Abstract
Background
Hepatitis B virus (HBV) infection increases the risk of liver disease and hepatocellular carcinoma. Small interfering RNA (siRNA) can be a potential new tool for HBV therapy. Given the high heterogeneity of HBV strains and the sensitivity towards sequences changes of siRNA, finding a potent siRNA inhibitor against the conservative site on the HBV genome is essential to ensure a therapeutic application.
Results
Forty short hairpin RNA (shRNA) expression plasmids were constructed to target conserved regions among nine HBV genotypes. HBV 1.3-fold genome plasmids carrying various genotypes were co-transfected with shRNA plasmids into either Huh7 cells or mice. The levels of various viral markers were examined to assess the anti-HBV efficacy of siRNA. Four (B245, B376, B1581 and B1789) were found with the ability to potently inhibit HBV RNA, DNA, surface antigen (HBsAg), e antigen (HBeAg) and core antigen (HBcAg) expression in HBV genotypes A, B, C, D and I (a newly identified genotype) in Huh7 cells and in mice. No unusual cytotoxicity or off-target effects were noted.
Conclusions
Such siRNA suggests an alternate way of inhibiting various HBV genotypes in vitro and in vivo, promising advances in the treatment of HBV.
Background
Worldwide, there are over 350 million people persistently infected with hepatitis B virus (HBV) [1]. Chronic HBV infections may have serious consequences, including acute hepatitis, as well as chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) [2]. Together, these are responsible for over 1 million deaths worldwide each year [3]. Current treatments for HBV infections are not only expensive and have significant side effects, but also only induce a partial response [4-6].
In eukaryotic cells, RNA interference (RNAi), a type of double-stranded (ds) RNA, initiates and directs sequence-specific, post-transcriptional silencing of homologous genes [7,8]. It has been demonstrated in previous studies that expression and replication of HBV can be suppressed by siRNA or shRNA with clinical implications [9-11]. However, the wide heterogeneity of HBV sequences may render RNAi inhibitors ineffective. To explore this further, 40 shRNA expression plasmids were constructed to target the sites that were conserved among HBV genotypes A through I. Their anti-HBV efficacy was then evaluated in vitro and in vivo.
Results
Screening for effective and broad anti-HBV shRNA
The shRNA plasmids co-transfected with two HBV 1.35 plasmids (N10 and Y1021) exhibited varying levels of extracellular HBsAg expression (Table 1). Of the forty shRNA plasmids, four plasmids (B245, B376, B1581 and B1789, Figure 1) were selected as candidates for further research based on their remarkable inhibitory ability and also relatively lower off-target probability (off-target score of above 30). The sequence conservation among the A to I genotypes for B245, B376, B1581 and B1789 were 95.1% (95%CI: 92.2~97.2), 88.7% (95%CI: 84.7~91.9), 97.3% (95%CI: 94.8~98.7), and 97.6% (95%CI: 95.2~98.9), respectively (Table 2). The data also shows that the target sequences of B245, B1581 and B1789 were more conserved than the target sequence of B376 (p < 0.05) in genotype B and C (Table 2).
Table 1.
