Abstract
Multizinc finger peptides are likely to reach increased prominence in the search for the “ideal” designer transcription factor for in vivo applications such as gene therapy. However, for these treatments to be effective and safe, the peptides must bind with high affinity and, more importantly, with great specificity. Our previous research has shown that zinc finger arrays can be made to bind 18 bp of DNA with picomolar affinity, but also has suggested that arrays of fingers also may bind tightly to related sequences. This work addresses the question of zinc finger DNA binding specificity. We show that by changing the way in which zinc finger arrays are constructed—by linking three two-finger domains rather than two three-finger units—far greater target specificity can be achieved through increased discrimination against mutated or closely related sequences. These new peptides have the added capability of being able to span two short gaps of unbound DNA, although still binding with picomolar affinity to their target sites. We believe that this new method of constructing zinc finger arrays will offer greater efficacy in the fields of gene therapy and in the production of transgenic organisms than previously reported zinc finger arrays.
Keywords: gene regulation, DNA binding site, zinc finger, polydactyl peptides
Zinc finger peptides are rapidly emerging as the most likely candidates for use as “designer” transcription factors. They have been successfully manipulated to bind desired target sequences (1–5), attached to transcription repression and activation domains (6), joined to enzymatic domains (7–10), and covalently linked to create multifinger peptides, which recognize extended DNA sequences with increased affinity (11–13).
It is desirable that a designer transcription factor for uses such as gene therapy should have the ability to target virtually unique sites within any genome. For complex genomes such as in humans, an address of at least 16 bp is required to specify a potentially unique DNA sequence. Hence, a six-zinc-finger peptide (with an 18-bp recognition sequence) could, in theory, be used for the specific regulation of a single gene within any genome. In addition, a significant increase in binding affinity might also be expected. In simple terms, if a three-finger peptide (with a 9-bp recognition sequence) binds DNA with nanomolar affinity, two tandemly linked three-finger peptides might be expected to bind an 18-bp sequence with an affinity of 10−15-10−18 M. Several polyzinc finger peptides of this length have now been described, but with the exception of one example in which a six-finger peptide was reported to bind >6,000-fold tighter than its three-finger components (11), the affinity enhancements have been modest (12, 13). The cause of this is thought to arise from the geometry of the interaction between amino acid side chains within the peptide and the bases within the DNA binding site. Various studies have now shown that the helical periodicity of the α-helix of a zinc finger does not exactly match that of B-form DNA. Hence, DNA is slightly unwound when a zinc finger peptide is bound (14, 15). It also has been shown that the canonical -TGEKP- linker sequence is fully extended when the transcription factor IIIA for the Xenopus 5S RNA gene (TFIIIA) and Zif268 bind DNA, but even then, it has been suggested that this sequence is fractionally too short to allow the fingers to bind in their optimal conformation (16, 17). The combination of these two factors means that the longer the array of zinc fingers, the greater the strain within the protein–DNA complex, and hence, the greater the loss in binding energy. In this paper we suggest a new approach to zinc finger peptide design that minimizes the strain when creating long arrays of zinc fingers.
An additional and perhaps more important consideration when employing polydactyl peptides for in vivo use is that of binding specificity. Our previous work suggests that individual zinc finger modules in an array can be repositioned when they do not have a spatially correlated binding site, to enable the remaining fingers to bind their correct recognition sequences (18). We believe that this phenomenon would allow the polydactyl peptides (based on tandemly arrayed three-finger domains) reported in previous studies to bind with relatively high affinity to related DNA sites containing various mutations and deletions. This would effectively mean that these peptides would not exclusively target the desired sequences within complex genomes.
In this work we address the issue of zinc-finger specificity by designing six-finger peptides that bind with high affinity only to their specific target sequence. We have created a series of six-finger peptides by joining two-finger units with slightly longer-than-canonical linkers (either by inserting -gly- or -gly, gly, ser- within the canonical linker sequence). We also created a six-finger peptide by joining two three-finger domains with the 9-aa linker -LRQKDGERP-, as used in a previous study (11). Our results clearly demonstrate that 3 × 2 finger peptides (which we will refer to as 3 × 2F peptides) are far more specific for their designed DNA binding sequence than similar 2 × 3F peptides. One such 3 × 2F peptide demonstrated a discrimination of almost 106-fold for its 18-bp target site over one of its constituent 9-bp half-site, whereas the similar 2 × 3F peptide discriminated between the same two sites by a factor of ≈103.
