Abstract
Background
Mass spectrometry and proteomic analyses have become powerful tools for the analysis of proteins and peptides. Investigation of proteins contained in the various layers of the avian eggshell has focused entirely on domesticated species. It has been widely assumed that this existing research can inform the study of wild bird species despite the fact that the vast majority of the diversity in avian species (~95%) exists outside the Orders to which domestic and poultry species belong. Museum collections offer a potentially valuable source of material for studying composition of wild avian eggshell matrix proteins. We used museum and fresh eggshells of common quails Coturnix coturnix to compare the protein composition of their organic matrices. Four eggs of domestic chickens were analysed simultaneously as a control for comparison to the fresh and museum quail eggs. The determination of the proteins was carried out using enzymatic cleavage followed by high-performance mass spectrometry.
Results
We found that some of the expected key eggshell proteins (3 out of 11) were not present in the samples of museum quail egg. These proteins were either entirely absent from the museum eggs or the technique was unable to detect them. There was no pattern in the absent proteins in the sense of protein function or where they are located within the eggshell.
Conclusion
We conclude it is likely that such studies on museum specimens using a proteomic approach will be limited in coverage of proteins and may, therefore, be misleading.
Background
Mass spectrometry and proteomic analyses have become powerful tools for the analysis of proteins and peptides [1]. The recent elucidation of the chicken genome has provided the opportunity to apply this technique to identify and then categorise the matrix proteome of the chicken eggshell [2,3]. The avian eggshell is a highly regulated crystalline biocomposite ceramic composed of multiple layers, predominantly the trigonal phase of calcium carbonate (CaCO3), known as calcite [3]. Eggshells provide a number of important functions such as protection against physical damage and microbial contamination, regulation of gas and water exchange during embryogenesis, and provision of calcium for embryonic development [3,4]. The general structure of the eggshell consists of the innermost membrane, a continuous calcified layer, and the outermost cuticle [2,4], and thus far, over 520 proteins have been identified from these three layers [2] by use of a proteomics approach.
With few exceptions, most studies on eggshell proteins have been associated with the commercial poultry industry or other domesticated species [5,6]. For example, Panheleux et al. [6] compared the biochemical characteristics of eggshells from seven domesticated species of birds. Their study showed the presence of common matrix components in the various domestic avian species, supporting the notion of universality of their distribution across species. However, their study also demonstrated some particularities, with not all of the domesticated species having the key eggshell proteins that were the focus of the study [6]. It had been widely assumed that this existing research can inform the study of wild bird species despite the fact that the vast majority of the diversity in avian species (~95%) exists outside the Orders to which domestic and poultry species belong. While the general structure of the eggshell is similar across all bird species, the thickness, form and size of the whole eggshell and its mineralised microstructure all vary between species [7]. Moreover, the organic and organomineral content of the eggshell matrix that is believed to regulate eggshell mineralization differs between the domestic species that have been studied so far. For example, non-proteomic approaches have shown that the eggs of ostriches (Struthio camelus), emus (Dromaius novaehollandiae), rheas (Rhea americana) and domestic ducks (Anas p. domestica) contain species-specific proteins that are not found in poultry but are thought to be homologous in function, at least, to a chicken protein [2,8,9]. Therefore, the little evidence available suggests that eggshell proteins may be present in different quantities and different forms in different species.
To answer key ecological questions as to why the structure and composition of the eggshell differ among species, it is thus vital to be able to characterise the eggshell matrix components from a number of bird species from a wide range of Orders. As a result of persecution, collecting fresh wild bird eggs was made illegal in the United Kingdom in 1954 (Wild Birds Protection Act). Therefore, museum egg specimens are an important repository of historical samples [10]. As such, museum egg specimens could provide an unprecedented opportunity to sample proteins from a wide range of wild bird species from a multitude of Orders.
