposed to the solvent, and the exchange rate is proportional to the stability of the structure that is protecting the amide against exchange. Earlier HX experiments showed that the exchange rates for individual amides change considerably with the addition of denaturant 22,36,37 (Fig. 6A). These experiments indicate that amide hydrogens in Cyt c exchange through three types of unfolding reactions 22,36-38: (1) local unfolding that involves unfolding of only the structure or hydrogen bond protecting that amide, (2) subglobal unfolding where individual protein regions unfold cooperatively, and (3) global unfolding where the entire protein is unfolded. Since local unfolding exposes little or no surface area, their ΔG is invariant with the denaturant concentration, because the slope or m-value is proportional to the surface area exposed upon unfolding. At low denaturant (GdmCl) concentrations, many residues exchange through local unfolding reactions (Fig. 6A), because of their lower stability compared to subglobal and global unfolding reactions. Increase in the denaturant concentration promotes larger unfoldings, such as subglobal and global unfolding reactions. This is because these unfoldings expose significant surface area upon unfolding and hence have larger m-values resulting in a larger decrease in the ΔG with increase in denaturant concentration. After a certain denaturant concentration, ΔG of subglobal and global unfoldings become lower than that of the local unfolding and the amides exchange through these less stable, larger unfolding reactions. Such HX experiments performed as a function of the denaturant concentration have identified individual subglobal cooperatively unfolding regions in Cyt c, known as foldons, where the amides in a single foldon exchanged with the same ΔG as the denaturant concentration is increased (Fig. 6A). In the oxidized state, at neutral pH and room temperature, four foldons were seen, colored in terms of their increasing stability: Red, Yellow, Green, and Blue foldons 30,36,37 (Fig. 1A). Individual foldons contain particular residues, known as stability markers, that exchange only through complete unfolding of the entire foldon. For example, residue Ile75 in the Red foldon exchanges only when the entire Red foldon unfolds. Hence, the stability measured by the residue Ile75 unfolding represents the stability of the Red foldon 36,37. Similarly, Lys60 & Leu64 are stability markers for the unfolding of the Yellow foldon. Leu68 is stability marker for the Green foldon, whereas Ala96, Tyr97, and Leu98 are stability markers for the Blue foldon. Here, we examined the effects of BA on such partial unfolding and foldon substructure of Cyt c.
Fig. 6.
Effect of BA on partial unfolding and foldon substructure of Cyt c. (A) ΔG’s of various residues determined using HX methods as a function of GdmCl concentration (Figure from ref. 22). (B) ΔG’s determined at various BA concentrations. Within a foldon, residues that exchange through local unfolding reactions with denaturant retained their local unfolding character with BA, and residues that exchange through larger unfolding reactions such as unfolding of the entire foldon (subglobal unfolding) with the addition of denaturant showed decreased stability with BA.
Exchange rates for individual residues were measured as a function of BA concentration over a period of 3 months, and converted to ΔGs using standard equations (see Methods) (Fig. 6B). Note that a decrease in ΔG by 0.6 kcal/mol correspond to an increase in the exchange rate by 2.7 times. Within a given foldon, the residues which exchange through local unfolding reactions at low denaturant (GdmCl) concentrations retained their local unfolding character with BA. For example, residues Lys7, Lys8, Ile9, Phe10, Val11, Gln12, Lys13, Cys14, Ala15, and His18 in the Blue N-terminal helix exchanged through local unfolding reactions at low GdmCl concentrations as well as at low BA concentrations (Compare Figs. 6A & B). Their ΔG of unfolding did not change with the addition of BA, indicating that their unfolding with BA did not expose significant surface area. Similar results were obtained for residues in other foldons, except a few residues like Met65 in the Green helix, which exchanges through local unfolding at low GdmCl (zero m-value), but exchanges through an unfolding reaction that exposes significant surface area at low BA concentrations as evident from its non-zero slope of ΔG variation with BA concentration. Similar to denaturant experiments (Fig. 6A), the stability markers for the individual foldons, Ile75 for the Red foldon, Lys60 and Lys64 for the Yellow foldon, and Lys68 for the Green foldon, exchanged with the highest ΔG and with a