This paper demonstrates another advantage of the 3 × 2F design. By extending the linker sequence between zinc finger pairs, we show that 3 × 2F peptides are able to accommodate two regions of unbound DNA within their recognition sequence, rather than one, as is the case for 2 × 3F peptides. Hence, these constructs also allow more flexibility in the selection of DNA target sequences for designer transcription factors.
Materials and Methods
Design, Construction, and Cloning of Zinc Finger Genes.
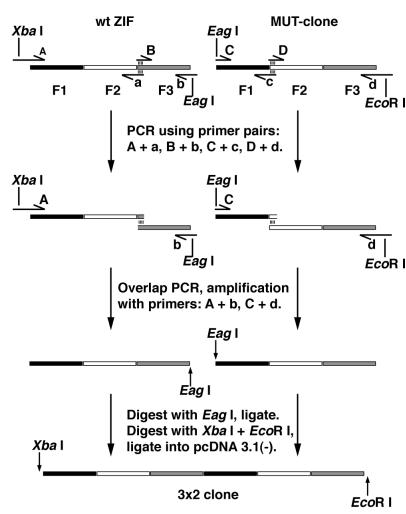
3 × 2F constructs were created by PCR amplification of the three-finger wild-type Zif268 and a three-finger mutant peptide of the three zinc fingers of murine transcription factor Zif268a, MUT, with primers A, a, B, b, and C, c, D, d respectively, as shown in Fig. 1. Overlap PCR was then used to reconstruct the three-finger units with modified linker sequences between fingers 2 and 3 of wild-type Zif268 and fingers 1 and 2 of the MUT clone (18). The six-finger constructs were then synthesized by digesting the PCR products with EagI and ligating at that site to create a -TGERP- linker peptide between the third and fourth fingers of the construct. These constructs were called 3 × 2F ZMS, 3 × 2F ZML, 3 × 2F ZMSL, and 3 × 2F ZMLS, depending on the sequence length of the modified linkers. The 2 × 3F ZM construct was synthesized by PCR amplification of the three-finger peptides Zif268 and MUT, by using primers that created EagI sites at the position to be joined, as above. The nomenclature and descriptions of the peptides described above are displayed in Table 1. All zinc-finger constructs were digested with XbaI and EcoRI restriction enzymes and inserted into the similarly digested, eukaryotic expression vector pcDNA 3.1(−) (Invitrogen). The sequences of all constructs were confirmed by dideoxy sequencing.
Figure 1.
Schematic representation of the PCR construction procedure used to create the 3 × 2F peptides.
Table 1.
The nomenclature and descriptions of the peptides used in the study
| Peptide name | Description |
|---|---|
| ZIF (Z) | The three fingers of Zif268 |
| MUT (M) | A three-finger mutant of Zif268 |
| 3 × 2F ZMS | A six-finger peptide of Z fused N-terminally to M using the linker sequence -TGERP-, which contains a single glycine insertion within the linkers between fingers 2 and 3 and between fingers 4 and 5 |
| 3 × 2F ZML | As above, but with -gly, ser, gly- insertions instead of a single glycine |
| 3 × 2F ZMSL | As above, but with a single glycine insertion in the linker between fingers 1 and 2, and a -gly, ser, gly- insertion between fingers 4 and 5 |
| 3 × 2F ZMLS | As above, but with a -gly, ser, gly- insertion between fingers 1 and 2 and a single glycine insertion between fingers 4 and 5 |
| 2 × 3F ZM | A six-finger peptide of Z fused N-terminally to M using the linker sequence -LRQKDGERP- between fingers 3 and 4 |
Template Preparation and Protein Expression.
Plasmids containing zinc finger peptides were purified and quantified as described (18). Peptide expression was performed in vitro by using the TNT Quick Coupled Transcription/Translation System (Promega), according to the manufacturer's instructions, except that the medium was supplemented with 500 μM ZnCl2.
Gel-Shift Assays.
All peptides were assayed by using 32P end-labeled synthetic oligonucleotide duplexes containing the required binding site sequences. The sequences of the binding sites used are shown in Tables 2 and 3.
Table 2.
