Abstract
Background
Human uterine leiomyomas (ULM) are characterized by dysregulation of a large number of genes and non-coding regulatory microRNAs. In order to identify microRNA::mRNA associations relevant to ULM pathogenesis, we examined global correlation patterns between the altered microRNA expression and the predicted target genes in ULMs and matched myometria.
Methodology/Principal Findings
Patterns of inverse association of microRNA with mRNA expression in ULMs revealed an involvement of multiple candidate pathways, including extensive transcriptional reprogramming, cell proliferation control, MAP kinase, TGF-β, WNT, JAK/STAT signaling, remodeling of cell adhesion, and cell-cell and cell-matrix contacts. We further examined the correlation between the expression of the selected target gene protein products and microRNAs in thirty-six paired sets of leiomyomas and matched myometria. We found that a number of dysregulated microRNAs were inversely correlated with their targets at the protein level. The comparative genomic hybridization (CGH) in eight ULM patients revealed that partially shared deletions of two distinct chromosomal regions might be responsible for loss of cancer–associated microRNA expression and could thus contribute to the ULM pathogenesis via deregulation of target mRNAs. Last, we functionally tested the repressor effects of selected cancer-related microRNAs on their predicted target genes in vitro.
Conclusions/Significance
We found that some but not all of the predicted and inversely correlated target genes in ULMs can be directly regulated by microRNAs in vitro. Our findings provide a broad overview of molecular events underlying the tumorigenesis of uterine ULMs and identify select genetic and regulatory events that alter microRNA expression and may play important roles in ULM pathobiology by positively regulating tumor growth while maintaining the non-invasive character of ULMs.
Introduction
Uterine leiomyomas (ULMs) are the most common benign smooth muscle tumors in women of reproductive age. About 40% of ULMs contain non-random chromosomal anomalies involving a small number of specific chromosomal regions [1]. Global gene expression profiling of ULMs revealed that hundreds of genes are dysregulated including those with functional roles in cell proliferation, differentiation and extracellular matrix production [2]. So far, only a few specific genes or cytogenetic aberrations have been identified to be associated with ULMs. While many of the dysregulated genes may function as either effectors or promoters of ULMs growth, they are likely secondarily induced and indirectly responsible for tumor growth into morbid and symptomatic ULMs.
MicroRNAs are a class of small, non-coding regulatory RNAs. In cells, transcribed microRNA precursors (pri-microRNAs) undergo multistep biogenesis to form mature microRNAs of 18–25 nucleotides in length [3]. MicroRNAs regulate a high number of biological processes including cell proliferation, differentiation and cell death during development by sequence-specific targeting of particular mRNAs [4] and are aberrantly expressed in many solid tumors [5], [6], [7], [8], [9], [10] including uterine ULMs [11], [12], [13]. According to miRBase (Release 15), a total of 940 human microRNAs have been identified [14]. Cloning approaches and computational predictions have indicated that there are possibly even more microRNA encoding loci in the human genome [15]. Largely based on ectopic expression experiments and computational algorithms for prediction of microRNA target sites in mRNA sequences, it is estimated that each microRNA may regulate hundreds of genes at the post-transcriptional and translational levels [16].
Recently, studies revealed that a subset of microRNAs are significantly dysregulated in ULMs compared to matched myometria [11], [12], [13]. Many of these microRNAs are also associated with other neoplasms, indicative of their roles in tumorigenesis in general [13]. It is thus important to establish the roles played by the highly dysregulated microRNAs in ULMs pathogenesis through regulation of specific target genes with key roles in tumorigenesis. We and others demonstrated that the let-7 microRNA family could functionally repress HMGA2 expression [13], [17], [18], [19]. These findings prompted us to explore the broader relationship between other dysregulated microRNAs and their target genes exhibiting aberrant expression in ULMs.
In this study, we compared global microRNA expression patterns with the expression of their predicted target genes, at both mRNA and protein levels, in paired sets of ULMs and matched myometria, focusing primarily on the inverse association between the levels of microRNAs dysregulated in ULMs and the expression of their predicted target genes. We found that in uterine ULMs the levels of the most dysregulated microRNAs show an inverse association with the expression levels of many predicted target genes, and that they may affect multiple homeostatic pathways and functions. Next, by correlating comparative genomic hybridization (CGH) results and microRNAs data, we found that dysregulation of certain microRNAs with established roles in cancer could be due to the underlying genomic alterations. Finally, we show that selected predicted target genes can be validated as functional targets of specific microRNAs in vitro. In summary, our study offers a catalogue of microRNA, mRNA and protein expression alterations genetically or functionally related to the pathogenesis of human uterine ULMs.
Materials and Methods
Patients and Tissue Samples
This study included 55 uterine ULMs of usual type from 41 patients, as published elsewhere [13]. All clinical information is well documented. Among 55 ULMs, 36 ULMs and matched myometria were collected to prepare high density tissue microarray (TMA) from formalin-fixed paraffin embedded tissue (FFPE), and 24 ULMs and matched myometria were collected to prepare total RNA from snap frozen tissue, and used for RT-PCR validation. Among them, eight were selected for comparative genomic hybridization and five cases were selected for gene expression analysis (see below). The patient and tissue sample information is summarized in Table 1. The study was approved by the New York University and Northwestern University institutional review boards.
Table 1. Summary of patient and tissue sample information.
| Case No. | Ethnic | Age (yrs) | Uterine weight (gm) | Tumor size (cm) | No. Tumors (n) | Profile | TMA | RT-PCR | ||
| miRNA* | mRNA | CGH | ||||||||
| C4 | Black | 45 | 750 | 10.5 | 5 | yes | yes | yes | yes | Yes |
| C7 | Black | 48 | 850 | 11 | 5 | yes | yes | yes | yes | |
| C18 | Black | 48 | 2500 | 12 | 30 | yes | yes | yes | yes | yes |
| C19 | Black | 48 | 3800 | 24 | 8 | yes | yes | yes | yes | yes |
| C21 | Black | 43 | 1000 | 9 | 11 | yes | yes | |||
| C32 | Black | 35 | 1300 | 17 | 6 | yes | yes | yes | yes | yes |
| C36 | Black | 42 | 1050 | 15 | 42 | yes | yes | yes | yes | yes |
| C41 | Black | 51 | 900 | 9 | 50 | yes | yes | yes | yes | |
| C51 | Black | 39 | 2210 | 14 | 105 | yes | yes | yes | yes | |
| C52 | Black | 52 | 5400 | 26 | 20 | yes | yes | yes | ||
| C56 | Black | 50 | 600 | 12 | 10 | yes | yes | yes | ||
| C57 | Black | 50 | 1140 | 8 | 102 | yes | yes | yes | ||
| C58 | Black | 50 | 440 | 6 | 52 | yes | ||||
| C59 | Black | 45 | 950 | 10 | 20 | yes | ||||
| C68 | Black | 50 | 840 | 8 | 10 | yes | ||||
| C6 | White | 52 | 1200 | 17 | 3 | yes | yes | yes | ||
| C9 | White | 38 | 450 | 12 | 2 | yes | yes | yes | ||
| C10 | White | 52 | 1200 | 11 | 10 | yes | yes** | yes | ||
| C13 | White | 44 | 475 | 11.5 | 3 | yes | yes | yes | ||
| C15 | White | 38 | 2100 | 14 | 5 | yes | yes | yes | ||
| C20 | White | 46 | 1200 | 11 | 2 | yes | yes** | yes | ||
| C22 | White | 53 | 1875 | 14 | 5 | yes | yes | yes | ||
| C26 | White | 46 | 650 | 8 | 25 | yes | yes | yes | ||
| C29 | White | 56 | 1500 | 11 | 5 | yes | yes | yes | ||
| C46 | White | 51 | 1300 | 11 | 51 | yes | yes | yes | ||
| C47 | White | 47 | 1150 | 12 | 15 | yes | yes | yes | ||
| C53 | White | 54 | 2400 | 20 | 3 | yes | yes | |||
| C61 | White | 48 | 1200 | 10 | 10 | yes | yes | |||
| C66 | White | 45 | 850 | 11 | 3 | yes | yes | |||
| C67 | White | 54 | 650 | 8 | 3 | yes | ||||
| C3 | Asian | 44 | 800 | 7 | 22 | yes | ||||
| C24 | Asian | 42 | 890 | 13 | 2 | yes | yes | |||
| C27 | Asian | 56 | 950 | 10.5 | 5 | yes | yes | |||
| C24 | Asian | 40 | 1100 | 13 | 12 | yes | ||||
| C40 | Asian | 41 | 950 | 8 | 25 | yes | yes | |||
| C42 | Asian | 51 | 900 | 12 | 5 | yes | yes | |||
| C1 | Hisp | 41 | 1100 | 11 | 10 | yes | yes | |||
| C5 | Hisp | 48 | 550 | 11 | 6 | yes | yes | |||
| C14 | Hisp | 48 | 1100 | 14 | 5 | yes | yes | |||
| C16 | Hisp | 50 | 2100 | 11 | 40 | yes | yes | |||
| C23 | Hisp | 47 | 450 | 8.5 | 2 | yes | ||||
*Wang et al. 2007.