steeper slope within that foldon with BA addition. This indicates that these residues unfold through larger unfolding reactions in the presence of BA, possibly through the unfolding of the individual foldons. The observation that individual amides exchange through similar unfolding reactions in the presence of denaturant as well as in the presence of BA indicates that at low concentrations, BA behaves like a denaturant. However, a closer examination of the results indicates the differences between an alcohol and a denaturant. ΔG of the Yellow and Red foldon stability markers (Lys60, Leu64, and Ile75), instead of showing a continuous linear decrease with BA, seemed to show a slight upward curvature at higher BA concentrations (Fig. 6B), which might be due to the formation of stable structure in these loop regions. HX experiments with BA could not be performed at higher BA concentrations because of its limited solubility in water (< 4% v/v). In addition, m values for individual foldons with BA do not follow the trend seen with denaturant. With GdmCl, the m values (kcal/mol/M [GdmCl]) of individual foldons increase in the order Red (1.5) < Yellow (2.3) < Green (3.1) < Blue (4.5), which lead to the hypothesis that these foldons unfold sequentially with the addition of denaturant 36 that was later confirmed by ‘stability labeling’ experiments 39. These earlier results indicate that unfolding of the Green foldon with denaturant includes unfolding of the Yellow and Red foldons. In contrary to GdmCl, m-values with BA (kcal/mol/% v/v BA) of individual foldons vary in the order Red (0.8) < Green (0.9) < Yellow (1.2) (We were unable to measure m-value of the Blue foldon because its stability markers did not exchange within 3 months of exchange time). That means, unfolding of the Green foldon with BA does not include unfolding of the Yellow foldon, since m-value, which is proportional to the amount of surface area exposed upon unfolding, of Yellow foldon is higher than that of the Green foldon. The above two observations indicate that BA does not behave exactly like a denaturant. Further examination of these HX results and the effects of alcohols on foldon substructure is needed and is in progress. It is possible that the foldon whose unfolding causes Cyt c aggregation is much smaller than the Red foldon, because the stability measured by Ile75 is much higher than that measured by the 695 nm absorbance that represents the unfolding of just the Met80 region (Fig. 4C). Alternatively, Cyt c aggregation might be controlled by the unfolding of the entire Red foldon, but the conformer that is populated in the presence of BA might protect amides in the Red foldon significantly despite lacking Met80 to heme ligation. In addition, the ΔG’s measured by HX do not correlate with the changes in the observed amide crosspeak volumes (shown in Fig. 5C), indicating that the partially unfolded state populated in the presence of BA has significant protection against HX.
Stabilizing the Met80 region decreases Cyt c aggregation
Our results indicate that BA induces local unfolding of the protein region around Met80 that leads to Cyt c aggregation. Therefore, stabilizing this local region should decrease the protein aggregation, or increase the temperature at which the protein aggregates. The best method that has been demonstrated in literature to very specifically stabilize the Met80 region is by reducing the heme group (Fe+3 to Fe+2), which increases the stability of the Met80-heme bond by an exceptionally large amount of 3.2 kcal/mol 39 with no change in Cyt c’s structure 40. As before, thermal scanning experiments were used to measure the solution turbidity changes due to protein aggregation of reduced Cyt c (Fig. 7A). When compared to the oxidized form, reduced Cyt c aggregated at higher temperatures at all BA concentrations (Compare Fig. 7A with Fig. 2A). In 0% v/v and 1% v/v BA, reduced Cyt c did not show a complete melt up to 95°C. Assuming that in 1% v/v BA, Cyt c aggregation melt reaches the same plateau value as that of 2% v/v and 3% v/v, its TmAgg value was determined. The TmAgg values for reduced Cyt c are given in Table 1 & Fig. 7 Inset, which decreased at a linear rate of 8°C/ % v/v BA. When compared to the oxidized state, the TmAgg values increased to the same extent (ΔTm ~ 20°C) at all BA concentrations. Also, the slope of TmAgg variation with BA (analogous to denaturant m value) for reduced Cyt c is of the same magnitude as that of oxidized Cyt c (Fig. 7B), indicating that the same region controls the aggregation of both forms of Cyt c. These stability labeling results further confirm the earlier conclusion that unfolding of the Met80 region by BA causes Cyt c aggregation.