The characterization and screening for multiplex anti-HBV siRNA
| ID | Sequence | Start Position | Off-target numbera | off-target scorea | Genome localization | Anti- Y1021 | Anti- N10b |
|---|---|---|---|---|---|---|---|
| B182 | GGACCCCTGCTCGTGTTACAG | 182 | 8 | 30 | S, P | ++ | + |
| B183 | GACCCCTGCTCGTGTTACAGG | 183 | 3 | 30 | S, P | - | - |
| B184 | ACCCCTGCTCGTGTTACAGGC | 184 | 3 | 30 | S, P | - | - |
| B243 | AGAGTCTAGACTCGTGGTGGA | 243 | 3 | 30 | S, P | + | + |
| B244 | GAGTCTAGACTCGTGGTGGAC | 244 | 9 | 30 | S, P | +++ | +++ |
| B245 | AGTCTAGACTCGTGGTGGACT | 245 | 4 | 30 | S, P | +++ | +++ |
| B246 | GTCTAGACTCGTGGTGGACTT | 246 | 4 | 30 | S, P | - | - |
| B250 | AGACTCGTGGTGGACTTCTCT | 250 | 10 | 35 | S, P | - | + |
| B251 | GACTCGTGGTGGACTTCTCTC | 251 | 7 | 35 | S, P | + | ++ |
| B252 | ACTCGTGGTGGACTTCTCTCA | 252 | 2 | 30 | S, P | ++ | ++ |
| B375 | GGATGTGTCTGCGGCGTTTTA | 375 | 1 | 25 | S, P | ++ | ++ |
| B376 | GATGTGTCTGCGGCGTTTTAT | 376 | 7 | 30 | S, P | +++ | +++ |
| B377 | ATGTGTCTGCGGCGTTTTATC | 377 | 5 | 35 | S, P | + | ++ |
| B379 | GTGTCTGCGGCGTTTTATCAT | 379 | 4 | 35 | S, P | + | + |
| B410 | ATCCTGCTGCTATGCCTCATC | 410 | 76 | 25 | S, P | - | - |
| B415 | GCTGCTATGCCTCATCTTCTT | 415 | 54 | 25 | S, P | + | ++ |
| B456 | AAGGTATGTTGCCCGTTTGTC | 456 | 2 | 30 | S, P | ++ | ++ |
| B457 | AGGTATGTTGCCCGTTTGTCC | 457 | 1 | 40 | S, P | - | + |
| B458 | GGTATGTTGCCCGTTTGTCCT | 458 | 7 | 35 | S, P | ++ | ++ |
| B459 | GTATGTTGCCCGTTTGTCCTC | 459 | 15 | 25 | S, P | ++ | ++ |
| B461 | ATGTTGCCCGTTTGTCCTCTA | 461 | 11 | 30 | S, P | + | + |
| B1260 | GCCGATCCATACTGCGGAACT | 1260 | 2 | 25 | EnhI, P | + | ++ |
| B1577 | GTGTGCACTTCGCTTCACCTC | 1577 | 13 | 30 | X, P, DR1 | +++ | ++ |
| B1579 | GTGCACTTCGCTTCACCTCTG | 1579 | 5 | 25 | X, P, DR1 | ++ | ++ |
| B1581 | GCACTTCGCTTCACCTCTGCA | 1581 | 15 | 30 | X, P, DR1 | +++ | +++ |
| B1583 | ACTTCGCTTCACCTCTGCACG | 1583 | 21 | 30 | X, P, DR1 | ++ | ++ |
| B1787 | GGAGGCTGTAGGCATAAATTG | 1787 | 4 | 30 | Pc, EnhII | ++ | ++ |
| B1788 | GAGGCTGTAGGCATAAATTGG | 1788 | 9 | 25 | Pc, EnhII | ++ | + |
| B1789 | AGGCTGTAGGCATAAATTGGT | 1789 | 5 | 30 | Pc, EnhII | +++ | +++ |
| B1880 | AAGCCTCCAAGCTGTGCCTTG | 1880 | 3 | 30 | Pc | + | - |
| B1881 | AGCCTCCAAGCTGTGCCTTGG | 1881 | 23 | 25 | Pc | - | - |
| B2389 | AGAAGAAGAACTCCCTCGCCT | 2389 | 42 | 25 | C, P | - | + |
| B2390 | GAAGAAGAACTCCCTCGCCTC | 2390 | 26 | 25 | C, P | - | + |
| B2391 | AAGAAGAACTCCCTCGCCTCG | 2391 | 29 | 25 | C, P | - | - |
| B2392 | AGAAGAACTCCCTCGCCTCGC | 2392 | 19 | 30 | C, P | - | + |
| B2393 | GAAGAAGAACTCCCTCGCCTC | 2393 | 18 | 30 | C, P | - | + |
| B2394 | AAGAACTCCCTCGCCTCGCAG | 2394 | 29 | 25 | C, P | - | + |
| B2395 | AGAACTCCCTCGCCTCGCAGA | 2395 | 14 | 35 | C, P | + | + |
| B2396 | GAACTCCCTCGCCTCGCAGAC | 2396 | 18 | 35 | C, P | - | + |
| B2397 | GATCCATACTGCGGAACTCCT | 2397 | 11 | 35 | C, P | - | - |
| L1254 | TGGCTACATTCTGGAGACATA | NA | NA | NA | luciferase | - | - |
NA, no application.