The binding site sequences used in gel-shift experiments with the 3 × 2F ZMS and 2 × 3F ZM peptides and the binding affinities obtained
| Binding site name | Binding site sequence | Apparent
Kd, pM
|
|
|---|---|---|---|
| 3 × 2F ZMS | 2 × 3F ZM | ||
| ZIF | GCG TGG GCG | 1.1 × 105 | 2,200 |
| 123456 | GCG GAC GCG GCG TGG GCG | 0.6 | 1.4 |
| 123///56 | GCG GAC ATC GCG TGG GCG | 890 | 15 |
| 123//56 | GCG GAC TC GCG TGG GCG | 270 | 14 |
| 123/56 | GCG GAC T GCG TGG GCG | 630 | 14 |
| 12356 | GCG GAC GCG TGG GCG | 2.2 × 104 | 360 |
Binding site residues that were mutated (and subsequently deleted) are underlined.
Table 3.
The binding site sequences used in gel-shift experiments with the 3 × 2F peptides and the binding affinities determined
| Binding site name | Binding site sequence* | Apparent
Kd, pM
|
|||
|---|---|---|---|---|---|
| 3 × 2F ZMS | 3 × 2F ZML | 3 × 2F ZMSL | 3 × 2F ZMLS | ||
| 123456 | GCG GAC GCG GCG TGG GCG | 0.6 | 0.9 | ND | ND |
| 12/34/56 | GCG GAC T GCG GCG T TGG GCG | 1.8 × 104 | 5 | 110 | 120 |
| 12//34//56 | GCG GAC TC GCG GCG TC TGG GCG | ND | 1.1 × 104 | 1.2 × 104 | 1.2 × 104 |
| 1234/56 | GCG GAC T GCG GCG TGG GCG | 54 | ND | 3 | 89 |
| 12/3456 | GCG GAC GCG GCG T TGG GCG | 77 | ND | 73 | 5 |
ND, not done, represents experiments for which Kds were not calculated.
Designed gaps in the target sequence are shown in bold.
DNA binding reactions were performed as described (18).
Active Peptide Concentration, Binding Affinity, and Specificity.
The concentration of active peptide, its binding affinity, and its DNA sequence specificity were determined as described (18).
Results
Design of the Six-Zinc Finger Fusion Peptides.
The goal was to create high-affinity six-finger fusion peptides with greater discrimination against nontarget sites than previously achieved. It was thought that within a three-finger unit the suboptimal binding of an individual finger would be better compensated for than within a two-finger unit. Therefore, by linking pairs of fingers together (with linkers slightly longer than canonical linkers), a more effective peptide for gene regulation may be generated. In other words, the entire zinc finger pair would contribute minimal binding energy to the peptide–DNA complex if one of the fingers has a suboptimal binding interaction. The design may also improve six-finger peptide–DNA interactions by allowing the peptide to adjust more regularly to the register of the DNA double helix, reducing the strain within the complex and enhancing the binding affinity. Creating six-finger constructs with two extended linker sequences also gave the opportunity to design extended zinc finger peptides that are capable of binding to composite targets with two regions of unbound DNA.
Pairs of zinc fingers were fused with linkers containing amino acid insertions of -gly- or -gly, gly, ser- after the glycine residue of their natural linker sequence. The six-finger peptides were based on a C-terminal fusion of the three-finger wild-type Zif268 peptide to the N terminus of the three-finger Zif268 mutant (MUT). MUT is a phage-selected variant of Zif268 that binds the DNA sequence 5′-GCG GAC GCG-3′ (19). The third finger of wild-type Zif268 was fused to the N-terminal finger of the MUT clone by using the canonical linker peptide -TGERP-. A number of 3 × 2F peptides were created by using a combination of the above insertions. The names of all of the peptides made and their descriptions can be seen in Table 1. To judge the success of the 3 × 2F design, a fusion of two three-finger units constructed from the same individual fingers was made for comparison. This 2 × 3F construct was created by fusing the C-terminal finger of wild-type Zif268 to the N-terminal finger of the three-finger MUT-clone, using the previously reported, extended linker peptide -LRQKDGERP- (11). The general structure of the 3 × 2F and 2 × 3F peptides and the type of binding site structure they are designed to target is demonstrated in Fig. 2. All six-finger peptides are expected to bind the same 9-bp half-sites and 18-bp contiguous DNA site 5′-GCGGACGCGGCGTGGGCG-3′. To test the specificity of the various six-finger peptides, they were targeted against binding sites containing 1- to 3-bp deletions (at the finger 4 recognition site) within the 18-bp target sequence, and also to sites that contained mutations to the sequence recognized by finger 4 of the peptides (Table 2). The 3 × 2F peptides also were targeted against sites comprising three 6-bp target subsites, in tandem order, which were either contiguous or separated by 1 or 2 bp of DNA (Table 3).