**Two tumors (Large and small).
Cell lines
Four uterine smooth muscle cell lines were used for the study. They include two immortalized uterine leiomyoma cell lines, ULM-3401 (obtained from an intramural leiomyoma of 45 year old African American woman), immortalized by the introduction of stable human telomere terminal transferase expression (hTERT) and UtLM, obtained from Dr. Dixon's lab (for details please see reference [20]). ULM-3401 exhibits a high level of HMGA2 expression. Two uterine leiomyosarcoma cell lines SK-LMS-1 and SK-UT-1 were obtained from ATCC.
Gene expression analysis
The mRNA expression profiles of 5 ULMs and matched myometria were examined. Affymetrix® HG-U133A GeneChips were used to generate expression data for 22,000 probe sets identifying 18,400 transcripts of 14,500 genes. ULMs and matched myometrial tissues were homogenized in lysis buffer (Ambion, Austin, TX). Following the manufacturer's protocol, total RNA extraction and clean-up were performed using an Ambion RNA purification kit (Ambion, Austin, TX). RNA samples were processed following the Affymetrix protocol (Affymetrix Inc., Santa Clara, CA). In brief, 8 µg of total RNA was amplified, biotin labeled, and hybridized to the Affymetrix HG-U133A GeneChips (Affymetrix Inc.). The gene expression data from ULMs previously published by Hoffman et al [21] were obtained from the NCBI Gene Expression Omnibus database under the accession number GSE593, with sample IDs GSM9093 through GSM 9102. This data set was used as reference to compare with our gene profiling data. The raw array data generated in this study are deposited in NCBI GEO database under the identifier GSE 23112.
Comparative genomic hybridization (CGH)
Eight ULMs and matched myometria from African American women were selected for array CGH analysis (Table 1). Genomic DNA from ULMs were used as the “test” DNA and matched myometria as the “reference” DNA during the hybridization reaction. Human Genome CGH 44K Microarrays (ID:014950) from Agilent Technologies were used according to the instructions of the array manufacturer. Briefly, the test and reference DNAs were digested with Alu I and Rsa I (Promega), and purified with the QIAprep Spin Miniprep kit (Qiagen). Test DNA (500 ng) and reference DNA (500 ng) l were labeled with either Cy3-dUTP or Cy5-dUTP (Perkin Elmer) using the Bioprime Array CGH Genomic Labeling kit (Invitrogen), and hybridized with 2× Hybridization buffer (Agilent, Palo Alto, CA), 10× blocking agent (Agilent,), and Human Cot-1 DNA (Invitrogen) in an Agilent SureHyb chamber for 24 hours at 65°C. After four washing steps, all slides were scanned by Agilent Scanner (G2565CA). Raw data were obtained by Agilent Feature extraction software 9.0, and then imported into Agilent CGH analytics 3.5 software for analysis.
DNA copy number changes were detected by CGH analytics software 3.5 (Agilent). The ADM-1 statistical algorithm was used with a sensitivity threshold of 6.0 and a moving average window of 1 Mb. In order to determine that there was a copy number change in a particular locus, three criteria had to be met. These were positive calls by the software, presence of 10 consecutive probes pointing in the same direction, and 1.5 fold average fold difference in the test DNA compared to the reference DNA.
Expression data and bioinformatics analyses
GeneSpring GX11 (Agilent Technologies, Palo Alto, USA) and TM4 Microarray Software Suite [22] were used to identify differentially expressed mRNAs in ULMs. The raw array intensities (CEL files) were normalized using robust multichip average (RMA) and filtered using Significance Analysis of Microarrays (SAM) with a false discovery rate of 5%. The TargetScan and PicTar microRNA target prediction algorithms were then used to identify, on the mRNAs that passed the statistical filtering, putative targets of the 10 most highly dysregulated microRNAs (top five up and top five down). The genes identified by both prediction algorithms were further analyzed by function and pathway using the Database for Annotation, Visualization and Integrated Discovery (DAVID), available from http://david.abcc.ncifcrf.gov/ [23]. Unsupervised hierarchical clustering was performed to visualize the correlations between microRNAs and target mRNA/protein levels. For visualization of global microRNA::mRNA interactions in ULMs, box plots were used as a convenient way of graphically depicting the regulation of the mRNA targets of the 5 most highly upregulated and 5 most downregulated microRNA. The arms of the box plot represent the smallest and largest observation, the box is delimited by the lower and upper quartile, and the line through the box represents the median of the observations. Gene set enrichment analysis (GSEA) in ULMs versus matched myometria was performed according to the authors' guidelines published at the Broad Institute web pages (http://www.broadinstitute.org/gsea/index.jsp), using C2 (curated gene sets) and C5 (Gene Ontology gene sets) collections using Signal2Noise ratios.
MicroRNA transfection
MicroRNA oligonucleotides were used at a concentration of 60 pmol/well for a 6-well plate. To estimate transfection efficiency, the negative control Block-iT (Fluorescent double-stranded random 22mer RNA from Invitrogen (Carlsbad, CA)) and positive control TSC2 siRNA (Invitrogen, CA) were used. Mature microRNAs mimics and inhibitors from let-7c and miR-296 were purchased from Dharmacon Inc. (Lafayette, CO). Cells receiving only the tagged random sequence (Block-iT) were used as non-specific references at all data points. Following transfection, cells were harvested and analyzed at the indicated times.
shRNA miR-200a infection
Human miR-200a shRNA in pGIPZ was prepared. Lentivirus expressing miR-200a shRNA were produced in HEK293T cells packaged by pMD2G and psPAX2. For stable infection, 4×104 cells/mL of UtLM cells were plated in each well of 6-well plates in 2 ml medium without antibiotics. After overnight incubation, media was replaced with 1 ml Opti-MEM® I Reduced-Serum Medium containing 12 µg/mL polybrene per well. 50 µL of concentrated lentiviral particles were added to each well. After 48 hours infection, fresh media was added containing 2 µg/ml puromycin. Fresh media with puromycin was replaced every 3–4 days. Single clones were picked after two weeks of puromycin selection. Stable miR-200a expression was validated by RT-PCR (see below).
Cellular proliferation assay
UtLM cells were seeded in 24-well plates in triplicate wells at densities of 1×104 per well. Cell proliferation was monitored at 24, 48, 72 and 96 hrs using the colorimetric MTS assay (CellTiter 96® Aqueous Assay, Promega).