"+" indicates weak inhibition (below 50%),
"++" indicates medium inhibition (above 50%, but below 90%),
"+++" indicates strong inhibition (above 90%),
"-" indicates no significant inhibition,
An underline represents the four candidates that were worthy for further research.
a: off-target effects were evaluated by the online SOS program http://rnai.cs.unm.edu/offTarget.
b: anti-HBV effects were evaluated by decreases in extracellular HBsAg level.
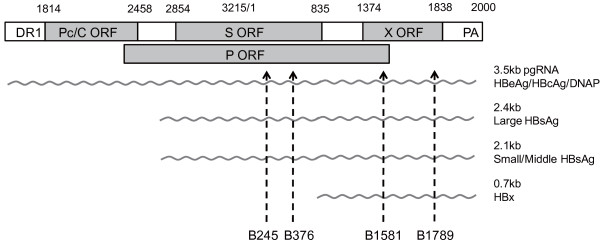
Figure 1.
A schematic diagram depicting the locations of siRNA targets in association with viral open reading frames and viral mRNAs within the HBV genome. The circular HBV genome is presented in a linear form. The coding regions for e/core, surface, polymerase, and X proteins are displayed and designated as Pc/C, S, P, and X, respectively. The relative locations of the target sites of B245, B376, B1581 and B1789 are also indicated by arrowheads.
Table 2.
Sequence conservation of four selected siRNA targets in 327 HBV strains
| Genotype (No. of HBV strains) |
Number of strains with identical sequence with siRNA(%) |
Subtype (No. of HBV strains) |
|||
|---|---|---|---|---|---|
| B245 | B376 | B1581 | B2379 | ||
| Genotype A (63) | 61(96.8) | 62(98.4) | 62(98.4) | 63(100) | Aa(45), Ac(9), Ae(9) |
| Genotype B(72) | 69(95.8) | 49(68.1)* | 71(98.6) | 70(97.2) | Bj(9), Ba(38), B3(7), B4(8), B5(4), B6(6) |
| Genotype C(58) | 53(91.4) | 46(79.3)* | 57(98.3) | 56(96.6) | C1(38), C2(13), C3(2), C4(2), C5(3) |
| Genotype D(30) | 29(96.7) | 29(96.7) | 28(93.3) | 28(93.3) | D1(11), D2(6), D3(8), D4(5) |
| Genotype E(34) | 33(97.1) | 34(100) | 33(97.1) | 33(97.1) | F1(4), F2(14) |
| Genotype F(18) | 15(83.3) | 18(100) | 18(100) | 18(100) | |
| Genotype G(17) | 17(100) | 17(100) | 15(88.2) | 16(94.1) | |
| Genotype H(13) | 13(100) | 13(100) | 12(92.3) | 13(100) | |
| Genotype I(22) | 21(95.5) | 22(100) | 22(100) | 22(100) | I1(10), I2(12) |
| Total (327) | 311(95.1) | 290(88.7)* | 318(97.3) | 319(97.6) | |
a: An asterisk represents a statistical difference of P < 0.05 in comparison with B376 and others.
Adverse side-effects evaluation for selected shRNA plasmids
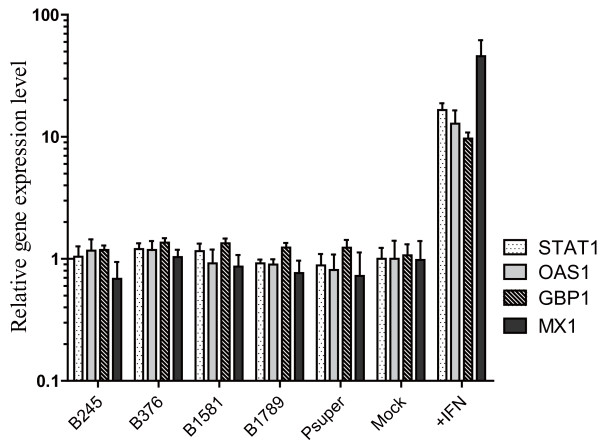
The B245, B376, B1581, and B1789 plasmids were transfected into Huh7 cells to determine cytotoxicity by the WST-8 assay. No significant siRNA-induced cytotoxicity was observed for these siRNA when compared to an empty pSUPER vector (p = 0.66, data not shown). The mRNA levels of four major interferon stimulated genes (STAT1, OAS1, GBP1 and MX1) in transfected cells were measured by quantitative realtime PCR with GAPDH mRNA acting as a control. As shown in Figure 2, between values 1 and 2, logarithmic increases for the IFN-stimulatable mRNAs were only observed in the IFN-treated cells, but not observed in any of the shRNA treated cells vs. untreated cells. From this, it can be concluded that an IFN response is not activated by these anti-HBV siRNAs.
