Abstract
Purpose
Aniridia and congenital cataract represent rare but severe developmental ocular conditions. We examined 33 probands from France for mutations in several transcription factors associated with these phenotypes, the forkhead box E3 (FOXE3), paired box gene 6 (PAX6), paired-like homeodomain transcription factor 2 (PITX2), and paired-like homeodomain transcription factor 3 (PITX3) genes.
Methods
Out of 33 probands, 27 were affected with congenital cataract while the remaining six were affected with aniridia (with or without cataract). The coding regions of FOXE3, PAX6, PITX2, and PITX3 were examined by direct DNA sequencing of gene-specific PCR products.
Results
A novel dominant mutation at the stop codon of FOXE3, c.959G>C (p.X320SerextX72), was identified in a patient with congenital cataract. Another novel FOXE3 sequence change, c.571–579dup (p.Tyr191_Pro193dup), was identified in a patient with aniridia, mild lens opacities, and some additional ocular defects; this patient was also found to carry a nonsense mutation in PAX6. PAX6 mutations were identified in two additional probands with aniridia and cataracts. None of the observed sequence alterations were found in normal controls. No mutations were identified in PITX2 or PITX3.
Conclusions
The p.X320SerextX72 mutation is only the fourth FOXE3 allele associated with a dominant phenotype since the majority of FOXE3 mutations appear to be recessive with no phenotype observed in heterozygous carriers. The encoded protein is predicted to contain a complete normal sequence followed by seventy-two erroneous amino acids; the position and effect of this mutation are similar to two of the previously reported dominant changes, suggesting a common mechanism for dominant alleles. The p.Tyr191_Pro193dup is predicted to result in an in-frame duplication of three amino acids; however, the contribution of this mutation to the phenotype is unclear since the affected patient also carries a nonsense mutation in PAX6 which acts upstream of FOXE3 in the molecular pathway. The identified PAX6 mutations correspond to the two most commonly observed mutant alleles and demonstrate phenotypes that are consistent with the previously reported spectrum.
Introduction
Aniridia and congenital cataract represent rare but severe ocular conditions reflecting abnormal development of the anterior segment of the eye. Aniridia is characterized by the congenital absence of the iris and is most commonly inherited as an autosomal-dominant disorder. Aniridia is a panocular disease often associated with additional ocular symptoms such as visual impairment and nystagmus [1]. Congenital cataract is an opacification of the ocular lens present at birth. Congenital cataract is usually hereditary and can be transmitted as a dominant or a recessive trait. Congenital cataracts are frequently accompanied by other ocular defects, particularly anterior segment anomalies including corneal limbal deficiency and glaucoma with goniodysgenesis, but also posterior segment anomalies such as foveal hypoplasia [2]. The complexity of anterior segment phenotypes is likely due to close interaction of corresponding tissues during development and/or sharing of genetic factors that underlie their proper differentiation. Mutations in transcription factors that play active roles in directing normal embryonic developmental patterning were shown to result in a spectrum of anterior segment anomalies including aniridia and cataracts.
Mutations in forkhead box E3 (FOXE3), a forkhead transcription factor expressed in the lens which is located at chromosome 1p32, have been associated with both recessive and dominant ocular disease. Dominant mutations in FOXE3 have been seen in patients with congenital cataract and/or anterior segment disease; three dominant disease-causing mutations have been reported to date [3-5]. Meanwhile, recessive mutations in FOXE3 have been identified in eight families with aphakia, microphthalmia, and sclerocornea as the principal phenotype [5-8]. Obligate carriers in all recessive families were unaffected with the exception of one parent who was reported to have unilateral ocular ‘cloudiness’ [7].