Luciferase transfection assays
Cell lines were transfected with 200 ng luciferase reporter PGL-3 control (Promega, Madison, WI), or pGL-3 HMGA2-3′UTR construct and 1ng of the pRLuc internal control plasmid (Biosignal, Montreal, QB). The luciferase expression was determined as recommended by Promega (Madison, WI).
Western blot analysis
The culture cell samples were homogenized at 4°C in a protein lysis buffer. Equal amounts of total protein from each sample were resolved through a 10% SDS–PAGE gel and then transferred to a PVDF membrane (Perkin Elmer Life Scientific Inc.). Development of the immunoblot with antisera against TSC2 (from Dr. Mizuguchi [24], [25]) was tested and a single specific HMGA2 band at 25 kDa was detected, as previously described.
RT-PCR
For detection of mature microRNAs, mirVana qRT-PCR Primers and the mirVana qRT-PCR Detection Kit (Ambion, Austin, TX) were used and optimized according to the abundance of microRNAs in the tested tissue samples. Primers for 11 let-7 and 5 miR-200 predicted target genes are summarized in Table S1. The abundance of cDNA products was normalized to the internal controls of small nuclear RNA U6 and α-Actin.
Tissue Microarrays, immunohistochemistry and scoring
All tissue sections were reviewed and cellular areas of tumor were selected for TMA analysis. Six 0.6 mm tissue cores were collected from each case, including three cores of ULMs and three cores of matched myometria. The technical details and reliability of the data have been described previously [26]. In brief, 432 tissue cores from 36 ULM with matched myometria were arrayed into one recipient paraffin block. The tissue cores were arrayed in a random distribution of cases.
Antibodies selected for this study included sixteen protein markers and are listed in Table S2. The detailed protocols and conditions used for each antibody followed the manufacturers' recommendations which were described previously [27]. In brief, the paraffin-embedded tissue array blocks were sectioned at 4 µm. Antigen retrieval was performed by either heat-induced epitope retrieval or by proteolytic enzyme digestion as previously described. All immunohistochemical staining procedures were performed on a Ventana Nexus automated system (Tucson, Arizona, USA).
Stained TMA slides were graded jointly by two pathologists using a visual semiquantification method (optical density of the immunoreactivity) depending on the staining characteristics of the antibody. The net gain or loss of immunoscores was calculated for a specific immunomarker from each individual ULM in comparison with matched myometrium (net value = tumor immunoscore − myometrial immunoscore). Each net value gave positive or negative scores.
Results
Largely based on ectopic expression studies and computational analyses, it has been predicted that microRNAs can regulate at least 30% of gene transcripts. Several recent studies have compared microRNA expression with the levels of corresponding predicted target genes in normal and tumor tissues at a global level. Our previous findings of microRNA dysregulation in ULMs led us to further examine whether these dysregulated microRNAs correlated with abnormal expression of their predicted target genes at both transcriptional and translational levels.
Genome-wide correlation of dysregulated microRNAs and their predicted mRNA targets
We had reported that a subset of 45 microRNAs were differentially expressed between ULMs and matched myometria [13]. To test whether altered expression of microRNAs in ULMs are responsible for dysregulation of the predicted target gene expression, we selected five ULMs and matched myometria (for which we had identified microRNA profiles, see Table 1) for gene expression analysis using Affymetrix U133A arrays. The five cases selected for the gene expression study were marked by the most significant microRNA dysregulation in ULMs and the highest magnitude of their modulation (Figure S1). In particular, we selected ULMs of large size (>10 cm) and from African American women. As previous stated by Aslan et al [2], the gene expression profiles in ULMs varied widely among studies, affected by the clinical setting, methods and platforms. We selected to also analyze the NCBI GEO GSE593 ULM profiling along with the data from this study due to the same sample size, patient age and the microarray platform used [21].
Applying significance analysis of microarray to the combined set of our 5 pairs of ULM and matched myometria and the GSE593 data, we identified 2674 probe sets (1,117 up- and 1,557 down-modulated) that were significantly different between ULMs and matched myometrium controls (Figure S1).
We used a combination of two microRNA prediction methods, TargetScan and PicTar, to search for all predicted gene targets of the 5 most highly upregulated (let-7s, miR-21, miR-23b, miR-27a and miR-30a) and downregulated (miR-29b, miR-32, miR-144, miR-197 and miR-212) microRNAs [13]. Both mRNA profiling data sets (ours and GSE593) were used for the microRNA target prediction.
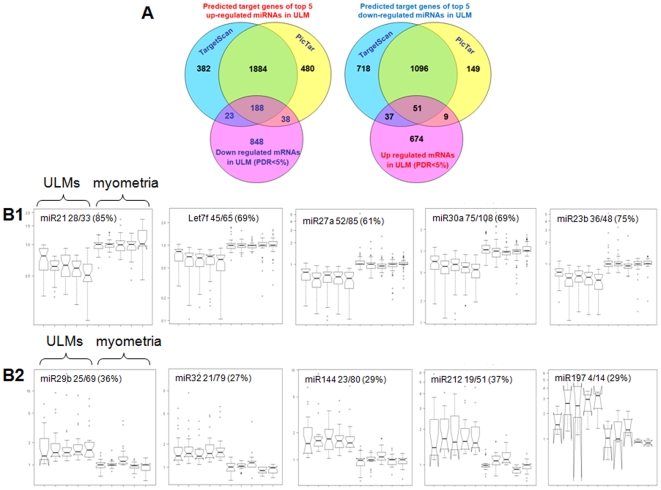
There were a total of 2995 and 2060 TargetScan and PicTar-predicted genes of the top 5 upregulated and 5 top downregulated microRNAs, respectively (Figure 1A). Utilizing the expression data from these five cases, we identified 2674 mRNAs that were significantly dysregulated in ULMs (SAM, FDR<5%, Figure 1A). Among these significantly dysregulated mRNAs, 249 downregulated mRNAs were predicted targets of the 5 upregulated microRNAs and 97 upregulated mRNAs were predicted targets of the 5 downregulated microRNAs (Figures 1B). Together, they represented 13% of 2674 genes found to be significantly dysregulated in ULMs. The Box Plot analysis and significance analysis revealed a trend of overall inverse association between the predicted target genes and either up- or down- regulated microRNAs (Figure 1B). Upregulated predicted gene targets of downregulated microRNAs were significantly enriched in comparison to the overall pool of significantly dysregulated targets (0.05>p>0.01). The findings suggest that the most up and down regulated microRNAs in ULMs may regulate the expression of their predicted target genes at the level of mRNA stability as previously reported [28].
Figure 1. Predicted target genes or microRNAs in uterine ULMs.
A Predicted target genes for the 5 most highly upregulated (left) and 5 most highly downregulated microRNAs (right). TargetScan (light blue) and PicTar (yellow) identified 1884 genes as predicted targets of the 5 most highly upregulated microRNAs (let-7s, miR-21, miR-23b, miR-27a and miR-30a). Among 1079 significantly downregulated genes in ULMs (pink circle), 188 (intersection of the three circles) are the best predicted targets of these upregulated microRNAs. B Differential expression of the predicted target genes for each of the 5 most highly upregulated microRNAs (B1) and downregulated microRNAs (B2) in ULMs and matched myometria. Each box plot represents the average level of predicted target gene expression. Expression of microRNA targets is plotted as RMA-normalized, median centered and log2-transformed, relative abundance levels (Y-axis), with a baseline = 1 corresponding to median centered normal myometrial samples.
At the level of individual matches between microRNAs and the inversely modulated, predicted targets, a rather small proportion of the target genes were inversely correlated with a considerable correlation coefficient (ranging from r = −0.5 to −0.95, see Table 2). Small sample size and important biological variables such as microRNA subcellular compartmentalization and the fact that levels of mature microRNAs do not necessarily reflect their functional impact may be the reason for the decreased rates of significant negative correlation of individual microRNA and mRNA expression patterns.