The sampling of museum eggs has been made possible through a collaboration between the Centre for Ornithology, University of Birmingham (UK) and the Natural History Museum (NHM, Tring, UK), which made a limited number of data-poor eggs available for destructive analysis. The NHM collection is believed to be the most comprehensive in the world with an estimated one million eggs [11]. However, it is not known if the eggshell proteins will degrade over time, and thus not be present in museum eggs of unknown age. That occurrence would make it difficult to draw conclusions as to whether a protein was simply never present in the eggshell, or absent through temporal degradation. Simple protein and DNA examination has been conducted on fossilised dinosaur eggshells [12,13], but no such work has yet been conducted on museum bird eggs which have potentially been treated with detergents and chemicals. Therefore, the aim of this study was to use both fresh and museum eggs of common quails Coturnix coturnix to investigate how the protein composition of the two eggshell matrices compared, and to ascertain whether museum eggs are suitable candidates for study using modern proteomic techniques.
Results
Four museum and four fresh quail eggs were analysed. Each egg came from a different clutch. For the four museum eggs, location and date of collection are; 1925 (Cornwall), between 1961-1963 (Aberdeen), 1901-1910 (England, exact location unknown) and 1911-1930 (North Yorkshire). To ensure the protein extraction procedure and protocol were effective at detecting the eggshell proteins consistently, four eggs of domestic chickens were analysed simultaneously as a control for comparison to the fresh and museum quail eggs. This ensured that any proteins not detected in the museum eggs was not a result of a methodological artefact. The fresh eggs of the chickens and quails were commercially obtained from local retailers. Four replicates were studied from each egg. As the four museum eggs were collected in the United Kingdom, it is assumed they all belong to the Coturnix coturnix nominate race. The four fresh quail eggs were from a quail breeder in north-Worcestershire (UK), and were also of the Coturnix coturnix nominate race.
Two protocols were used for protein extraction. The preparation of the various insoluble layers followed the previously published method, described in detail in Mann et al. [2]. Briefly the eggshells were treated with 5% EDTA and then washed extensively with distilled water to facilitate mechanical removal of the membrane. In protocol one, the crushed calcified eggshell layer was demineralised with 10% acetic acid. Acid-insoluble material was removed by centrifugation and the supernatant, containing the soluble matrix, was dialysed against 5% acetic acid and lyophilised. A second extraction protocol was based loosely on that of Mann et al [14]. The shell fragments were dissolved in 8 M urea, the solution was diluted to 2 M urea with 0.2 M ammonium bicarbonate, and the samples lyophilised. As we were not attempting to quantify proteins, sub samples of the shells were used.
After lyophilisation and reconstitution in a known volume, an OD (optical density) 280nm was performed to ascertain protein concentration. Fixed volumes were used for digestion as opposed to protein concentration. The samples were dried down to 0.5 ml, and the pH adjusted with 5M NaOH to pH 7.0. Added to this solution was 50 μl 10 mM dithiothreitol (DTT), prior to the samples being incubated at 60°C for 30 mins. Samples were then cooled to room temperature for a further 5-10 mins and cysteines alkylated by addition of 50μl 50mM iodoacetamide, mixed and incubated at room temperature in the dark for 45 mins. For the first protocol, 50 μl of trypsin gold (Promega, Southampton, Hampshire, UK, 6 ng/μl) was subsequently added to the samples, which were then incubated at 37°C overnight (c. 14 hours). For protocol two, 4 μl of lysyl endopeptidase was added to the solution, and left for 14 h at 23°C. The reaction mixture was then diluted to 2 M urea and trypsin added as above. The peptides were extracted and washed using millipore C18 ZipTips. Briefly, tips were prepared by pre-wetting in 100% acetonitrile and rinsed in 2 × 10 μl 0.1% formic acid. Samples were repeat pipetted throughout the volume of the samples five times. The tip was then washed with 3 × 10μl 0.1% formic acid to remove excess salts and undigested protein before elution of peptides with 10-20μl of 50% acetonitrile/water/0.1% formic acid. Samples were dried down to remove the acetonitrile, and then re-suspended in 0.1% formic acid solution in distilled water. All chemicals were purchased from Sigma (Gillingham, Dorset, UK) or Fisher Scientific (Loughborough, Leicestershire, UK). A 7 Tesla LTQ FT ultra mass spectrometer (ThermoFisher Scientific, Germany) were used to perform data-dependent scanning [15]. Data acquisition was controlled by Xcalibur 2.0 software. The mass spectrometer alternated between a full FT-MS scan (m/z 380 - 2000) and subsequent collision-induced dissociation (CID) MS/MS scans of the three most abundant ions. Survey scans were acquired in the ICR cell with a resolution of 100 000 at m/z 400. Precursor ions were isolated and subjected to CID in the linear ion trap. Collision activation for the experiment was performed in the linear trap using helium gas at collision energy normalized to precursor m/z of 35% and qexcite = 0.25. The width of the precursor isolation window was 2 m/z and only multiply-charged precursor ions were selected for MS/MS.