Fig. 7.
Effect of stability labeling of Met80 region on Cyt c aggregation. (A) Thermal aggregation of reduced Cyt c monitored by changes in the optical density at 695 nm. Three curves were shown for each BA concentration. Black, Red, Green, and Yellow symbols represent the values at the four BA concentrations: 0%, 1%, 2%, and 3% v/v respectively. Reduced form aggregates at a much higher temperature compared to its less stable oxidized form (Fig. 2A) at all BA concentrations. Inset shows that TmAgg decreased at a rate of 8.2 ± 0.1°C/ % v/v BA. Individual TmAgg values are in Table 1. For 0% v/v BA, no melting transition was observed, hence only the lower limit of TmAgg was shown. For 1% v/v BA, TmAgg was determined assuming that it reaches the same plateau value as that of 2% and 3% v/v BA. (B) Variation in the TmAgg of oxidized and reduced Cyt c. Both decrease at the same rate with the addition of BA.
Discussion
Alcohol-induced protein unfolding
Alcohols have been used as ‘co-solvents’ to study the effects of various physical parameters of solvents, such as hydrophobicity, on the structure and stability of proteins 41-43. Addition of alcohols weakens nonlocal hydrophobic interactions and enhances local polar interactions 42-44. This results in destabilization of the protein’s native hydrophobic core and increased formation of local hydrogen bonds resulting in extended secondary structures. However, none of these studies have examined the alcohol effects on partial protein unfolding or foldon substructure of proteins. Our results on Cyt c show that increasing concentrations of BA have minimal effect on the global structure of Cyt c (Figs. 3A, 4A, 5A & 5B), except an increase in the α-helical content (Fig. 3B&C). However, BA specifically unfolds a local protein region around Met80 (Figs. 1C, 2A, 3A & 4C). It is important to note that the BA concentrations employed in this study ranged from 1% to 3% v/v, much lower than other alcohols used in previous studies (typical concentrations of TFE were about 15 – 70% v/v). For example, in a recent study on alcohol effects on Cyt c’s native structure 45, methanol concentrations of up to 80% v/v were used that resulted in a slight increase in the native α-helical structure with a magnitude similar to that observed here for 3% v/v BA (Fig. 3). A possible reason for the significant effects of BA at such low concentrations may be its hydrophobicity (dielectric constants of BA and water are 13.5 46 and 80.1 respectively). Increase in hydrophobicity is a major factor in enhancing the denaturing efficiency of alcohols 42,47-49, besides other factors. In relation, it is notable that BA is not miscible in water at concentrations above 4% v/v.
We have also probed the effect of alcohols on foldon substructure of Cyt c (Fig. 6B). This is particularly important because of the demonstrated role of foldons in controlling many protein behaviors which include protein folding and protein function 22,28-30,50-52, and understanding alcohol effects can explain how alcohols influence protein function. No such study was performed earlier to understand the alcohol effects on partial protein unfolding and foldon substructure. Our results indicate that the Cyt c retains its foldon substructure in the presence of BA, and the individual amides exchange through similar unfolding reactions as that observed with denaturant. However, a closer examination indicate that BA does not behave exactly like denaturant in terms of its effects on m-values of individual foldons and the formation of structure in Yellow and Red foldons. The HX experiments were done under low concentrations of BA (up to 3% v/v) due to its limited miscibility with water (max 4% v/v), and the results will be further examined with other alcohols.
Alcohol-induced protein aggregation
Recent studies indicate that alcohols induce aggregation of both disease and non-disease proteins. Experimental results in literature so far indicate that TFE accelerates the aggregation of Alzheimer’s β-protein 53 and superoxide dismutase 8, ethanol accelerates the aggregation of insulin 9, whereas in the case of non-disease proteins, TFE induces the aggregation of α-chymotrypsin 54,55 and barstar 56. In the case of BA, it induces the aggregation of interferon-γ 16, interleukin-1 receptor antagonist 10, and human granulocyte colony stimulating factor 17. The molecular mechanisms underlying such alcohol-induced protein aggregation were not understood. In particular, it was not clear whether there exits any direct correlation between alcohol-induced protein unfolding and aggregation.