Table 2. Correlation analyses of top 10 most highly dysregulated miRNAs and their predicted target genes in 5 large ULM of black women based on global gene expression profiles.
| miRNA | Symbol | Target gene Name | AFFY ID | Correlation coefficient (n = 5) |
| let7c | TRIB1 | tribbles homolog 1 (Drosophila) | 202241_at | −0.437 |
| let7f-2 | PLCB4 | phospholipase C, beta 4 | 203896_s_at | −0.648 |
| miR-21 | BRD1 | bromodomain containing 1 | 215460_x_at | −0.878 |
| SKI | v-ski sarcoma viral oncogene homolog (avian) | 204270_at | −0.435 | |
| miR-27a | ANK2 | ankyrin 2, neuronal | 202921_s_at | −0.942 |
| RAB11FIP | RAB11 family interacting protein 1 (class I) | 219681_s_at | −0.869 | |
| GATA2 | GATA binding protein 2 | 209710_at | −0.890 | |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 202768_at | −0.662 | |
| PPARG | peroxisome proliferative activated receptor, gamma | 208510_s_at | −0.692 | |
| miR-30a-5p | SMARCD2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 | 201827_at | −0.760 |
| SLC29A3 | solute carrier family 29 (nucleoside transporters), member 3 | 219344_at | −0.604 | |
| HLF | hepatic leukemia factor | 204755_x_at | −0.817 | |
| MAP3K5 | mitogen-activated protein kinase kinase kinase 5 | 203836_s_at | −0.881 | |
| TNXA | tenascin XA pseudogene | 216339_s_at | −0.598 | |
| miR-23b | PPARG | peroxisome proliferative activated receptor, gamma | 208510_s_at | −0.766 |
| HIVEP2 | human immunodeficiency virus type I enhancer binding protein 2 | 212641_at | −0.828 | |
| GATA2 | GATA binding protein 2 | 209710_at | −0.863 | |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 202768_at | −0.641 | |
| miR-29b | BMP1 | bone morphogenetic protein 1 | 202701_at | −0.942 |
| RARB | retinoic acid receptor, beta | 205080_at | −0.625 | |
| FAM131B | family with sequence similarity 131, member B | 205368_at | −0.814 | |
| NASP | nuclear autoantigenic sperm protein (histone-binding) | 201969_at | −0.940 | |
| TGFB3 | transforming growth factor, beta 3 | 209747_at | −0.644 | |
| miR-32 | NFIB | nuclear factor I/B | 211467_s_at | −0.785 |
| FLI1 | Friend leukemia virus integration 1 | 210786_s_at | −0.805 | |
| FHL3 | four and a half LIM domains 3 | 218818_at | −0.799 | |
| miR-212 | PRKD1 | protein kinase D1 | 205880_at | −0.308 |
| miR-144 | PTHLH | parathyroid hormone-like hormone ; parathyroid hormone-like hormone | 211756_at | −0.239 |
| miR-197 | RNPC1 | RNA-binding region (RNP1, RRM) containing 1 | 212430_at | −0.803 |
| TNRC5 | trinucleotide repeat containing 5 | 217931_at | −0.566 |
Correlation of microRNA expression with protein expression in pair-matched tissue samples of ULMs and myometria
To further investigate the correlation of microRNAs and predicted targets in ULMs, we examined the predicted target gene products by immunohistochemistry analysis to establish the effects of microRNAs on the synthesis of target proteins. We utilized ULMs and matched myometria from 36 cases, from which global microRNA expression profiles had been obtained [13] (Table 1). A total of 16 proteins (Table S2) that were known to be significantly dysregulated in ULMs [27], [29] were selected for immunohistochemical analysis. The relative expression level of each protein was scored in a semiquantitative manner in ULMs and in matched myometrial controls (see Methods). The net changes of the selected gene products (Figure S3) were analogous to our previously published studies in ULMs [27], [29].
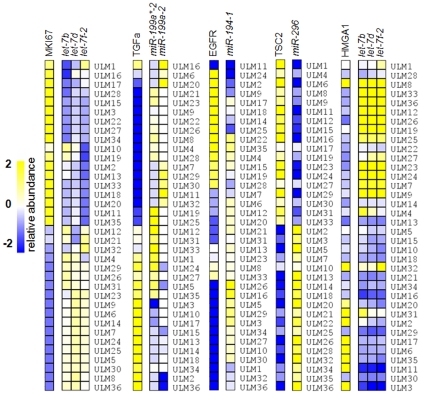
The Ki-67 index in ULMs in comparison to the matched myometria can be used to evaluate tumor growth rate. Correlation of let-7 microRNA levels in 36 ULMs established an inverse association with the Ki-67 index (MKI67 protein expression) (Figure 2).
Figure 2. Correlation of the selected microRNAs and their predicted target gene products (proteins) in 36 ULMs.
The negative correlation of microRNA and the target proteins are illustrated by inverse correlation of regulating miRNAs and their select targets. The gene expression levels are depicted by the intensity of yellow (overexpression), white (no change) and blue (underexpression) color. In each panel, correlated microRNAs and the target proteins are shown on the top and tumor IDs are on the right.
Moderate individual negative correlations with microRNAs were found for EGFR and miR-194-1, TGFα and miR-199a-2 (r = −0.39 and r = −0.58, respectively), however, the overall pattern appeared entirely inversely correlated (Figure 2). Most other gene products, the patterns of which correlated with their targeting microRNAs, exhibited similar negative correlation (r = −0.2 to −0.4) (data not shown). These findings validate the targets and prediction-based correlative analysis of inverse associations of microRNA and tumor-associated protein expression levels.
Genomic alteration and microRNA expression
To evaluate whether selected dysregulated microRNAs in ULMs associated with specific genomic alterations, we performed comparative genomic hybridization (CGH). We selected 8 cases of ULMs for the study (Table 1). By comparison to matched myometria, few genomic alterations were found (Table 3, Figure S2). In general, the loss of genomic material was the only finding in all eight cases. Chromosomal regions of 1p36, 3p11, 3q22 and 6p were the most commonly altered, with 1p36.33-p36.23 and 3q26.1-q27.2 established as regions of deletion overlap between ULMs derived from patients B4, B9 and B1, B4, respectively. Interestingly, members of cancer-inhibitory miRNA family miR-200a, miR-200b, miR-429 and miR-551a are located in the region of loss at 1p36 while the 3q26-27 region harbors miR-15b, miR-16-2 and other microRNAs including miR-1263, miR-720, miR-551b, miR-569, miR-1224. We also found that additional members of the oncogenic miR15/16 family (miR-15a and miR-16) as well as the members of the miR-17-92 polycistron were lost with the 13q12.12-q33.2 region of a single patient B4 (Table 3).
Table 3. Comparative genomic hybridization analysis and associated microRNAs in eight large leiomyomas of black women.
| Patient | Chr. bands | Size (Mb) | Aberrations | MiRNAs in regions of loss* |
| B1 | 3p11.2-p11.1 | 0.55 | loss | - |
| 3q13.31-q21.2 | 9.90 | loss | miR-198 | |
| 3q22.2-q27.2 | 49.60 | loss | miR-15b, miR-16-2, miR-1263, miR-720, miR-551b, miR-569, miR-1224 | |
| B2 | No change | - | - | - |
| B4 | 1p36.33-p34.3 | 38.66 | loss | miR-200b, miR-200a , miR-429, miR-551a, miR-34a, miR-1290, miR-1256, miR- 552 |
| 3q26.1-q29 | 37.68 | loss | miR-15b, miR-16-2 , miR-1263,miR- 720, miR-551b,miR- 569, miR-1224, miR-1248, miR-28, miR-944, miR-570, miR-922 | |
| 6q13-q24.3 | 76.21 | loss | miR-30c-2, miR-30a, miR-2113, miR-587, miR-548b, miR-588, miR-548a-2 | |
| 13q12.12-q33.2 | 82.49 | loss | miR-320d-1, miR-621, miR-16-1, miR-15a , miR-1297, miR-622, miR-17 , miR-18a, miR-19a, miR-20a , miR-19b, miR-92a-1 , miR-623 | |
| B5 | No change | - | - | - |
| B6 | No change | - | - | - |
| B7 | 22q11.1-q13.33 | 35.09 | loss | miR-648, miR-185 , miR-1306, miR-1286, miR-649, miR-301b , miR-130b , miR-650, miR-548j, miR-658, miR-659, miR-1281, miR-33a, miR-1249, let-7a-3, let-7b |
| B8 | No change | - | - | - |
| B9 | 1p36.33-p36.23 | 7.09 | loss | miR-200b , miR-200a, miR-429, miR-551a |
| 3p12.3-p11.1 | 10.24 | loss | - | |
| 6p25.3-p22.2 | 24.79 | loss | miR-548a-1 | |
| 6p12.3-p11.1 | 11.61 | loss | miR-206 , miR-133b |
*: bold = miRNA detected as downregulated in the patient.