Based on Mann et al. [2,16], 11 key eggshell proteins were selected to test for their presence in both fresh and museum quail egg specimens (Fig. 1, Additional File 1). Database searches (Swissprot and NCBInr) were conducted using the MASCOT (Matrix Science, London, UK). Variable modifications were N-acetyl and oxidation (M). The peptide tolerance was 5 ppm and the MS/MS tolerance was 0.5Da. One missed cleavage was allowed. BLAST analysis of identified proteins was performed with the programme provided by the NBCI against the non-redundant database for all organisms. Protein identifications provided by MASCOT software were accepted if the data set contained at least two peptides with a MASCOT score > 25 at P > 0.01. Subsequent reverse searches of all data showed the number of false positives to be less than 1%.
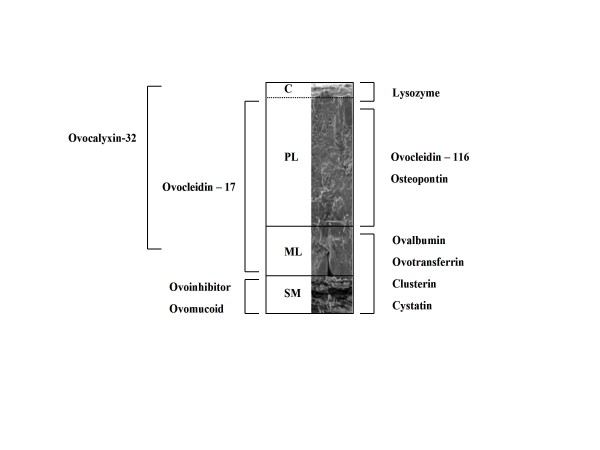
Figure 1.
Distribution of protein types in the avian eggshell. Differential localised pattern for eggshell matrix proteins within the avian eggshell. Data from different studies (see Hincke et al [3]) are summarised to demonstrate the localisation of the eggshell proteins through the 3 main layers; PL (pallisade layer), ML (mamillary layer) and SM (shell membrane), plus the C (cuticle). The scanning electron microscope image is taken from Hincke et al [3].
Peptide sequences and identified proteins were very similar within each species, and as such, the data presented in Table 1 is for one egg of each of the species and samples (chicken, fresh quail, museum quail). The criteria for which egg is presented, was that it contained all peptide sequences that were present in the other three eggs for each species, plus any additional peptides that were identified in that egg alone. For each of the four runs, all 11 of the key eggshell proteins were identified in the fresh eggs of chickens and quails (Table 1, Additional File 2). The four museum eggs were consistent in their results, with ovocalyxin-32, ovoinhibitor and ovotransferrin consistently being totally absent (Table 1). Cystatin and ovocleidin-17 were only identified in the museum quail eggs using the urea extraction protocol, all other proteins were identified using both urea and acetic acid techniques. In the chicken, ovalbumin, lysozyme and ovocleidin-116 had the highest Mascot scores. For both the fresh and museum quails, the three proteins with the highest Mascot scores were ovomucoid, ovalbumin and ovocleidin-116 (Table 1).
Table 1.