We have used Cyt c to understand the alcohol-induced protein aggregation mechanisms, because of the reason that various optical and structural probes are available to monitor its global and partial unfolding. BA aggregates Cyt c (Figs. 1 & 2), similar to other proteins. BA decreased the temperature at which Cyt c aggregates. Proteins aggregate at higher temperatures because of the enhancement of hydrophobic effect due to increase in the disorder of solvent molecules. It is therefore likely that BA accelerates the aggregation kinetics or makes the protein to aggregate at lower temperatures because it increases the hydrophobicity of the solvent.
To establish the relationship between BA-induced protein unfolding and aggregation, we first studied the effects of BA on Cyt c partial unfolding. BA did not cause global unfolding of the protein, but unfolded a local protein region around Met80 (Figs. 3 & 4). The temperature at which Cyt c aggregates decreased linearly with the decrease in the melting temperature of the Met80 region (Fig. 2C). When this region is stabilized, the aggregation temperature shifted to higher values (Fig. 7). This clearly demonstrates that a mechanism by which BA induces Cyt c aggregation is by partially unfolding the native protein rather than global unfolding. However, since the aggregation is much slower at room temperature even when this partially unfolded state is half populated at equilibrium at higher BA concentrations (Figs. 1B, 2A & 4C), the aggregation is possibly limited by a free energy barrier or the formation of a transient species. It will be interesting to probe the role of such partial unfolding in alcohol-induced protein aggregation with other alcohols and proteins to arrive at general principles of how alcohols induce protein aggregation.
Role of intra-protein stability variation and foldon substructure in protein aggregation
The results presented here show that the decrease in the stability of local protein regions (foldons) in a native protein increases protein aggregation. Increasing concentrations of BA populates a partially unfolded form where protein region around Met80 is unfolded, which increases the protein’s propensity to aggregate. That means, if we determine the stability of local regions in a native protein under aggregating conditions, it might be possible to identify the potential ‘hot-spot’s for protein aggregation. It is interesting to note that the same region around Met80 plays an important role in Cyt c’s folding and function. For example, it is one of the first units to unfold 52,57 and is linked to Cyt c’s alkaline transition 29,58.
Our experimental results can be used to validate computational methods that predict the aggregation ‘hot-spot’s in proteins. Aggregation of proteins is controlled by a few physical properties 59-61, mainly hydrophobicity, secondary structure propensity, electrostatics and hydrogen bonding. Prediction methods developed based on these physical properties, in particular, TANGO 60 (http://tango.crg.es/) and AGGRESCAN 62 (http://bioinf.uab.es/aggrescan/) point to Met80 region as one of the possible aggregation ‘hot-spots (Supplementary Figure). In addition, the 3D profile method ZipperDB (http://services.mbi.ucla.edu/zipperdb/) 63 that scans the crystal structure of the cross-β spine of the hexapeptide NNQQNY against the protein sequence indicates Met80 region as one of the regions that can form intermolecular cross-β structure. In relation, FT-IR spectra of Cyt c aggregates do indicate that these aggregates contain significant intermolecular β-sheet (data not shown). Comparing the results from these computational methods, all the three point to a single region, the hexapeptide Met80-Ile81-Phe82-Ala83-Gly84-Ile85, as the common aggregation ‘hot-spot’. It is interesting to note that the three residues Met80, Phe82, and Ile85 that are part of this hexapeptide became more dynamic with the addition of 1% v/v BA compared to all other residues in Cyt c (Fig. 5C).