Candidate role of the loss of miR-200 family and miR-15 and miR16 in ULMs
Loss of miR-200 family is associated with epithelial and mesenchymal transition (EMT) and aggressive tumor phenotypes of ovarian cancer [30], [31]. We observed that miR-200a and miR-200b were significantly down regulated in 51 ULMs (net loss of −0.36±0.11 and −0.45±0.06, respectively). To further explore the potential significance of the deletion overlap involving the miR-200 family, we examined gene expression in patients and found that a number of TargetScan-predicted targets of either miR-200a or miR-200b were collectively upregulated in the tumor compared to unaffected pair-matched myometrium (∼180 genes ranging from 1.25–10.3 fold upregulation). Among these were genes with established roles in cancer and cell death regulation: MAF (v-maf musculoaponeurotic fibrosarcoma oncogene), CTBP2 (C-terminal binding protein 2), antiapoptotic BCL2 (B-cell CLL/lymphoma 2), CITED2 (Cbp/p300-interacting transactivator), LASS6 (LAG1 homolog, ceramide synthase 6), PHF21A (PHD finger protein 21A), TSC22D1 (TSC22 domain family, member 1), ATXN1 (ataxin 1), JUN (jun oncogene) and NFIB (nuclear factor I/B). Focused pathway analysis (using GO, KEGG, Biocarta and Panther databases) of the predicted miR-200 family targets that are consistently upmodulated in ULMs including patient B4 (Table S3) implicates categories of regulation of transcription proliferation and cell cycle control, actin cytoskeleton and adherens, tight, gap and focal adhesion junction remodeling, as well as cancer related signaling pathways (MAPK, RAS, WNT, NOTCH, TGF-β, VEGF).
Similarly, loss of miR-15/miR-16 cluster is associated with aggressive tumor growth [32]. The findings indicated alteration of these two genomic regions may be related to the tumorigenesis of ULMs. We have evaluated the expression levels of TargetScan predicted mRNA targets and found that important transcriptional, signaling and other regulators of cell growth and survival linked to cancer were also collectively upmodulated in patient B4, among them FOXO1A (forkhead box O1A (rhabdomyosarcoma)), BCL2 (B-cell CLL/lymphoma 2), TGFBR3 (transforming growth factor, beta receptor III (betaglycan, 300kDa)), MAP3K4 (mitogen-activated protein kinase kinase kinase 4), VEGF (vascular endothelial growth factor), TCF3 (transcription factor 3), EIF4E (eukaryotic translation initiation factor 4E), JARID2 (Jumonji, AT rich interactive domain 2), EVI5 (ecotropic viral integration site 5), IGF1 (insulin-like growth factor 1), WNT5B (wingless-type MMTV integration site family, member 5B). Detailed pathway analysis of the upregulated targets of the lost miR-15/16 family identified biological categories of pathways in cancer, endometrial cancer, transcription, melanoma, apoptosis, signaling pathways including insulin, MAPK, mTOR, VEGF, ErbB, JAK/STAT signaling, and cell-cell adhesion and cytoskeleton remodeling (see Supplemental Table 3)
We also identified a subset of upregulated genes that are predicted as targets of both miR-200a/b and miR-15/16 microRNAs from the regions of deletion overlap on Chr1 and Chr3 (Table 3), including ACTR1A, BACH2, BCL2, CDC14B, CLASP1, CYP26B1, E2F3, EVI5, FUBP1, IKBKB, IRS2, IRS2, LRIG1, OTUD4, PCDH9, PCDH9, PELI2, PHF21A, PPAP2B, SLC2A3, SNTB2, TMCC1 and TUBB.
These findings indicated that alteration (loss) of two overlapping genomic regions (7.09 Mb of Chr1 1p36.33-p36.23 and 24.56 Mb of Chr3 3q26.1-q27.2 harboring cancer related miRNAs may be related to the tumorigenesis of a subset of ULMs via deregulation of the miR200a/b and miR-15/16 gene targets and in part this process may be due to the loss of convergent inhibitory action of the miR-200 family and miR-15 and miR-16 on a small group of the same downstream target genes.
Functional correlation of microRNAs and mRNA expression in cultured leiomyoma cell lin
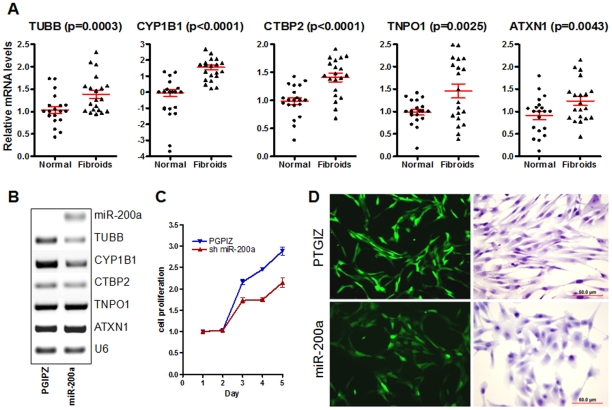
As shown above, downregulation of miR-200 family members appears a candidate event in leiomyomas in select patients due to loss of corresponding genomic DNA loci and broader reduction of miR-200 expression in ULMs (Table 3 and Figure S2). We selected 5 target genes of miR-200a that were all significantly upregulated in ULMs (Figure 3A) for functional testing as direct targets and for their roles in cell phenotype changes. In the UtLM cell line with stable miR-200a overexpression, 3 of 5 mRNAs (TUBB, CYP1B1 and CTBP2 were suppressed by miR-200a in vitro (Figure 3B). Importantly and as predicted by pathway analysis (Table S3), overexpression of miR-200a in UtLM cells led to growth inhibition compared to mock infected controls (Figure 3C), and reverted the fibroblastoid morphology towards more pronounced epithelial phenotype (Figure 3D), consistent with the established role of miR-200 family in epithelial-mesenchymal transition [33]. Collectively, these findings suggest that the loss of miR-200 family, identified by miRNA profiling and a CGH analysis, together with upregulation of its target genes may contribute to the tumor growth and underlie the mesenchymal character of ULMs.
Figure 3. MiR-200 predicted target gene analysis in uterine ULMs.
A Scatter plot analysis of relative mRNA expression in five miR-200 predicted target genes in 10 ULMs and matched myometria (our data and GSE593). Red bars indicate the mean and standard error of measurement. B RT-PCR analysis of expression of five miR-200 predicted target genes in ULMs cell line UtLM with stable miR-200a expression (see Methods) and control (vector PGIPZ only). Repression of TUBB, CYP1B1 and CTBP2 can be readily appreciated. C Growth curves illustrate significantly reduced proliferative rate in UtLM cell line with miR-200a overexpression in comparison UtLM cell line with vector control only (PGPIZ). D. Photomicrographs illustrate stable viral control (upper panels) and miR-200a (lower panels) expression in UtLM cell lines. The stromal to epithelial morphology transition in UtLM cell line with miR-200a overexpression is evident (lower panels).