Proteins detected in fresh chicken and quail eggs in comparison to museum stored quail eggs of unknown age.
| Chicken | Fresh Quail | Museum Quail | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | Present | No. Pept. | Mascot | % Cov. | Present | No. Pept. | Mascot | % Cov. | Present | No. Pept. | Mascot | % Cov. |
| Clusterin | ✓ | 7 | 201 | 23 | ✓ | 5 | 145 | 12 | ✓ | 1 | 39 | 1 |
| Cystatin | ✓ | 5 | 175 | 39 | ✓ | 5 | 134 | 31 | ✓* | 1 | 40 | 9 |
| Lysozyme | ✓ | 12 | 227 | 80 | ✓ | 11 | 315 | 75 | ✓ | 5 | 5 | 34 |
| Osteopontin | ✓ | 1 | 59 | 5 | ✓ | 2 | 48 | 5 | ✓ | 2 | 61 | 5 |
| Ovalbumin | ✓ | 17 | 1997 | 78 | ✓ | 17 | 1053 | 42 | ✓ | 9 | 378 | 20 |
| Ovocalyxin-32 | ✓ | 3 | 61 | 44 | ✓ | 2 | 129 | 4 | ✗ | - | - | - |
| Ovocleidin-17 | ✓ | 7 | 434 | 54 | ✓ | 2 | 82 | 33 | ✓* | 2 | 85 | 11 |
| Ovocleidin-116 | ✓ | 24 | 2017 | 44 | ✓ | 18 | 1214 | 42 | ✓ | 5 | 308 | 12 |
| Ovoinhibitor | ✓ | 3 | 139 | 9 | ✓ | 3 | 38 | 10 | ✗ | - | - | - |
| Ovomucoid | ✓ | 3 | 151 | 25 | ✓ | 14 | 497 | 88 | ✓ | 7 | 247 | 38 |
| Ovotransferrin | ✓ | 24 | 1036 | 43 | ✓ | 2 | 64 | 3 | ✗ | - | - | - |
A tick mark (✓) denotes presence and a cross (✗) absence. Number of unique peptides is abbreviated to "No. Pept.". Mascot refers to the Mascot score. Further information on peptide sequences is located in Additional File 2. The asterix (*) refers to proteins that were detected only through the urea extraction technique.
Discussion
The concurrent runs of the fresh eggs of chickens and quails demonstrate the technique used was sufficient to identify the 11 key eggshell proteins in fresh eggs, suggesting those not detected in the museum specimens were either no longer present in the shell or were undetectable using this proteomic approach. Osteopontin was found in the least frequency from a coverage perspective (Additional File 2), but again, was present in all three species eggs suggesting number of peptides and % coverage is not an issue relating to egg storage or time. Ovocleidin-17 was detected in quail eggs for the first time and was identified in both the fresh and museum specimens in the present study (see Additional File 3 for spectra). Panheleux et al [[6], see also [17]] had detected a band of 17 kDa through Western blotting when stained with Coomassie blue suggesting the possible presence of ovocleidin-17 in quails, which the current study has subsequently confirmed. Prior to storage, museum eggs can be chemically treated. The collection we were working with was provided by private collectors, who may use a combination of different mechanical and chemical techniques to remove egg contents. This treatment, plus the potential accumulation of exogenous particulates in storage may affect degradation of proteins [18]. However, if this was to occur, it would be expected to happen in either the cuticle and palisade layer (PL) or the membrane (SM) of the eggshell. A 'missing' protein in the museum eggs such as ovotransferrin, for example, is predominantly found in the mammillary layer (ML), however (Fig. 1). Also, egg collectors typically prefer eggs that have been freshly laid, and will therefore not require extensive chemical treatment, as the contents of the egg will be liquid and easily removed through blowing [19].
A review of techniques used to remove embryos, blow eggs and treat museum eggshells found that pepsin, trypsin and wine vinegar were the most damaging to the eggshell [19]. We are unable to know exactly what treatment the museum quail eggs in the study received, particularly prior to admittance to the museum collection. From a proteomics perspective, trypsin and wine vinegar would be the most problematic. Wine vinegar would cause acid hydrolysis of the proteins. If the eggshells had been treated with trypsin or pepsin, it is not implausible that proteins may have been partially digested in the preparation process. This partial digestion may also cause protein folding, which could prevent the trypsin from acting effectively in the present study, and result in the proteins not being detected. However, unless different proteins respond differently to partial trypsin digestion, there seems no basis why some proteins would have degraded and some not. Again, this effect would be predicted to be most prevalent on the outside layers such as the PL and SM, yet most of the proteins absent from the museum eggs reside in the ML. Bada et al. [20] noted that in prehistoric dinosaur bone, certain proteins didn't degrade as fast as expected because of their close association with bone material. Other factors such as environmental temperature, humidity and the type of matrix in which the molecules are contained are also known to affect the extent of protein degradation [21]. In general, however, the absence of no clear pattern of missing proteins with respect to the layer they are located within the shell (Fig. 1) suggests factors such as storage temperature did not affect protein degradation of one specific layer of the eggshell differently. Therefore, it is likely that degradation of protein in the eggshell is a function of time, rather than storage conditions, at least for the eggs at the Natural History Museum. There are other potential problems for future proteomic work on both fresh and museum eggshells. One is the lack of sequences for more exotic species, which will inevitably lead to missed proteins or mis-identification. This in part will be why all proteomic studies on avian eggshells thus far have focused on domesticated species. The use of data-poor museum eggshells could also present concerns over the provenance of the eggs used, and potentially limit the conclusions that can be drawn from research on such samples.