Methods to inhibit alcohol-induced protein aggregation
Alcohols such as BA are often used at low levels in multi-dose protein formulations and drug delivery systems to prevent microbial growth 11,64. However, these alcohols have been shown to induce protein aggregation 65. Here, we show that a fundamental physical mechanism underlying such alcohol-induced protein aggregation is partial unfolding of local protein regions. We also show that stabilizing such local regions by site-specific mutations reduces protein aggregation. This might be a useful strategy to adopt in the case of pharmaceutical proteins to counteract alcohol-induced protein aggregation or, in general, any aggregation, i.e., identify the aggregation ‘hot-spot’s and stabilize them using protein engineering methods.
Supplementary Material
Supplementary Figure: Prediction of aggregation ‘hot-spots’ by three computational methods, TANGO (http://tango.crg.es/), AGGRESCAN (http://bioinf.uab.es/aggrescan/), and ZipperDB (http://services.mbi.ucla.edu/zipperdb/). In the case of TANGO and AGGRESCAN, protein regions with positive values represent aggregation ‘hot-spots’, whereas in the case of ZipperDB, regions with negative free energy represent segments with high fibrillation propensity. All the three programs predict Met80 region as the common aggregation ‘hot-spot’.
Acknowledgements
We acknowledge the help of Brooke Hirsch and Shaun Bevers (Biophysics facility) and David Jones (NMR facility). We thank John Carpenter, Walter Englander, David Eisenberg, and David Bain for helpful discussions.
This work was funded by the School of Pharmacy, University of Colorado Denver. Regina Hutchings was supported by a NIH training grant in Pharmaceutical Biotechnology (T32GM008732).
Abbreviations
- BA
benzyl alcohol
- CD
circular dichroism
- Cyt c
Cytochrome c
- FT-IR
Fourier transform infrared spectroscopy
- GdmCl
Guanidinium chloride
- HX
hydrogen exchange
- NMR
nuclear magnetic resonance
- SEC
Size exclusion chromatography
References
- 1.Forman MS, Trojanowski JQ, Lee VM-Y. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nature Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.De Llano JJM, Manning JM. Properties of a recombinant human hemoglobin double mutant: Sickle hemoglobin with Leu-88(β) at the primary aggregation site substituted by Ala. Protein Sci. 1994;3:1206–1212. doi: 10.1002/pro.5560030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrington DJ, Adachi K, Royer WE., Jr. The high resolution crystal structure of deoxyhemoglobin S. J Mol Biol. 1997;272:398–407. doi: 10.1006/jmbi.1997.1253. [DOI] [PubMed] [Google Scholar]
- 5.Ratner RE, Phillips TM, Steiner M. Persistent cutaneous insulin allergy resulting from high molecular weight insulin aggregates. Diabetes. 1990;39:728–733. doi: 10.2337/diab.39.6.728. [DOI] [PubMed] [Google Scholar]
- 6.Thornton CA, Ballow M. Safety of intravenous immunoglobulin. Arch Neurol. 1993;50:135–136. doi: 10.1001/archneur.1993.00540020013009. [DOI] [PubMed] [Google Scholar]
- 7.Fradkin AH, Carpenter JF, Randolph TW. Immunogenicity of aggregates of recombinant human growth hormone in mouse models. J Pharm Sci. 2009;98:3247–3264. doi: 10.1002/jps.21834. [DOI] [PubMed] [Google Scholar]
- 8.Stathopulos PB, Rumfeldt JAO, Scholz GA, Irani RA, Frey HE, Hallelwell RA, Lepock JR, Meiering EM. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc Natl Acad Sci USA. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzwolak W, Grudzielanek S, Smirnovas V, Ravindra R, Nicolini C, Jansen R, Loksztejn A, Porowski S, Winter R. Ethanol-perturbed amyloidogenic self-assembly of insulin: Looking for origins of amyloid strains. Biochemistry. 2005;44:8948–8958. doi: 10.1021/bi050281t. [DOI] [PubMed] [Google Scholar]