Paired box gene 6 (PAX6) is a transcriptional regulator located at chromosome 11p13. Mutations or deletions of PAX6 were first identified in patients with aniridia [9,10]. While aniridia remains the predominant phenotype associated with PAX6 mutations [11], related ocular phenotypes including anterior segment malformations such as Peters’s anomaly [12], keratitis [13], foveal hypoplasia [14], congenital cataract [15], and optic nerve defects [16] were also shown to be associated with PAX6 deficiency. Nonocular defects include abnormal brain structure, central auditory function, and olfactory bulb development [17-19]. Two patients with Gillespie-like syndrome (iris hypoplasia/aniridia, tremor/ataxia, and learning disability/mental retardation) were also found to have mutations in PAX6 [20,21]. In addition, WAGR syndrome (Wilms tumor, Aniridia, Genitourinary anomalies, and mental Retardation) is associated with a contiguous gene deletion which includes PAX6 and the WT1 gene (WT1) [22]. A recent review of PAX6 mutations in the Human PAX6 Mutation Database [23] found that mutations which lead to a premature termination codon are generally associated with aniridia while missense mutations typically result in other related ocular phenotypes [11]. This review also identified four CpG dinucleotides which are mutation hotspots accounting for 21% of mutations in the database and all resulting in aniridia [11].
Paired-like homeodomain transcription factor 2 (PITX2) is a homeodomain transcription factor at chromosome 4q25. Mutations in PITX2 were first reported in patients with Axenfeld-Rieger syndrome [24]. Subsequently, the associated phenotypes have been expanded to include partial aniridia [25], Peters’ anomaly [26], iris hypoplasia/iridogoniodysgenesis syndrome [27,28], and ring dermoid of the cornea [29]. A wide range of mutations have been reported including missense, nonsense, splice-site, and intragenic deletions/duplications, along with whole gene deletions and translocations upstream of PITX2; the majority of these mutations result in Axenfeld-Rieger syndrome [30].
Mutations in paired-like homeodomain transcription factor 3 (PITX3), another homeodomain transcription factor located at chromosome 10q25, are associated with autosomal dominant congenital cataract with or without anterior segment dysgenesis [31]. A recurrent 17-bp insertion (c.657_673dup17) in the COOH-terminal region in the gene is the most common mutation, resulting primarily in congenital posterior polar cataract along with highly variable anterior segment mesenchymal dysgenesis (ASMD) in some individuals [31-35]. Two additional heterozygous mutations associated with isolated cataract have been reported [31,32,36]; one of these mutations, c.650delG, was also seen in homozygous form in one family, resulting in severe microphthalmia with neurologic deficits [36].
In this study, we screened thirty-three unrelated patients affected with congenital cataract and/or aniridia for mutations in FOXE3, PAX6, PITX2, and PITX3. Causative mutations were identified in FOXE3 and PAX6 but not PITX2 or PITX3.
Methods
The human studies were approved by Paris 7 University Hospital and the Institutional Review Boards of the Children’s Hospital of Wisconsin and the University of Iowa Hospitals and Clinics and informed consent was obtained for every subject. We screened a population of 33 probands from France: 27 were affected with congenital cataract while the remaining six were affected with aniridia (with or without cataract).
Gene-specific products were generated using the previously reported primers and conditions for FOXE3 [3], PAX6 [37], PITX2 [24], and PITX3 [31]. Briefly, PCR was conducted in 30 μl reactions containing 40 ng DNA, Biolase Reaction Buffer (Bioline, Taunton, MA), 0.25 mmol/l each dNTP, 1.5 units Biolase DNA polymerase (Bioline) and 0.2 micromol/l of each oligodeoxynucleotide primer. Cycling profile included one cycle of 94 °C for 5 min followed by 30 cycles of 94 °C for 45 s, 55-60 °C (please see Table 1 for annealing temperature and other details) for 45 s, and 72 °C for 45 s, and one cycle of 72 °C for 10 min. The coding regions of FOXE3, PAX6, PITX2, and PITX3 were examined by direct DNA sequencing of PCR products, as previously described [38]. Briefly, PCR products were sequenced in both directions using Big Dye Terminator v3.1 (Applied Biosystems, Foster City, CA) using a 3730XL DNA Analyzer (Applied Biosystems). Chromatograms were examined manually and all initially identified changes were confirmed by additional independent PCR and sequencing experiments. One hundred eighty two samples from healthy individuals of Caucasian decent were examined for the changes observed in FOXE3 and PAX6 using SSCP (single strand conformational polymorphism) analysis along with the corresponding patient samples as positive controls. In addition to this, the previously reported FOXE3 normal variation data [7] and PAX6 variation summarized in the Human PAX6 Mutation Database [23] were used for comparisons.