Figure 2.
The expression profile of four major interferon stimulated genes (ISGs) in shRNA plasmids transfected cells. Cytoplasmic RNAs, from Huh7 cells treated with or without IFNα-2a or transfected with either pSUPER vector or shRNA plasmids, were analysed by realtime RT-PCR for IFN stimulated genes STAT1, OAS1, GBP1 and MX1. The values on the figure, plotted as "Relative gene expression level" on the y-axis, were calculated as the mRNA levels of ISGs divided by the GAPDH (control) mRNA level. Student t test was used to assess the difference between shRNA plasmids (including empty pSUPER vector) of transfected cells and non-transfected cells (mock). No significant difference was observed.
ShRNA inhibit gene expression of HBV strains with different genotypes in vitro
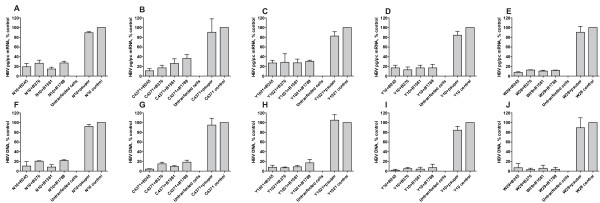
The levels of cytoplasmic HBV pg/pc RNA (3.5 kb) and HBV DNA in cultured supernatants were determined by realtime RT-PCR/PCR and presented in Figure 3. The pg/pc RNA level of five HBV strains with different genotypes were reduced by 58%~93% in B245(69%~93%), B376(59%~91%), B1581(67%~90%) and B1789(58%~88%) treatments, while the HBV DNA level observed in supernatants was decreased by 77%~99% in these shRNA plasmid treatments (B245: 83%~99%, B376: 79%~99%, B1581:88%~98%, B1789: 77%~99%).
Figure 3.
SiRNAs inhibit RNA and DNA expression of HBV strains with different genotypes in Huh7 cells. The histogram show the cytoplasmic HBV pg/pc RNA levels (A, B, C, D, E) and extracellular HBV DNA (F, G, H, I, J) of five HBV strains with genotypes Ae(N10), Ba(C4371), C1(Y1021), D1(Y10) and I1(W29) in treated shRNA plasmids, treated pSUPER vector, and non-treated Huh7 cells.
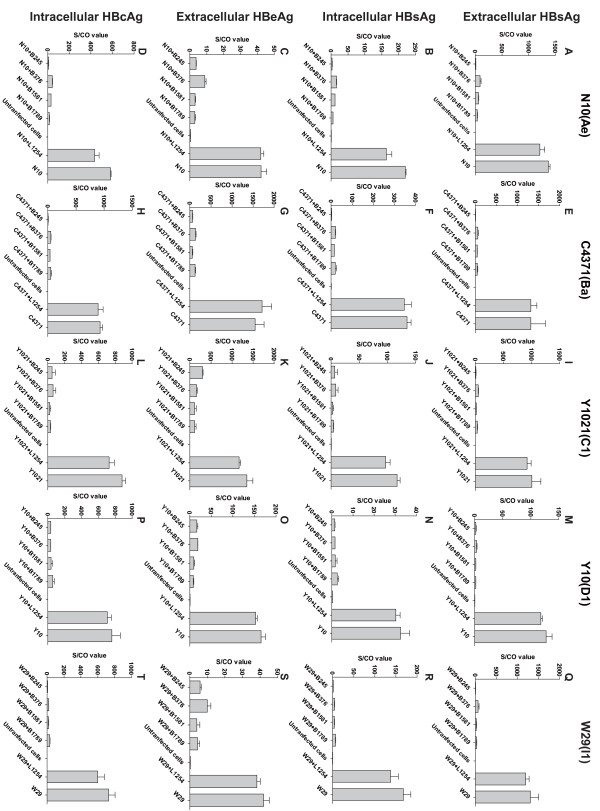
In addition, the extracellular and intracellular antigen levels in Huh7 cells that were co-transfected with HBV and shRNA plasmids were also determined (Figure 4). In the shRNA-treated Huh7 cells, the average extracellular HBsAg expression level of all five HBV strains decreased by 1.66 ± 0.36 logs. The average intracellular HBsAg expression level decreased by 1.47 ± 0.33 logs, while the extracellular HBeAg levels decreased by 1.04 ± 0.23 logs, and the intracellular HBcAg levels by 1.71 ± 0.49 logs. The effect of the siRNA treatment on HBeAg levels was weaker than that on the HBsAg or HBcAg levels (P < 0.001, Figure 5).