- 10.Roy S, Katayama D, Dong A, Kerwin BA, Randolph TW, carpenter JF. Temperature dependence of benzyl alcohol - and 8-anilinonaphthalene-1-sulfonate - induced aggregation of recombinant human interleukin-1 receptor antagonist. Biochemistry. 2006;45:3898–3911. doi: 10.1021/bi052132g. [DOI] [PubMed] [Google Scholar]
- 11.Akers MJ. Considerations in selecting antimicrobial agents for parental product development. Pharm Technol. 1984;8:36–46. [Google Scholar]
- 12.Gordon LM, Sauerheber RD, Esgate JA, Dipple I, Marchmont RJ, Houslay MD. The increase in bilayer fluidity of rat liver plasma membranes achieved by the local anesthetic benzyl alcohol affects the activity of intrinsic membrane enzymes. J Biol Chem. 1980;255:4519–4527. [PubMed] [Google Scholar]
- 13.Regev R, Assaraf YG, Eytan GD. Membrane fluidization by ether, other anesthetics, and certain agents abolishes P-glycoprotein ATPase activity and modulates efflux from multidrug-resistant cells. Eur J Biochem. 1999;259:18–24. doi: 10.1046/j.1432-1327.1999.00037.x. [DOI] [PubMed] [Google Scholar]
- 14.de Marco A, Vigh L, Diamant S, Goloubinoff P. Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol-overexpressed molecular chaperones. Cell Stress & Chaperones. 2005;10:329–339. doi: 10.1379/CSC-139R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair B. Final report on the safety assessment of benzyl alcohol, benzoic acid, and sodium benzoate. Int J Toxicol. 2001;20(4):23–50. doi: 10.1080/10915810152630729. [DOI] [PubMed] [Google Scholar]
- 16.Tobler SA, Holmes BW, Cromwell MEM, Fernandez EJ. Benzyl alcohol-induced destabilization of interferon-γ. J Pharm Sci. 2004;93:1605–1617. doi: 10.1002/jps.10589. [DOI] [PubMed] [Google Scholar]
- 17.Thirumangalathu R, Krishnan S, Brems DN, Randolph TW, Carpenter JF. Effects of pH, temperature, and sucrose on benzyl alcohol - induced aggregation of recombinant human granulocyte colony stimulating factor. J Pharm Sci. 2006;95:1480–1497. doi: 10.1002/jps.20619. [DOI] [PubMed] [Google Scholar]
- 18.Lam XM, Patapoff TW, Nguyen TH. The effect of benzyl alcohol on recombinant human interferon-γ. Pharm Res. 1997;14:725–729. doi: 10.1023/a:1012190120061. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Roy S, Jones LS, Krishnan S, Kerwin BA, Chang BS, Manning MC, Randolph TW, Carpenter JF. Mechanism for benzyl alcohol-induced aggregation of recombinant human interleukin-1-receptor antagonist in aqueous solution. J Pharm Sci. 2004;93:3076–3089. doi: 10.1002/jps.20219. [DOI] [PubMed] [Google Scholar]
- 20.Fink AL. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Folding Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 21.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 22.Maity H, Maity M, Krishna MMG, Mayne L, Englander SW. Protein folding: The stepwise assembly of foldon units. Proc Natl Acad Sci USA. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore GR, Pettigrew GW. Cytochromes c: Evolutionary, structural and physicochemical aspects. Springer-Verlag; Berlin Heidelberg, Germany: 1990. [Google Scholar]
- 24.Pettigrew GW, Moore GR. Cytochromes c. Biological aspects. Springer-Verlag; Berlin Heidelberg, Germany: 1987. [Google Scholar]
- 25.Krishna MMG, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez KM, Schlamadinger DE, Gable JE, Kim JE. Förster resonance energy transfer and conformational stability of proteins. An advanced biophysical module for physical chemistry students. J Chem Ed. 2008;85:1253–1256. doi: 10.1021/ed085p1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell BS, Melenkivitz R, Bren KL. NMR investigation of ferricytochrome c unfolding: detection of an equilibrium unfolding intermediate and residual structure in the denatured state. Proc Natl Acad Sci USA. 2000;97:8312–8317. doi: 10.1073/pnas.150239397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishna MMG, Lin Y, Rumbley JN, Englander SW. Cooperative omega loops in cytochrome c: Role in folding and function. J Mol Biol. 2003;331:29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 29.Maity H, Rumbley JN, Englander SW. Functional role of a protein foldon - An Ω-loop foldon controls the alkaline transition in ferricytochrome c. Proteins: Struct Funct Bioinform. 2006;63:349–355. doi: 10.1002/prot.20757. [DOI] [PubMed] [Google Scholar]