Table 1. PCR primers and conditions.
| Set | Forward primer | Reverse primer | Gene region | Annealing temperature (°C) | Product size (bp) |
|---|---|---|---|---|---|
|
FOXE3 primers [3] | |||||
| 1 |
tgtccatataaagcgggtcg |
atgtacgagtagggcggctt |
Exon 1 (N-terminus) |
56 |
298 |
| 2 |
ttctctggcttccctgccct |
tcggtgatgaagcggtagat |
Exon 1 (N-terminus, forkhead domain) |
57 |
271 |
| 3 |
aagccgccctactcgtacat |
tcgttgagcgtgagattgtg |
Exon 1 (Forkhead domain) |
56 |
170 |
| 4 |
ttcatcaccgaacgctttgc |
aggaagctgccgttgtcgaa |
Exon 1 (Forkhead domain) |
56 |
185 |
| 5 |
aagggcaactactggacgct |
tagctccggctgcaggttca |
Exon 1 (Forkhead domain, C-terminus) |
55 |
267 |
| 6 |
tctgttcagcgtcgacagc |
acaggtcgcacaggtgcct |
Exon 1 (C-terminus) |
55 |
352 |
|
PAX6 primers [37] | |||||
| 1 |
ctcatttcccgctctggttc |
aagagtgtgggtgaggaagt |
Exon 1 |
60 |
197 |
| 2 |
ttatctctcactctccagcc |
aagcgagaagaaagaagcgg |
Exon 2 |
60 |
276 |
| 3 |
tcagagagcccatcgacgtat |
ctgtttgtgggttttgagcc |
Exon 3 |
60 |
193 |
| 4 |
ttgggagttcaggcctacct |
gaagtcccagaaagaccaga |
Exon 4 |
60 |
153 |
| 5 |
cctcttcactctgctctctt |
atgaagagagggcgttgaga |
Exon 5 |
60 |
257 |
| 5a |
tgaaagtatcatcatatttgtag |
gggaagtggacagaaaacca |
Exon 5a |
55 |
237 |
| 6 |
gtggttttctgtccacttcc |
aggagagagcattgggctta |
Exon 6 |
60 |
299 |
| 7 |
caggagacactaccatttgg |
atgcacatatggagagctgc |
Exon 7 |
60 |
252 |
| 8 |
gggaatgttttggtgaggct |
caaagggccctggctaaatt |
Exon 8 |
60 |
371 |
| 9 |
gtagttctggcacaatatgg |
gtactctgtacaagcacctc |
Exon 9 |
60 |
206 |
| 10 |
gtagacacagtgctaacctg |
cccggagcaaacaggtttaa |
Exon 10 |
60 |
243 |
| 11 |
ttaaacctgtttgctccggg |
ttatgcaggccaccaccagc |
Exon 11 |
60 |
208 |
| 12 |
gctgtgtgatgtgttcctca |
tgcagcctgcagaaacagtg |
Exon 12 |
60 |
227 |
| 13 |
catgtctgtttctcaaaggga |
gaacaattaacttttgctggcc |
Exon 13 |
55 |
957 |
|
PITX2 primers [24] | |||||
| 1a |
ttggctcctaagtgcccc |
ccagactcgcattatctcac |
Exon 1a |
56 |
596 |
| 1b |
cttgacacttctctgtcagg |
aagcgggaatgtctgcagg |
Exon 1b |
56 |
667 |
| 2 |
tagtctcatctgagccctgc |
cactggcgatttggttctga |
Exon 2 |
56 |
282 |
| 3 |
acgcctctctccgcacgt |
ttcttgcgctttcgcccga |
Exon 3 |
56 |
258 |
| 4 |
cagctcttccacggcttct |
ttctctcctggtctacttgg |
Exon 4 |
56 |