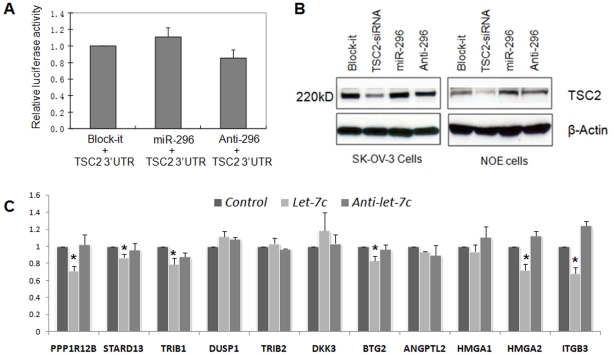
We previously found that the product of TSC2 gene (tuberin) was significantly down regulated in ULMs [27], [34]. Downregulation of TSC2 was also found in this study (Figure S3). By correlation analysis, as illustrated in Figure 2, we found that predicted regulatory miR-296 was inversely correlated with TSC2 protein in 36 ULMs. TSC2 contains 41 exons with a very short 3′ untranslated region (3′UTR, <110 nt). The short TSC2 3′UTR may prevent microRNA regulation. However, in the TSC2 3′UTR immediately adjacent to the stop codon, there is a highly conserved sequence that harbors the complementary sites of miR-296 and a few other microRNAs. To study whether TSC2 is the target of miR-296, we prepared a TSC2 3′UTR reporter construct and examined the luciferase activity by treated cells with control, miR-296 mimic and inhibitor. There was no reduction of luciferase expression in cell treated with miR-296 (Figure 4A). We further examined whether miR-296 could inhibit TSC2 protein production. In comparison to TSC2 siRNA, no significant protein reduction was noted in cells treated with miR-296 (Figure 4B). The findings indicated that TSC2 was not a direct target of miR-296. The inverse correlation of TSC and miR-296 levels may thus be related to indirect or unrelated molecular mechanisms participating in ULM tumorigenesis.
Figure 4. Analysis of the miR-296 predicted target gene TSC2 and 11 let-7 predicted target genes in vitro.
A Transient transfection analysis for luciferase reporter expression with TSC2 3′UTR in the presence and absence of miR-296. B Immunoblotting analysis of transient transfection analysis of miR-296 for TSC2 expression. TSC2 siRNA was used as a positive control antagonizing TSC2 expression. Block-iT = nonfucntional small RNA control. β-Actin was used as protein loading control. C Relative expression of let-7 predicted target genes (listed above) (y-axis) in transient transfection of nonfunctional small RNA (Block-iT (Controls), let-7c mimic and let-7 inhibitor (Anti-let-7). The relative expression levels were obtained in three cell lines of immortalized ULM cell line (ULM-3401), leiomyosarcoma cell lines (LMS-1, UT-1) (see Methods). T-bars indicate standard error of measurement. * = p value<0.05.
We had reported that most members of the let-7 family were dysregulated in ULMs [13]. To test whether let-7 microRNAs could regulate other predicted target genes in addition to HMGA2 in ULMs, we examined the expression of several predicted let-7 target genes in the ectopically induced presence or absence of let-7 microRNAs in uterine ULMs and leiomyosarcoma cell lines in vitro. TargetScan and PicTar databases [35], [36] predicted 65 target genes of let-7 mRNAs that are significantly dysregulated in ULMs (Figure 1B1), of which 45 (69%) were downregulated. Of those, we selected 11 candidate targets involved in cell proliferation and extracellular matrix regulation that might be functionally associated with the pathogenesis of ULMs. By transient transfection and RT-PCR analysis (Table S1), we found that 6 of them can be repressed by let-7s (Figure 4C). Thus our findings indicate that a subset of the predicted target genes can be significantly repressed in experimental conditions. Importantly, this set of experiments indicates that follow-up biochemical functional studies are warranted to validate the prediction- and inverse correlation-based analyses of individual microRNA::mRNA relationships.
Interpretation of focused miRNA::mRNA relationship compared to genome wide pathway analysis
We asked whether inverse correlation of the top 5 up- and down-regulated miRNA profiles with their target genes is sufficiently representative to identify top categories in comparison to broad data mining of the ULM mRNA profiles. To that end, we performed gene set enrichment analysis (GSEA, Figure 5) using the combined expression data from our and the GSE593 data and compared the results of top categories with the DAVID analysis of pooled 346 top miRNA targets (Fig 1B) presented in Table 4. A number of themes showed overlap, such as cell cycle and DNA replication control, proliferation control indicated by pathways such as MAPK, JAK/STAT, developmental signaling (WNT, TGF-β), and modulation of cellular processes associated with morphological and migratory properties of the tumors (cell-cell junction, cell adhesion and cytoskeleton remodeling) found in miRNA target analysis are complemented by MET, WNT and TGF-β signaling, major regulators of invasion and motility, found in both types of analysis. We thus propose that the reduced information contents resulting from functional interpretation of the relationships between a limited number of top regulated miRNAs and their inversely correlated mRNA targets sufficiently identifies key functional themes, with results comparable to the global, genome-wide mRNA analysis.
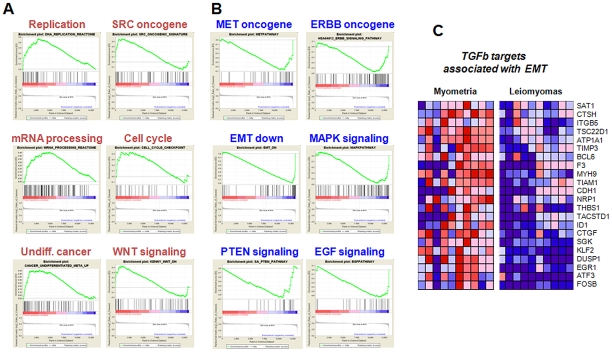
Figure 5. Gene Set Enrichment Analysis (GSEA) of mRNA profiling results from primary ULMs and matched myometria.
The histograms show the distribution of select top GSEA molecular signatures from predefined C2 and C5 categories (accessible from the MolSig database at http://www.broadinstitute.org/gsea/msigdb/index.jsp). The leading edge (most significant genes) are shown as vertical bars accumulated either left and below the peak of green enrichment score plot (A) or right of the valley of the green plot (B), indicating the respective up- or down-regulated genes of each shown GSEA characterized by the highest enrichment score. C The leading edge (genes with the highest enrichment score) of the EMT-down category (shown in B) is shown as individual gene expression-based heat map and indicates downregulation of the TGF-β signal and of its canonical targets in ULMs.