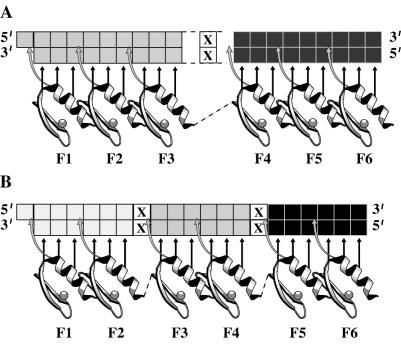
Figure 2.
The general structure of the six-finger arrays used in this study and potential regions of nonbound DNA marked with an X. Shown are 2 × 3F peptides with 9-bp subsites indicated (A) and 3 × 2F peptides with 6-bp subsites indicated (B).
Binding Specificity and Affinity of the 3 × 2F Peptides.
Peptide samples from the same transcription and translation system used to calculate active peptide concentration were used to determine the binding affinity of all of the zinc finger peptides for the various target sequences used. A preliminary experiment was conducted with the three-finger Zif268 peptide against its 9-bp binding site as a form of “protocol calibration.” This gave a value for the dissociation constant (Kd) of Zif268 of 0.45 nM, which is within the range expected for this peptide. To determine the binding specificity of different styles of six-finger peptides, the 3 × 2F ZMS and 2 × 3F ZM peptides were first used in gel-shift experiments with the 9-bp Zif268 half-site and the full 18-bp 123456 binding site. These results show that the 3 × 2F ZMS and 2 × 3F ZM peptides bind their full-length target site with similar affinities of 0.6 and 1.4 pM, respectively (Table 2). However, their affinities for the Zif268 half-site were dramatically different. The 2 × 3F ZM peptide bound with an affinity of approximately 2.2 nM (which is within the range expected), but the 3 × 2F ZMS peptide bound with an affinity of about 110 nM. This affinity was so weak that it was difficult to quantify with this system. From these data, it can be seen that the 3 × 2F peptide discriminates between the two sites over 100-fold more strongly than the 2 × 3F peptide.
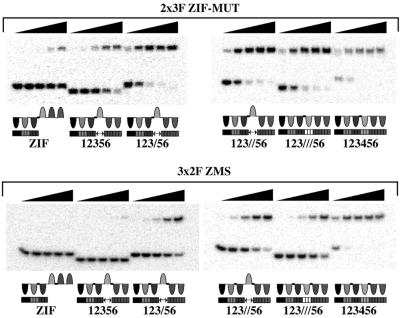
To further study the specificity of the two constructs, the 3 × 2F and 2 × 3F peptides were targeted against binding sites that had been mutated in the region normally bound by finger 4. These results show that the 3 × 2F ZMS peptide binds to the site with a 3-bp region mutated, 123///56, with an affinity of 890 pM. Meanwhile, it binds to a site with this 3-bp region deleted, 12356, with an affinity of 22 nM (see Table 3). Its affinities for sites with 1- or 2-bp deletions were 270 and 630 pM, respectively. Hence, the affinities of 3 × 2F ZMS for these mutant sequences are between 450- and 37,000-fold weaker than for the correct binding sequence. In contrast, the 2 × 3F ZM peptide binds 123///56, 123//56, and 123/56 with affinities of 15, 14, and 14 pM respectively. This is just 10-fold weaker than that for its correct binding site. The 2 × 3F ZM peptide shows a further reduction in affinity for the 12356 binding site, but this sequence is still bound more than 60 times stronger than it is bound by 3 × 2F ZMS. The gel-shift data in Fig. 3 demonstrate the relative binding affinities of the 2 × 3F ZM and 3 × 2F ZMS peptides for these binding sites. All these data serve to emphasize the enhanced specificity of the 3 × 2F construct for sequences that resemble its correct target site. The gel-shift data of Fig. 2 demonstrate the relative affinities of the 3 × 2F ZMS and 2 × 3F ZM peptides for the target sites above.
Figure 3.
A selection of DNA binding studies by gel-shift assay. The gels are designed to give a comparison between the binding affinities of the 2 × 3F ZM and 3 × 2F ZMS peptides and are not necessarily the gels used to quantify binding affinity. For example, the amount of 123456 binding site shifted by each peptide is limited by protein concentration rather than by Kd. (Top) Shown are 5-fold dilutions of 2 × 3F ZM (from 800 pM to 1.3 pM) against 2 pM binding sites. (Bottom) Shown are 5-fold dilutions of 3 × 2F ZMS (from 700 pM to 1.1 pM) against 2 pM binding sites. The proposed binding modes of the zinc finger peptides for each binding site is illustrated under each gel image. The lengths of oligonucleotides containing each binding site were as follows: 12356 and 123///56, 28 bp; ZIF, 123/56 and 123//56, 32 bp; and 123456, 34 bp.