Conclusion
In summary, we found that some of the expected proteins (3 out of 11) were entirely absent from samples of museum quail egg. Although the availability of museum eggs is a potential source for the comparative study of proteins using modern proteomic techniques, we caution that such studies will likely be limited in their coverage of proteins and may therefore be misleading.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SJP executed all experiments, performed the data analysis and wrote the manuscript. HJC and PC assisted with design and interpretation of the experiments and advised on the study. CGZ and LLW conducted sample preparation and analysis. All authors read and approved the final manuscript.
Supplementary Material
Eggshell Protein Types. Summary of the function of the key eggshell proteins.
Peptide Sequences Summary Table. Tables showing all peptides sequences identified for the 11 key eggshell proteins, from fresh chicken eggs, and museum and fresh quail eggs.
Spectra for Ovocleidin-17. Full spectra for ovocleidin-17 from both fresh and museum quail eggs.
Contributor Information
Steven J Portugal, Email: S.Portugal.1@bham.ac.uk.
Helen J Cooper, Email: H.J.Cooper@bham.ac.uk.
Cleidiane G Zampronio, Email: C.G.Zampronio@bham.ac.uk.
Laine L Wallace, Email: L.L.Wallace@bham.ac.uk.
Phillip Cassey, Email: P.Cassey@bham.ac.uk.
Acknowledgements
We are grateful to Golo Maurer, Douglas Russell and Judith White for assistance with the museum quail eggshells. Thank you to Kerry Hollands, Andrew Jones and Richard Tudor for advice on data analysis, and Cyrielle Arlot and Andrew Bowler for assistance with data collection. The LTQ FT Ultra used in this research was obtained through the Birmingham Science City Translational Medicine: Experimental Medicine Network of Excellent project, with support from Advantage West Midlands (AWM). We are also grateful to three anonymous reviewers for their input, advice and suggestions. This work was funded by a small project grant from NESTA, UK, while SJP was a Leverhulme postdoctoral research fellow.
References
- Aebersold R, Mass M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Mann K, Maćek B, Olsen J. Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics. 2006;6:3801–3810. doi: 10.1002/pmic.200600120. [DOI] [PubMed] [Google Scholar]
- Hincke MT, Wellman-Labadie O, McKee MD, Gautron J, Nys Y, Mann K. In: Biosynthesis and structural assembly of eggshell components, in Egg Bioscience and Biotechnology. Mine Y, editor. Wiley-Interscience, John Wiley and Sons publication, New Jersey, USA; [Google Scholar]
- Ahlborn GJ, Clare DA, Sheldon BW, Kelly RW. Identification of eggshell membrane proteins and purification of ovotransferrin and beta-NAGase from hen egg white. Prot J. 2006;34:71–81. doi: 10.1007/s10930-006-0010-8. [DOI] [PubMed] [Google Scholar]
- Hincke MT, Bernard AM, Lee ER, Tsang CPW, Narbaitz R. Soluble protein constituents of the domestic fowl's eggshell. Brt Poultry Sci. 1992;33:495–506. doi: 10.1080/00071669208417489. [DOI] [PubMed] [Google Scholar]
- Panheleux M, Bain M, Fernandez MS, Morales I, Gautron J, Arias SE, Solomon SE, Hincke M, Nys Y. Organic matrix composition and ultrastructure of eggshell: a comparative study. Brt Poult Sci. 1999;40:240–252. doi: 10.1080/00071669987665. [DOI] [PubMed] [Google Scholar]
- Ar A, Pagnanelli CV, Reeves RB, Greene DG, Rahn H. Avian egg: water-vapour conductance, shell thickness and functional pore area. Condor. 1974;76:153–158. doi: 10.2307/1366725. [DOI] [Google Scholar]