374 |
| 5a |
gtaatctgcactgtggcatc |
ctgtgggtgcggctcaca |
Exon 5 |
56 |
627 |
| 5b |
ctgagactgaaagcaaagca |
ctcccatgaaataaaacacattt |
Exon 5 |
56 |
787 |
|
PITX3 primers [31] | |||||
| 1 |
cctggtctgccataaagtga |
attctcgacctgttcccaag |
Exon 1 |
55 |
343 |
| 2 |
acgcagccccagctttac |
aagccagcgcatattctcc |
Exon 2 |
55 |
395 |
| 3 |
gtgcaggacataacagcttc |
gagcagaggctggaggttg |
Exon 3 |
55 |
528 |
| 4a |
ctctagccacctcatctcg |
aggcataagggcaggacac |
Exon 5 |
55 |
524 |
| 4b | agacctttccattcgccttc | agtcaaaatgaccccagtcc | Exon 5 | 55 | 483 |
Results
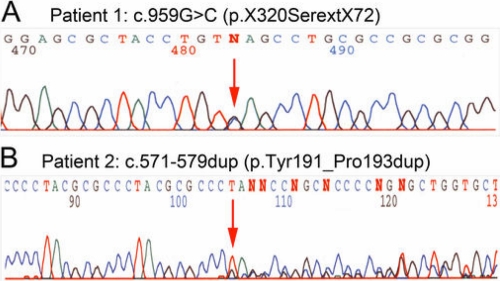
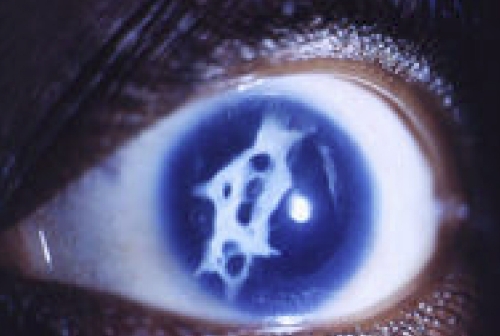
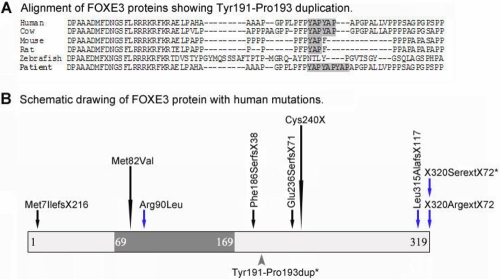
Screening of FOXE3 identified a mutation in the stop codon of FOXE3, c.959G>C (p.X320SerextX72), in Patient 1 with congenital cataract (Figure 1A). There was no family history of ocular disease and no other family members were available for testing. The mutation results in a change of the termination codon into serine leading to the addition of 72 erroneous amino acids to the end of the protein. No other changes in FOXE3 were identified in this patient. A second sequence alteration in FOXE3 was identified in Patient 2 affected with aniridia, corneal limbal insufficiency, nystagmus, severe axile myopia, mild lens opacities, and development of a fibrous posterior capsular reaction after lens surgery (Figure 2). This FOXE3 variant, c.571–579dup (p.Tyr191_Pro193dup), results in an in-frame duplication of 3 amino acids (Figure 1B and Figure 3A). The same patient was later found to also carry a PAX6 mutation (see below). the parents of Patient 2 appear to be unaffected but were not available for testing. Neither of the identified FOXE3 variants was seen in 182 Caucasian control individuals nor the previously reported 332 controls of mixed ethnicity [7].
Figure 1.
Novel FOXE3 mutations. Fragments of DNA sequence trace chromatograms corresponding to the c.959G>C (p.X320SerextX73) mutation in Patient 1(A) and the c.571–579dup (p.Tyr191_Pro193dup) variant in Patient 2 (B). Mutation sites indicated with arrows.
Figure 2.
Patient photograph. Photographic image of the eye of Patient 2.
Figure 3.