Binding Noncontiguous Sequences.
A second set of binding studies was conducted to demonstrate the ability of the 3 × 2F peptides to accommodate one or more regions of unbound DNA within their recognition sequence. First the 3 × 2F ZMS and ZML peptides were titrated against the 12/34/56 (three 6-bp subsites separated by 1 bp, which is represented by a single “/” in the binding site name) and 12//34//56 (three 6-bp subsites separated by 2 bp) binding sites. The results showed that the 3 × 2F ZMS peptide, which is designed to target only the contiguous 123456 site, was unable to accommodate either 1- or 2-bp gaps between the two-finger subsites. The 3 × 2F ZML peptide, however, bound the 12/34/56 site with an affinity of ≈5 pM, but also was unable to bind tightly to the site with 2-bp gaps. Next, the 3 × 2F ZMSL and 3 × 2F ZMLS peptides were targeted against the three noncontiguous sequences: 1234/56, 12/3456, and 12//34//56. These sites were bound by the 3 × 2F ZMSL peptide with affinities of approximately 3 pM, 73 pM, and 12 nM, which is in accordance with the binding of 6, 4, and 2 fingers, respectively. Also, 3 × 2F ZMLS showed a similar trend in binding affinities. These experiments demonstrate that 3 × 2F peptides can bind contiguous 18-bp sites, but also are unique amongst the six-finger peptides reported to date in being able to bind sequences with two regions of unbound DNA with high affinity.
Discussion
A successful application of designer transcription factors for the control of endogenous genes requires the transcription factor used to be specific for a unique sequence within the target genome. In a complex genome such as that of a human, a recognition sequence of at least 16 bp is required to specify a potentially unique sequence. Hence, a zinc-finger strategy is likely to employ a tandem array of six zinc fingers (with an 18-bp recognition sequence) to target a single gene in vivo. However, our previous work suggests that long arrays of zinc fingers may bind truncated or mutated binding site sequences without much reduction in binding affinity (18). This effect would, of course, cause a significant problem with the specificity of polydactyl peptides.
To address the issue of polydactyl zinc finger specificity, we decided to redesign six-finger arrays to achieve greater specificity. By linking pairs of zinc fingers together, rather than triplets, we thought that the array would be more sensitive to mutations in their binding site. Our results clearly demonstrate this fact. It appears that the more rigid nature of the 2 × 3F ZM peptide means that a mutation in the binding site of one finger is “felt” only by that finger, so that the 123///56 site is bound with the extremely high affinity of 15 pM. In contrast, the results suggest that the more sensitive design of the 3 × 2F peptides mean that a mutation in the binding sequence of a single finger weakens the entire two-finger unit. Thus, the 3 × 2F ZMS peptide binds the same site with an affinity of 890 pM. The large reduction in affinity of the 3 × 2F ZMS peptide for the Zif268 half-site must be attributed to the extended linker sequence between fingers 2 and 3. Presumably this linker reduces the cooperative binding effect of the adjacent fingers such that finger 3 of the peptide adds nothing to the binding of the half-site. Meanwhile, the unbound fingers probably “drag” on the complex to help pull the peptide off the DNA. The higher affinity of the 3 × 2F peptides for other sites that are bound by only two fingers (such as the 3 × 2F ZMS peptide against the 12/34/56 site) presumably arises because there are three separate two-finger binding sites present in the sequence.
A further advantage of the 3 × 2F design is the ability of these peptides to specifically target DNA sites with two gaps of unbound DNA. This property may be particularly useful in giving more flexibility in selecting target sequences for designer transcription factors.