- Mann K, Siedler F. Ostrich (Struthio camelus) eggshell matrix contains two different C-type lectin-like proteins. Isolation, amino acid sequence, and posttranslational modification. Biochem Biophys Acta. 2004;1696:41–50. doi: 10.1016/j.bbapap.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan R, Joseph JS, Kini R, Valiyaveettil S. Eggshell Matrix Protein Mimics: Designer Peptides to Induce the Nucleation of Calcite Crystal Aggregates in Solution. Biomacromol. 2005;6:741–751. doi: 10.1021/bm049423+. [DOI] [PubMed] [Google Scholar]
- Green RE, Scharlemann PW. Egg and skin collections as a resource for long-term ecological studies. Bull B O C. 2003;123a:165–176. [Google Scholar]
- Knox AG, Walters MP. Under the skin: the bird collections of the Natural History Museum. Bull Brit Orn Club. 1992;112a:169–190. [Google Scholar]
- Asara JM, Schweitzer MH, Freimark LM, Phillips M, Cantley LC. Protein Sequences from Mastodon and Tyrannosaurus Rex Revealed by Mass Spectrometry. Science. 2007;316:280. doi: 10.1126/science.1137614. [DOI] [PubMed] [Google Scholar]
- Schweitzer MH, Chiappe L, Garrido AC, Lowenstein JM, Pincus S. Molecular preservation in Late Cretaceous sauropod dinosaur eggshells. Proc Roy Soc B. 2005;272:775–764. doi: 10.1098/rspb.2004.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Olsen JV, Maćek B, Gnad F, Mann M. Phosphoproteins of the chicken eggshell calcified layer. Proteomics. 2007;7:106–155. doi: 10.1002/pmic.200600635. [DOI] [PubMed] [Google Scholar]
- Peterman SM, Dufresne CP, Horning SJ. The Use of a Hybrid Linear Trap/FT-ICR Mass Spectrometer for On-Line High Resolution/High Mass Accuracy Bottom-Up Sequencing. Biomolec Tech. 2005;16:112–124. [PMC free article] [PubMed] [Google Scholar]
- Mann K. Identification of the major proteins of the organic matrix of emu (Dromaius novaehollandiae) and rhea (Rhea americana) eggshell calcified layer. Br Poultry Sci. 2004;45:483–490. doi: 10.1080/00071660400001157. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan R, Loh XJ, Gayathri S, Sindhu S, Banerjee Y, Kini RM, Valiyaveettil S. Formation of transient amorphous calcium carbonate precursor in quail eggshell mineralisation: An in vitro study. Biomacromol. 2006;7:3202–3209. doi: 10.1021/bm0605412. [DOI] [PubMed] [Google Scholar]
- Garibaldi JA. Factors in egg white which control growth of bacteria. Food Res. 1960;25:337–344. [Google Scholar]
- Scharleman JP. PhD thesis, University of Cambridge, Cambridge, UK; 2002. [Google Scholar]
- Bada JL, Wang XS, Hamilton H. Preservation of key biomolecules in the fossil record: current knowledge and future challenges. Phil Trans. 1999;354:77–87. doi: 10.1098/rstb.1999.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MJ, Gernaey AM, Nielsen-Marsh CM, Vermeer C, Westbroek P. Slow rates of degradation of osteocalcin: Green light for fossil bone protein? Geology. 2000;28:1139–1142. doi: 10.1130/0091-7613(2000)28<1139:SRODOO>2.0.CO;2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Eggshell Protein Types. Summary of the function of the key eggshell proteins.
Peptide Sequences Summary Table. Tables showing all peptides sequences identified for the 11 key eggshell proteins, from fresh chicken eggs, and museum and fresh quail eggs.
Spectra for Ovocleidin-17. Full spectra for ovocleidin-17 from both fresh and museum quail eggs.