- 30.Englander SW, Mayne L, Krishna MMG. Protein folding and misfolding: Mechanism and Principles. Quarterly Rev Biophysics. 2007;40:287–326. doi: 10.1017/S0033583508004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandi S, Bowler BE. Probing the bottom of a folding funnel using conformationally gated electron transfer reactions. J Am Chem Soc. 2008;130:7540–7541. doi: 10.1021/ja801941r. [DOI] [PubMed] [Google Scholar]
- 32.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 33.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 34.Feng Y, Roder H, Englander SW, Wand AJ, Stefano DLD. Proton resonance assignments of horse ferricytochrome c. Biochemistry. 1989;28:195–203. doi: 10.1021/bi00427a027. [DOI] [PubMed] [Google Scholar]
- 35.Dong A, Huang P, Caughey WS. Protein secondary structures in water from second-derivative amide I infrared spectra. Biochemistry. 1990;29:3303–3308. doi: 10.1021/bi00465a022. [DOI] [PubMed] [Google Scholar]
- 36.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milne JS, Xu Y, Mayne LC, Englander SW. Experimental study of the protein folding landscape: Unfolding reactions in cytochrome c. J Mol Biol. 1999;290:811–822. doi: 10.1006/jmbi.1999.2924. [DOI] [PubMed] [Google Scholar]
- 38.Maity H, Lim WK, Rumbley JN, Englander SW. Protein hydrogen exchange mechanism: local fluctuations. Protein Sci. 2003;12:153–160. doi: 10.1110/ps.0225803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Mayne L, Englander SW. Evidence for an unfolding and refolding pathway in cytochrome c. Nature Struct Biol. 1998;5:774–778. doi: 10.1038/1810. [DOI] [PubMed] [Google Scholar]
- 40.Banci L, Bertini I, Huber JG, Spyroulias GA, Turano P. Solution structure of reduced horse heart cytochrome c. J Biol Inorg Chem. 1999;4:21–31. doi: 10.1007/s007750050285. [DOI] [PubMed] [Google Scholar]
- 41.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 42.Thomas PD, Dill KA. Local and nonlocal interactions in globular proteins and mechanisms of alcohol denaturation. Protein Science. 1993;2:2050–2065. doi: 10.1002/pro.5560021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buck M. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Quarterly Rev Biophysics. 1998;31:297–355. doi: 10.1017/s003358359800345x. [DOI] [PubMed] [Google Scholar]
- 44.Dill KA, Bromberg S, Yue K, Fiebig KM, Yee DP, Thomas PD, Chan HS. Principles of protein folding - A perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamatari YO, Konno T, Kataoka M, Akasaka K. The methanol-induced globular and expanded denatured states of cytochrome c: A study by CD Fluorescence, NMR and small-angle X-ray scattering. J Mol Biol. 1996;259:512–523. doi: 10.1006/jmbi.1996.0336. [DOI] [PubMed] [Google Scholar]
- 46.Wohlfarth C. Landolt-Börnstein - Group IV Physical Chemistry Supplement to IV/6. Volume 17. Springer; Berlin Heidelberg: 2008. Dielectric constant of benzyl alcohol; p. 396. [Google Scholar]
- 47.Nelson JW, Kallenbach NR. Stabilization of the ribonuclease S-peptide α-helix by trifluoroethanol. Proteins Struct Funct Genet. 1986;1:211–217. doi: 10.1002/prot.340010303. [DOI] [PubMed] [Google Scholar]
- 48.Hirota N, Mizuno K, Goto Y. Cooperative alpha-helix formation of beta-lactoglobulin and melittin induced by hexafluoroisopropanol. Protein Sci. 1997;6:416–421. doi: 10.1002/pro.5560060218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong D-P, Hoshino M, Kuboi R, Goto Y. Clustering of fluorine-substituted alcohols as a factor responsible for their marked effects on proteins and peptides. J Am Chem Soc. 1999;121:8427–8433. [Google Scholar]