Summary of FOXE3 mutations. A: Alignment of FOXE3 proteins showing region of p.Tyr191-Pro193 duplication. Please note two YAP amino acid motifs present in normal human and cow FOXE3 proteins and an additional insertion of this motif identified in Patient 2. B: Schematic drawing of FOXE3 protein with human mutations. Forkhead domain is shown in dark gray. Mutations resulting in recessive phenotypes are indicated with black arrows while mutations causing dominant disease- with blue arrows. The variant identified in Patient 2 is indicated with a gray arrowhead below the protein. Recurrent mutations are denoted with extended arrows. Positions of mutations identified in this study are shown with asterisks.
Mutations in PAX6 were identified in three unrelated probands with aniridia and cataract. Patient 2 (also carrying FOXE3 mutation, please see above) was found to have a c.718C>T (p.Arg240X) mutation in PAX6. Patient 3, affected with aniridia and congenital cataract, was found to carry the same mutation. Finally, Patient 4, with aniridia and mild cataract, was found to carry a c.607C>T (p.Arg203X) mutation. The mother of Patient 4 also has aniridia and cataract but was not available for testing. Both of these mutations were previously reported in patients with aniridia (The Human PAX6 Mutation Database) [11]. No mutations in PITX2 or PITX3 were identified in this group of patients.
Discussion
Mutation analysis of DNA obtained from patients affected with congenital cataracts (27) and aniridia with or without cataracts (6) identified two novel variants in FOXE3. One of these variants, p.X320SerextX72, apparently represents a causative mutation responsible for a phenotype of dominant congenital cataract, while the other variant, p.Tyr191_Pro193dup, is of unknown significance. Mutations in FOXE3 appear to explain ~4% of congenital cataract cases in this population.
The p.X320SerextX72 mutation in FOXE3 is only the fourth mutant allele associated with a dominant phenotype since the majority of FOXE3 mutations appear to be recessive with no phenotype observed in heterozygous carriers (Figure 3B) [3-8]. The location and the predicted effect of this mutation are consistent with two of the previously described dominant mutations in FOXE3: p.Leu315AlafsX117 [3] and p.X320ArgextX72 [5] (Figure 3B). All three mutations are predicted to result in the addition of numerous erroneous amino acids to the most distant end of the FOXE3 protein. Two of these mutations, p.X320SerextX72 (reported here) and p.X320ArgextX72 [5], affect the stop codon and therefore are predicted to retain the entire normal FOXE3 sequence. The similarity between these mutations suggests a possible common mechanism for FOXE3 dominant alleles. An additional dominant mutation which affects the DNA-binding domain of FOXE3 has also been reported (p.Arg90Leu [4]).
The congenital cataract phenotype observed in the patient carrying the p.X320SerextX72 allele (Patient 1) is consistent with other dominant FOXE3 mutations. The initially reported p.Leu315AlafsX117 mutation was found in a patient and her mother with cataracts and posterior embryotoxon; the proband was also affected with myopia [3]. The p.X320ArgextX72 mutation was identified in a family with a highly variable phenotype: the proband was affected with bilateral microphthalmia with aphakia and sclerocornea in the right eye and Peters’s anomaly and congenital cataract in the left while her mother and uncle had bilateral early onset cataract (age of diagnosis not given) with iris colobomas also seen in the uncle. A maternal aunt had a cataract extraction in her 20s and the maternal grandmother had a cataract extraction in her 40s. All of the above family members were found to carry the mutation while an unaffected maternal uncle did not carry the mutation [5]. Finally, the p.Arg90Leu dominant mutation affecting the DNA-binding domain of FOXE3 was associated with Peters’ anomaly and glaucoma, but not cataract [4]. There was a family history of the disease consistent with dominant inheritance but no other family members were available for testing. In general, the phenotype associated with dominant mutations is milder than the recessive FOXE3 phenotype (microphthalmia with aphakia in many cases), with the exception of the proband reported by Iseri et al. [5].