Table 4. KEGG/Biocarta analysis of functions of predicted target genes of miRNAs in ULM.
| PATHWAY | miRNAs: | Up | Down | Gene Symbols |
| mRNAs: | Down | Up | ||
| MAPK SIGNALING ACVR1B, MEF2C | 9 | RPS6KA3, DUSP1, PDGFRB, RRAS2, MAPK14, MAP3K5, | ||
| FOCAL ADHESION GAP JUNCTION | 7 | MET, CCND2, TNXB, PDGFRB, RRAS2, ITGB3, CVR1B | ||
| REGULATION OF ACTIN CYTOSKELETON | 6 | ADCY9, ADCY3, CSNK1A1, GJA1, PDGFRB, RRAS2, | ||
| CYTOKINE-CYTOKINE | 6 | RDX, MYH9, PDGFRB, RRAS2, ARHGEF6, ITGB3, | ||
| RECEPTOR INTERACTION | 5 | MET, CSF1, KITLG, PDGFRB, ACVR1B, | ||
| ECM-RECEPTOR INTERACTION | 5 | TNXB, ITGB3, DTPRECK, TIMP3, | ||
| CELL CYCLE | 5 | CCND2, CCNH, RB1, ABL1, KITLG, | ||
| CALCIUM SIGNALING | 4 | ADCY9, ADCY3, PDGFRB, EDNRB, | ||
| JAK-STAT SIGNALING | 4 | SPRY1, STAT3, CCND2, STAT5B, | ||
| NFAT AND HYPERTROPHY OF THE HEART | 4 | CSNK1A1, MAPK14, HBEGF, MEF2C, | ||
| PPARá SIGNALING | 3 | RB1, DUSP1,STAT5B, | ||
| NFκB ACTIVATION | 3 | NR3C1,DUSP1,MAPK14, | ||
| TGF-â SIGNALING | 3 | CHRD, ACVR1B, SMAD7, | ||
| ADHERENS JUNCTION | 2 | MET, ACVR1B, | ||
| TIGHT JUNCTION | 2 | MYH9, RRAS2, | ||
| WNT SIGNALING | 2 | CCND2, CSNK1A1, | ||
| CELL ADHESION | 3 | NCAM1, CDH2, IGSF4, | ||
| INSULIN SIGNALING | 3 | SOCS2, PPARGC1A, FOXO3A, | ||
| CYTOKINE-CYTOKINE RECEPTOR INTERACTION | 2 | CXCL12, PDGFC, | ||
| FOCAL ADHESION | 2 | COL2A1, PDGFC, |
Discussion
Although it has been shown that individual microRNAs may regulate multiple genes, the current knowledge for microRNA::mRNA regulation has been largely obtained from the studies of the disruption of microRNA::mRNA interactions on a single gene basis. Recently, several attempts to examine microRNA::mRNA interactions globally have been pursued [37], [38], [39]. In our previous study of TGF-β-directed epithelial to mesenchymal transition, we explored a global approach to study the dynamics of microRNA::mRNA interactions [40]. The application of this genome-wide analysis of microRNA::mRNA expression in ULMs allows us to gain insight into the role of microRNAs in the regulation specific mRNAs and their protein products. In a correlation analysis between dysregulated mRNAs and the most highly of dysregulated microRNAs in ULMs, an overall negative correlation was established (Table 2 and Table S1, Figure 1).
Among many dysregulated microRNAs and target genes in leiomyomas, correlation of HMGA2 and the let-7 family has been well characterized [13], [17], [18], [19] and shown as biologically significant for leiomyoma growth [13], [41]. We expected to see broader dysregulation of additional genes in uterine leiomyomas due to regulation by specific microRNAs. In the selected predicted targets which we examined at the mRNA and protein levels, we found that some, but not all of them, appear to be regulated by microRNAs (Figures 2, 3, and 4). Our findings further support the notion that the selection of the predicted microRNA targets relies on the rank (strength) of the target prediction scores (Table S1) and that the functional regulation relies on the level of microRNA expression in the cell and convergent, synergistic effects of multiple microRNAs on one target gene. Varying functional validation outcomes of multiple genes tested as miRNA targets (Figures 3B and 4) importantly demonstrate the complexity of target gene regulation by microRNAs and indicate the need for detailed validation in prediction- and inverse correlation-based studies.
Uterine ULMs are the most common benign neoplasms, exhibiting complex patterns of gene dysregulation [1], [2]. The dysregulated genes are involved in many cellular and molecular functions, such as defects in angiogenesis [42], nuclear receptors and local growth factors [2], dysfunction of extracellular matrix [43] and TGF-β signaling [44]. The molecular mechanisms leading to the dysregulation of these genes has been largely unknown. As illustrated in Table 4, several tumorigenic pathways seem to be regulated by the most highly dysregulated microRNAs in ULMs. For example, at least nine predicted target genes of upregulated microRNAs in the Mitogen-activated protein kinase (MAPK) signaling pathway were downregulated. This finding was validated by the GSEA analysis of mRNA expression data (Figure 5B). MAPK signaling has been shown to play a significant role in tumorigenesis by controlling tumor growth, proliferation, differentiation, migration and apoptosis [45]. Activation of MAPK signaling is very common in many malignant neoplasms, allowing tumors to gain the capability of independent growth, insensitivity to anti-growth signals, unlimited replicative potential and the ability to invade and metastasize [45]. ULMs are relatively slow growing smooth muscle tumors and rarely, if ever, progress to malignancy. Repression of MAPK pathway genes by microRNAs in ULMs may represent a protective mechanism from fast growth and potential tumor progression.
Downregulation of many target genes of upregulated microRNAs including genes involved in focal adhesion (seven), gap junction (six), actin cytoskeleton (six) and extracellular matrix formation (five) have been identified (Table 4). These categories are related to cell remodeling, migration, growth and tumor progression [2], [43]. Thus, certain upregulated microRNAs may prevent aggressive tumor growth through repression of the target genes that are responsible for tumor cell communication and extracellular matrix formation and may in part play a protective role in avoiding aggressive behavior of ULMs. Conversely, loss of miR-200 shown to modulate growth as well as the UtLM morphology (Figure 4C,D), may lead to upregulation of genes (some of them convergent targets of lost miR-15/16) that contribute to the progression of ULM tumorigenesis.
Key components of the TGF-β and of MET signaling pathways are downmodulated in ULMs (Figure 5B and Table 4), both having a role in cell growth control, tumor progression, and oncogenic effects associated with invasive growth and extracellular matrix remodeling [44], [46]. Despite the downregulation of established TGF-β targets CTGF and THBS1 in ULMs (see Figure 5C, listing significant genes from EMT down category in Fig 5B), future studies will be required to determine whether microRNAs participate in TGF-β-mediated regulation of the profibrogenic extracellular matrix deposition in ULMs.
In conclusion, our global correlation analysis of gene product levels and microRNA expression provide a comprehensive and biologically meaningful insight into tumorigenesis of ULMs. This is documented by the relationship between the five most highly overexpressed microRNAs and a downregulated group of target genes that are related to cellular structure, extracellular matrix and cell proliferation, a signature that can be related to the slow growth and non aggressive behavior of uterine ULMs.
Supporting Information
MicroRNA and mRNA expression profiles in five large uterine leiomyomas. microRNA and mRNA expression profiles in five large uterine leiomyomas from black women (Table 1). A. Unsupervised hierarchical clustering (HCL) of 206 human microRNAs in 5 large ULMs (>10 cm) normalized to the matched myometria is shown. B. HCL illustrating a concordance of total of 2674 significantly dysregulated genes between 5 ULMs from this study (left panel) and the NCBI GEO GSE593 data set (right panel), normalized to matched myometria. yellow = upregulation; blue = downregulation.
(0.69 MB JPG)
Regions of common loss in ULMs detected by array CGH. A, B. Genome browser mapping of the regions of common loss of genomic material as detected by a CGH in this study. Patient IDs are shown as yellow rectangles. Cancer-related microRNAs of interest in the deleted regions are listed under the chromosomes and highlighted by red arrowheads. C. Detailed genetic summary of the common regions of loss.
(0.60 MB JPG)
TMA immunohistochemical analysis of selected proteins in ULMs. Immunohistochemical analysis of selected proteins in 36 ULMs. A. Photomicrograph illustrating tissue microarray (TMA) sections with hematoxylin and eosin (H&E) stain (upper panel) and immunostaining of ER (bottom panel). Triplicate tissue cores from controls (matched myometrium) and tumors (ULMs) are indicated on the right. B. Differential expression of the selected target gene products is shown as mean and standard error of measurements (bars and t-bars, respectively). The net change for each gene product was calculated based on relative immunoreactivity in ULMs against matched myometrium. Red = gain in protein levels; Blue = reduction of protein levels.
(0.61 MB JPG)
Primers and PCR results for the predicted target genes of let-7 and miR-200s.
(0.03 MB DOC)
Antibodies used in this study.