- 50.Krishna MMG, Lin Y, Mayne L, Englander SW. Intimate view of a kinetic protein folding intermediate: Residue-resolved structure, interactions, stability, folding and unfolding rates, homogeneity. J Mol Biol. 2003;334:501–513. doi: 10.1016/j.jmb.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 51.Krishna MMG, Englander SW. The N-terminal to C-terminal motif in protein folding and function. Proc Natl Acad Sci USA. 2005;102:1053–1058. doi: 10.1073/pnas.0409114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishna MMG, Maity H, Rumbley JN, Lin Y, Englander SW. Order of steps in the cytochrome c folding pathway: Evidence for a sequential stabilization mechanism. J Mol Biol. 2006;359:1411–1420. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 53.Fezoui Y, Teplow DB. Kinetic studies of amyloid β-protein assembly. Differential effects of α-helix stabilization. J Biol Chem. 2002;277:36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 54.Pallarés I, Vendrell J, Avilés FX, Ventura S. Amyloid fibril formation by a partially structured intermediate state of α-chymotrypsin. J Mol Biol. 2004;342:321–331. doi: 10.1016/j.jmb.2004.06.089. [DOI] [PubMed] [Google Scholar]
- 55.Rezaei-Ghaleh N, Ebrahim-Habibi A, Moosavi-Movahedi AA, Nemat-Gorgani M. Role of electrostatic interactions in 2,2,2-trifluoroethanol-induced structural changes and aggregation of α-chymotrypsin. Arch Biochem Biophys. 2007;457:160–169. doi: 10.1016/j.abb.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 56.Kumar S, Udgaonkar JB. Structurally distinct amyloid protofibrils form in separate pathways of aggregation of a small protein. Biochemistry. 2009;48:6441–6449. doi: 10.1021/bi900682w. [DOI] [PubMed] [Google Scholar]
- 57.Hoang L, Bédard S, Krishna MMG, Lin Y, Englander SW. Cytochrome c folding pathway: Kinetic native-state hydrogen exchange. Proc Natl Acad Sci USA. 2002;99:12173–12178. doi: 10.1073/pnas.152439199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bandi S, Baddam S, Bowler BE. Alkaline conformational transition and gated electron transfer with a Lys 79 → His variant of iso-1-cytochrome c. Biochemistry. 2007;46:10643–10654. doi: 10.1021/bi700992y. [DOI] [PubMed] [Google Scholar]
- 59.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 60.Fernandez-Escamilla A-M, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nature Biotech. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 61.de Groot NS, Pallarés I, Avelés FX, Vendrell J, Ventura S. Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct Biol. 2005;5:18. doi: 10.1186/1472-6807-5-18. doi:10.1186/1472-6807-1185-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conchilli-Solé O, de Groot NS, Avilés FX, Vendrell J, Daura X, Ventura S. AGGRESCAN: A server for the prediction and evaluation of “hot-spots” of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldschmidt L, Teng PK, Riek R. Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer BK, Ni A, Hu B, Shi L. Antimicrobial preservative use in parenteral products: Past and present. J Pharm Sci. 2007;96:3155–3167. doi: 10.1002/jps.20976. [DOI] [PubMed] [Google Scholar]
- 65.Maa YF, Hsu CC. Aggregation of recombinant human growth hormone induced by phenolic compounds. Int J Pharm. 1996;140:155–168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure: Prediction of aggregation ‘hot-spots’ by three computational methods, TANGO (http://tango.crg.es/), AGGRESCAN (http://bioinf.uab.es/aggrescan/), and ZipperDB (http://services.mbi.ucla.edu/zipperdb/). In the case of TANGO and AGGRESCAN, protein regions with positive values represent aggregation ‘hot-spots’, whereas in the case of ZipperDB, regions with negative free energy represent segments with high fibrillation propensity. All the three programs predict Met80 region as the common aggregation ‘hot-spot’.