The effect of the p.Tyr191_Pro193dup mutation in FOXE3 is unclear since Patient 2 also carries the nonsense mutation in PAX6. The patient does demonstrate mild anterior segment dysgenesis and lens opacities, but these features are not inconsistent with the PAX6 mutation [11]. Since the identified PAX6 mutation has been previously reported as causative in many families and since Pax6 has been shown to act upstream of Foxe3 in the ocular pathway [39], the phenotype of this patient is apparently caused by the PAX6 deficiency. The effect of the p.Tyr191_Pro193dup mutation, if any, is most likely masked by this earlier molecular defect. It is also possible that this variant does make a contribution to the observed phenotype that is difficult to dissect in the presence of the PAX6 mutation, or that it represents a very rare polymorphism. This case emphasizes the importance of the exclusion of mutations in major known genes in mutation screens to ensure accurate identification of the causative allele.
The identified mutations in PAX6 are consistent with the associated phenotypes. These mutations, c.718C>T (p.Arg240X) and c.607C>T (p.Arg203X), are two of the most common mutations, both affecting a CpG mutational hotspot [11]. Of the 31 patients with the p.Arg240X mutation reported in the Human PAX6 Mutation Database, 26 have isolated aniridia while the remaining five have associated ocular anomalies (four with cataract). Of the 23 patients with the p.Arg203X mutation in the database, 18 are reported to have isolated aniridia and the remaining five have associated ocular anomalies (one with cataract).
The absence of PITX2 mutations in examined patients is not surprising given its limited contribution to aniridia and a strong association with syndromic versus isolated anterior segment phenotypes. As for PITX3, which is linked primarily with cataracts with or without anterior segment defects, our findings suggest that it is not a major cause of congenital cataract.
Acknowledgments
We are grateful to the families for their participation in this study and to our funding sources. This project was supported by awards EY015518 and EY013606 from the National Eye Institute and a grant from the Children’s Research Institute Foundation at Children’s Hospital of Wisconsin to EVS. We also thank the Robert Debre DNA bank and team for their support of this project.
References
- 1.Lee H, Khan R, O'Keefe M. Aniridia: current pathology and management. Acta Ophthalmol. 2008;86:708–15. doi: 10.1111/j.1755-3768.2008.01427.x. [DOI] [PubMed] [Google Scholar]
- 2.Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–49. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semina EV, Brownell I, Mintz-Hittner HA, Murray JC, Jamrich M. Mutations in the human forkhead transcription factor FOXE3 associated with anterior segment ocular dysgenesis and cataracts. Hum Mol Genet. 2001;10:231–6. doi: 10.1093/hmg/10.3.231. [DOI] [PubMed] [Google Scholar]
- 4.Ormestad M, Blixt A, Churchill A, Martinsson T, Enerback S, Carlsson P. Foxe3 haploinsufficiency in mice: a model for Peters' anomaly. Invest Ophthalmol Vis Sci. 2002;43:1350–7. [PubMed] [Google Scholar]
- 5.Iseri SU, Osborne RJ, Farrall M, Wyatt AW, Mirza G, Nurnberg G, Kluck C, Herbert H, Martin A, Hussain MS, Collin JR, Lathrop M, Nurnberg P, Ragoussis J, Ragge NK. Seeing clearly: the dominant and recessive nature of FOXE3 in eye developmental anomalies. Hum Mutat. 2009;30:1378–86. doi: 10.1002/humu.21079. [DOI] [PubMed] [Google Scholar]