The fact that 3 × 2F peptides contain two extended linkers rather than one also may help polydactyl peptides to achieve a more optimal DNA binding mode and perhaps even greater binding affinities. Various studies have now shown that the helical periodicity of the arrayed α-helices of a zinc finger peptide does not exactly match that of the DNA double helix (the exact periodicity of which is sequence-dependent). Hence, DNA is slightly unwound when a zinc finger peptide is bound (14, 15). It also has been shown that the canonical -TGEKP- linker sequence is fully extended when TFIIIA and Zif268 bind DNA, but even then, this sequence is fractionally too short to allow the fingers to bind in their optimal conformation (16, 17). The combination of these two factors means that the longer the array of zinc fingers, the greater the strain within the protein–DNA complex and, hence, the greater the loss in binding energy. By employing two extended linkers in a six-finger array, the peptide is able to realign with the DNA more regularly, thereby reducing the build-up of strain. Another contributing factor to the slightly higher affinity of the 3 × 2F peptides over the 2 × 3F peptide may be attributable to the sequence of the modified linker peptide. Previous studies have demonstrated that mutations in the canonical linker sequence of TFIIIA can reduce binding affinity by up to 20-fold (19). More recently, it has been shown that the canonical linker peptide forms important α-helix C-capping interactions when a zinc finger binds DNA (20). These interactions are thought to be significant in terms of peptide stability and DNA binding affinity. Unlike our 3 × 2F constructs, the -LRQKDGERP- linker sequence used in this study, following that by Kim and Pabo (11) does not maintain the helix-capping motifs or sequence conservation, which may be important to achieve optimal binding affinity (21).
Polyzinc finger peptides, with their ability to bind with high affinity to long (18-bp) DNA target sequences, are likely to be used more and more in the search for gene therapy treatments and applications such as transgenic plants and/or animals. However, for such applications to be effective and safe it is crucial that high-affinity zinc finger peptides are also highly specific. This is of particular importance given the extremely slow off-rates observed for extended zinc finger arrays (11). In this work we have better satisfied both these requirements by creating a design of six-finger peptides that not only gives a slightly higher affinity than a comparable 2 × 3F peptide but, more importantly, with far greater specificity for its full-length target. The two-finger units used also allow greater flexibility in the selection of target sites by allowing one or two gaps of nonbound DNA and reduce the library size required to select specific binding domains by techniques such as phage display. We believe that 3 × 2F peptides will greatly enhance the application of zinc finger arrays for the in vivo control of gene expression.
Acknowledgments
We thank Mark Isalan for critical comments on this manuscript and Armin Sepp for advice on data processing. This work was funded by the Medical Research Council.
Abbreviation
- MUT
three-finger mutant peptide of the three zinc fingers of murine transcription factor Zif268
- 3 × 2F peptides
3 × 2 finger peptides
- 2 × 3F peptides
2 × 3 finger peptides
References
- 1.Desjarlais J R, Berg J M. Proteins Struct Funct Genet. 1992;12:101–104. doi: 10.1002/prot.340120202. [DOI] [PubMed] [Google Scholar]
- 2.Rebar E J, Pabo C O. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 3.Jamieson A C, Kim S H, Wells J A. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 4.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Yang W-P, Barbas C F., III Proc Natl Acad Sci USA. 1995;92:344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerli R R, Dreier B, Barbas C F., III Proc Natl Acad Sci USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. . (First Published January 31, 2000; 10.1073/pnas.040552697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahon E, Reveh D. Nucleic Acids Res. 1998;26:1233–1239. doi: 10.1093/nar/26.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrasegaran S, Smith J. Biol Chem. 1999;380:841–848. doi: 10.1515/BC.1999.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu G-L, Bestor T H. Nat Genet. 1997;17:376–378. doi: 10.1038/ng1297-376. [DOI] [PubMed] [Google Scholar]
- 10.Beretta G L, Binaschi M, Zagni E, Capuani L, Capranico G. Cancer Res. 1999;59:3689–3697. [PubMed] [Google Scholar]
- 11.Kim J-S, Pabo C O. Proc Natl Acad Sci USA. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q, Segal D J, Ghiara J B, Barbas C F., III Proc Natl Acad Sci USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiuchi T, Abe E, Imanishi M, Kaji T, Nagaoka M, Sugiura Y. Biochemistry. 1998;37:13827–13834. doi: 10.1021/bi9811112. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y, Berg J M. Biochemistry. 1996;35:3845–3848. doi: 10.1021/bi952384p. [DOI] [PubMed] [Google Scholar]
- 15.Nekludova L, Pabo C O. Proc Natl Acad, Sci USA. 1994;91:6948–6952. doi: 10.1073/pnas.91.15.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wuttke D S, Foster M P, Case D A, Gottesfeld J M, Wright P E. J Mol Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 17.Elrod-Erickson M, Rould M A, Nekludova L, Pabo C O. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 18.Moore M, Choo Y, Klug A. Proc Natl Acad, Sci USA. 2001;98:1432–1436. doi: 10.1073/pnas.98.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choo Y, Klug A. Proc Natl Acad, Sci USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choo Y, Klug A. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laity J H, Gippert G P, Soman K V, Case D A, Wright P E. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]