(0.03 MB DOC)
Predicted target genes and pathways downstream of miR-200s amd miR-15/16.
(0.81 MB DOC)
Acknowledgments
We thank Yutong Zhang of the NYU Cancer Institute Genomics Facility for expert assistance with the IVT labeling protocol for gene expression profiling. We thank Drs. Luis Chiriboga and Herman Yee of NYU department of pathology for help in immunohistochemistry.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by Department of Defence Tuberous Sclerosis Complex Research Program (TSCRP) award W81XWH-09-1-0166 and National Institutes of Health Small Grant Program (NIH RO3) grant R03HD057380. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Flake GP, Andersen J, Dixon D. Etiology and pathogenesis of uterine leiomyomas: a review. Environ Health Perspect. 2003;111:1037–1054. doi: 10.1289/ehp.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arslan AA, Gold LI, Mittal K, Suen TC, Belitskaya-Levy I, et al. Gene expression studies provide clues to the pathogenesis of uterine leiomyoma: new evidence and a systematic review. Hum Reprod. 2005;20:852–863. doi: 10.1093/humrep/deh698. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 4.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 9.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2006;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Marsh EE, Lin Z, Yin P, Milad M, Chakravarti D, et al. Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil Steril. 2007;89:1771–1776. doi: 10.1016/j.fertnstert.2007.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med. 2007;12:227–240. doi: 10.1111/j.1582-4934.2007.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Wang T, Zhang X, Obijuru L, Laser J, Aris V, et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46:336–347. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carney SA, Tahara H, Swartz CD, Risinger JI, He H, et al. Immortalization of human uterine leiomyoma and myometrial cell lines after induction of telomerase activity: molecular and phenotypic characteristics. Lab Invest. 2002;82:719–728. doi: 10.1097/01.lab.0000017499.51216.3e. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman PJ, Milliken DB, Gregg LC, Davis RR, Gregg JP. Molecular characterization of uterine fibroids and its implication for underlying mechanisms of pathogenesis. Fertil Steril. 2004;82:639–649. doi: 10.1016/j.fertnstert.2004.01.047. [DOI] [PubMed] [Google Scholar]
- 22.Saeed AI, Sharov V, White J, Li J, Liang W, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 24.Mizuguchi M, Kato M, Yamanouchi H, Ikeda K, Takashima S. Tuberin immunohistochemistry in brain, kidneys and heart with or without tuberous sclerosis. Acta Neuropathol (Berl) 1997;94:525–531. doi: 10.1007/s004010050746. [DOI] [PubMed] [Google Scholar]
- 25.Mizuguchi M, Kato M, Yamanouchi H, Ikeda K, Takashima S. Loss of tuberin from cerebral tissues with tuberous sclerosis and astrocytoma. Ann Neurol. 1996;40:941–944. doi: 10.1002/ana.410400621. [DOI] [PubMed] [Google Scholar]
- 26.Wei JJ, Chiriboga L, Arslan AA, Melamed J, Yee H, et al. Ethnic differences in expression of the dysregulated proteins in uterine leiomyomata. Hum Reprod. 2006;21:57–67. doi: 10.1093/humrep/dei309. [DOI] [PubMed] [Google Scholar]
- 27.Wei JJ, Chiriboga L, Arslan AA, Melamed J, Yee H, et al. Ethnic differences in expression of the dysregulated proteins in uterine leiomyomata. Hum Reprod. 2005;21:57–67. doi: 10.1093/humrep/dei309. [DOI] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Wei JJ, Chiriboga L, Mittal K. Expression profile of the tumorigenic factors associated with tumor size and sex steroid hormone status in uterine leiomyomata. Fertil Steril. 2005;84:474–484. doi: 10.1016/j.fertnstert.2005.01.142. [DOI] [PubMed] [Google Scholar]
- 30.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. J Oncol. 2010;2010:821717. doi: 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 33.Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010;116:117–125. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei J, Chiriboga L, Mizuguchi M, Yee H, Mittal K. Expression profile of tuberin and some potential tumorigenic factors in 60 patients with uterine leiomyomata. Mod Pathol. 2005;18:179–188. doi: 10.1038/modpathol.3800283. [DOI] [PubMed] [Google Scholar]
- 35.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 37.Liu T, Papagiannakopoulos T, Puskar K, Qi S, Santiago F, et al. Detection of a microRNA signal in an in vivo expression set of mRNAs. PLoS ONE. 2007;2:e804. doi: 10.1371/journal.pone.0000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Z, Jian Z, Shen SH, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic Acids Res. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ. Transforming growth factor-beta and microRNA:mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs. 2007;185:157–161. doi: 10.1159/000101316. [DOI] [PubMed] [Google Scholar]
- 41.Peng Y, Laser J, Ye G, Mittal K, Lee P, et al. Antiproliferative effects by let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2007;6:663. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 42.Weston G, Trajstman AC, Gargett CE, Manuelpillai U, Vollenhoven BJ, et al. Fibroids display an anti-angiogenic gene expression profile when compared with adjacent myometrium. Mol Hum Reprod. 2003;9:541–549. doi: 10.1093/molehr/gag066. [DOI] [PubMed] [Google Scholar]
- 43.Catherino WH, Leppert PC, Stenmark MH, Payson M, Potlog-Nahari C, et al. Reduced dermatopontin expression is a molecular link between uterine leiomyomas and keloids. Genes Chromosomes Cancer. 2004;40:204–217. doi: 10.1002/gcc.20035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chegini N, Verala J, Luo X, Xu J, Williams RS. Gene expression profile of leiomyoma and myometrium and the effect of gonadotropin releasing hormone analogue therapy. J Soc Gynecol Investig. 2003;10:161–171. doi: 10.1016/s1071-5576(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 45.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 46.Luo X, Ding L, Xu J, Chegini N. Gene Expression Profiling of Leiomyoma and Myometrial Smooth Muscle Cells In Response to TGF-{beta}. Endocrinology. 2004;146:1097–1118. doi: 10.1210/en.2004-1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MicroRNA and mRNA expression profiles in five large uterine leiomyomas. microRNA and mRNA expression profiles in five large uterine leiomyomas from black women (Table 1). A. Unsupervised hierarchical clustering (HCL) of 206 human microRNAs in 5 large ULMs (>10 cm) normalized to the matched myometria is shown. B. HCL illustrating a concordance of total of 2674 significantly dysregulated genes between 5 ULMs from this study (left panel) and the NCBI GEO GSE593 data set (right panel), normalized to matched myometria. yellow = upregulation; blue = downregulation.
(0.69 MB JPG)
Regions of common loss in ULMs detected by array CGH. A, B. Genome browser mapping of the regions of common loss of genomic material as detected by a CGH in this study. Patient IDs are shown as yellow rectangles. Cancer-related microRNAs of interest in the deleted regions are listed under the chromosomes and highlighted by red arrowheads. C. Detailed genetic summary of the common regions of loss.
(0.60 MB JPG)
TMA immunohistochemical analysis of selected proteins in ULMs. Immunohistochemical analysis of selected proteins in 36 ULMs. A. Photomicrograph illustrating tissue microarray (TMA) sections with hematoxylin and eosin (H&E) stain (upper panel) and immunostaining of ER (bottom panel). Triplicate tissue cores from controls (matched myometrium) and tumors (ULMs) are indicated on the right. B. Differential expression of the selected target gene products is shown as mean and standard error of measurements (bars and t-bars, respectively). The net change for each gene product was calculated based on relative immunoreactivity in ULMs against matched myometrium. Red = gain in protein levels; Blue = reduction of protein levels.
(0.61 MB JPG)
Primers and PCR results for the predicted target genes of let-7 and miR-200s.
(0.03 MB DOC)
Antibodies used in this study.
(0.03 MB DOC)
Predicted target genes and pathways downstream of miR-200s amd miR-15/16.
(0.81 MB DOC)