- 6.Valleix S, Niel F, Nedelec B, Algros MP, Schwartz C, Delbosc B, Delpech M, Kantelip B. Homozygous nonsense mutation in the FOXE3 gene as a cause of congenital primary aphakia in humans. Am J Hum Genet. 2006;79:358–64. doi: 10.1086/505654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis LM, Tyler RC, Schneider A, Bardakjian T, Stoler JM, Melancon SB, Semina EV. FOXE3 plays a significant role in autosomal recessive microphthalmia. Am J Med Genet A. 2010;152A:582–90. doi: 10.1002/ajmg.a.33257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anjum I, Eiberg H, Baig SM, Tommerup N, Hansen L. A mutation in the FOXE3 gene causes congenital primary aphakia in an autosomal recessive consanguineous Pakistani family. Mol Vis. 2010;16:549–55. [PMC free article] [PubMed] [Google Scholar]
- 9.Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie ND, Meijers-Heijboer H, Drechsler M. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–74. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 10.Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet. 1992;1:328–32. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- 11.Tzoulaki I, White IM, Hanson IM. PAX6 mutations: genotype-phenotype correlations. BMC Genet. 2005;6:27. doi: 10.1186/1471-2156-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson IM, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ, Punnett HH, van Heyningen V. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat Genet. 1994;6:168–73. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 13.Mirzayans F, Pearce WG, MacDonald IM, Walter MA. Mutation of the PAX6 gene in patients with autosomal dominant keratitis. Am J Hum Genet. 1995;57:539–48. [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–72. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- 15.Azuma N, Yamaguchi Y, Handa H, Hayakawa M, Kanai A, Yamada M. Missense mutation in the alternative splice region of the PAX6 gene in eye anomalies. Am J Hum Genet. 1999;65:656–63. doi: 10.1086/302529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azuma N, Yamaguchi Y, Handa H, Tadokoro K, Asaka A, Kawase E, Yamada M. Mutations of the PAX6 gene detected in patients with a variety of optic-nerve malformations. Am J Hum Genet. 2003;72:1565–70. doi: 10.1086/375555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, Stevens JM, Kendall BE, Shorvon SD, Hanson IM, Moore AT, van Heyningen V. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet. 2001;28:214–6. doi: 10.1038/90042. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell TN, Free SL, Williamson KA, Stevens JM, Churchill AJ, Hanson IM, Shorvon SD, Moore AT, van Heyningen V, Sisodiya SM. Polymicrogyria and absence of pineal gland due to PAX6 mutation. Ann Neurol. 2003;53:658–63. doi: 10.1002/ana.10576. [DOI] [PubMed] [Google Scholar]
- 19.Bamiou DE, Musiek FE, Sisodiya SM, Free SL, Davies RA, Moore A, van Heyningen V, Luxon LM. Deficient auditory interhemispheric transfer in patients with PAX6 mutations. Ann Neurol. 2004;56:503–9. doi: 10.1002/ana.20227. [DOI] [PubMed] [Google Scholar]
- 20.Ticho BH, Hilchie-Schmidt C, Egel RT, Traboulsi EI, Howarth RJ, Robinson D. Ocular findings in Gillespie-like syndrome: association with a new PAX6 mutation. Ophthalmic Genet. 2006;27:145–9. doi: 10.1080/13816810600976897. [DOI] [PubMed] [Google Scholar]
- 21.Graziano C, D'Elia AV, Mazzanti L, Moscano F, Guidelli Guidi S, Scarano E, Turchetti D, Franzoni E, Romeo G, Damante G, Seri M. A de novo nonsense mutation of PAX6 gene in a patient with aniridia, ataxia, and mental retardation. Am J Med Genet A. 2007;143A:1802–5. doi: 10.1002/ajmg.a.31808. [DOI] [PubMed] [Google Scholar]
- 22.Brémond-Gignac D, Gerard-Blanluet M, Copin H, Bitoun P, Baumann C, Crolla JA, Benzacken B, Verloes A. Three patients with hallucal polydactyly and WAGR syndrome, including discordant expression of Wilms tumor in MZ twins. Am J Med Genet A. 2005;134:422–5. doi: 10.1002/ajmg.a.30646. [DOI] [PubMed] [Google Scholar]
- 23.Brown A, McKie M, van Heyningen V, Prosser J. The Human PAX6 Mutation Database. Nucleic Acids Res. 1998;26:259–64. doi: 10.1093/nar/26.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–9. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 25.Saadi I, Semina EV, Amendt BA, Harris DJ, Murphy KP, Murray JC, Russo AF. Identification of a dominant negative homeodomain mutation in Rieger syndrome. J Biol Chem. 2001;276:23034–41. doi: 10.1074/jbc.M008592200. [DOI] [PubMed] [Google Scholar]
- 26.Doward W, Perveen R, Lloyd IC, Ridgway AE, Wilson L, Black GC. A mutation in the RIEG1 gene associated with Peters' anomaly. J Med Genet. 1999;36:152–5. [PMC free article] [PubMed] [Google Scholar]
- 27.Alward WL, Semina EV, Kalenak JW, Heon E, Sheth BP, Stone EM, Murray JC. Autosomal dominant iris hypoplasia is caused by a mutation in the Rieger syndrome (RIEG/PITX2) gene. Am J Ophthalmol. 1998;125:98–100. doi: 10.1016/s0002-9394(99)80242-6. [DOI] [PubMed] [Google Scholar]
- 28.Kulak SC, Kozlowski K, Semina EV, Pearce WG, Walter MA. Mutation in the RIEG1 gene in patients with iridogoniodysgenesis syndrome. Hum Mol Genet. 1998;7:1113–7. doi: 10.1093/hmg/7.7.1113. [DOI] [PubMed] [Google Scholar]
- 29.Xia K, Wu L, Liu X, Xi X, Liang D, Zheng D, Cai F, Pan Q, Long Z, Dai H, Hu Z, Tang B, Zhang Z, Xia J. Mutation in PITX2 is associated with ring dermoid of the cornea. J Med Genet. 2004;41:e129. doi: 10.1136/jmg.2004.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tümer Z, Bach-Holm D. Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur J Hum Genet. 2009;17:1527–39. doi: 10.1038/ejhg.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–70. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- 32.Berry V, Yang Z, Addison PK, Francis PJ, Ionides A, Karan G, Jiang L, Lin W, Hu J, Yang R, Moore A, Zhang K, Bhattacharya SS. Recurrent 17 bp duplication in PITX3 is primarily associated with posterior polar cataract (CPP4). J Med Genet. 2004;41:e109. doi: 10.1136/jmg.2004.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finzi S, Li Y, Mitchell TN, Farr A, Maumenee IH, Sallum JM, Sundin O. Posterior polar cataract: genetic analysis of a large family. Ophthalmic Genet. 2005;26:125–30. doi: 10.1080/13816810500229124. [DOI] [PubMed] [Google Scholar]
- 34.Burdon KP, McKay JD, Wirth MG, Russell-Eggit IM, Bhatti S, Ruddle JB, Dimasi D, Mackey DA, Craig JE. The PITX3 gene in posterior polar congenital cataract in Australia. Mol Vis. 2006;12:367–71. [PubMed] [Google Scholar]
- 35.Summers KM, Withers SJ, Gole GA, Piras S, Taylor PJ. Anterior segment mesenchymal dysgenesis in a large Australian family is associated with the recurrent 17 bp duplication in PITX3. Mol Vis. 2008;14:2010–5. [PMC free article] [PubMed] [Google Scholar]
- 36.Bidinost C, Matsumoto M, Chung D, Salem N, Zhang K, Stockton DW, Khoury A, Megarbane A, Bejjani BA, Traboulsi EI. Heterozygous and homozygous mutations in PITX3 in a large Lebanese family with posterior polar cataracts and neurodevelopmental abnormalities. Invest Ophthalmol Vis Sci. 2006;47:1274–80. doi: 10.1167/iovs.05-1095. [DOI] [PubMed] [Google Scholar]
- 37.Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–9. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 38.Reis LM, Tyler RC, Abdul-Rahman O, Trapane P, Wallerstein R, Broome D, Hoffman J, Khan A, Paradiso C, Ron N, Bergner A, Semina EV. Mutation analysis of B3GALTL in Peters' Plus syndrome. Am J Med Genet A. 2008;146A:2603–10. doi: 10.1002/ajmg.a.32498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dimanlig PV, Faber SC, Auerbach W, Makarenkova HP, Lang RA. The upstream ectoderm enhancer in Pax6 has an important role in lens induction. Development. 2001;128:4415–24. doi: 10.1242/dev.128.22.4415. [DOI] [PubMed] [Google Scholar]