Figure 4.
SiRNAs inhibit viral antigens expression of HBV strains with different genotypes in Huh7 cells. (A, B, C, D) Extracellular HBsAg, intracellular HBsAg, extracellular HBeAg, and intracellular HBcAg expression levels of HBV N10(Ae), respectively. (E, F, G, H) Extracellular HBsAg, intracellular HBsAg, extracellular HBeAg, and intracellular HBcAg expression levels of HBV C4371(Ba), respectively. (I, J, K, L) Extracellular HBsAg, intracellular HBsAg, extracellular HBeAg, and intracellular HBcAg expression levels of HBV Y1021(C1), respectively. (M, N, O, P) Extracellular HBsAg, intracellular HBsAg, extracellular HBeAg, and intracellular HBcAg expression levels of HBV Y10(D1), respectively. (Q, R, S, T) Extracellular HBsAg, intracellular HBsAg, extracellular HBeAg and intracellular HBcAg expression levels of HBV W29(I1), respectively.
Figure 5.
Comparing the RNAi-induced silencing effect on different viral markers. Data were displayed the average antigen level of the 4 siRNAs reduced for five HBV strains. "Ex" = Extracellular and "In" = Intracellular. The Mann-Whitney test was used to assess the difference. An asterisk represents a statistical difference of P < 0.01 in comparison with the other markers (Ex HBeAg vs. Others P < 0.001, Ex HBsAg vs. In HBsAg P = 0.05, Ex HBsAg vs. In HBcAg P = 0.82, In HBsAg vs. In HBcAg P = 0.10.)
Inhibition of gene expression of HBV strains with different genotypes in vivo
Using the mouse model of acute hepatitis B virus infection [12], the profiles of serum HBsAg and HBeAg were used to evaluate the effect of shRNA over nine days (Figure 6). All HBV plasmids expressed detectable HBsAg and HBeAg in mice sera (Figure 6). As compared to the control mice (HBV+L1254), B245 and B376 treatments reduced HBsAg expression by over 99% in all five HBV genotypes. Furthermore, B1581 and B1789 treatments suppressed HBsAg by over 99% in mice infected with HBV genotypes A, B, C and D. In a novel W29 strain representing genotype I however, B1581 and B1789 treatments only reduced HBsAg expression by about 90%. With regards to serum HBeAg for genotypes A, B, C, D and I, B245, B376, B1581 and B1789 treatments suppressed HBeAg by 96%~99%, 79%~99%, 94%~99%, and 89%~99%, respectively. The overview of the results shows that B245 is the most potent agent.
Figure 6.
Kinetics of serum HBV antigen (HBsAg and HBeAg) of various HBV genotypes in RNAi-treated mice. For each group (each line in the figure), the experiment was repeated using two different groups of five mice. Due to limited serum resources, each sample was diluted 10-fold. (A) Genotype Ae (N10 group), (B) Genotype Ba (C4371 group), (C) Genotype C1 (Y1021 group), (D) Genotype D1 (Y10 group), (E) Genotype I1 (W29 group).
Discussion
Activated RNAi pathway can silence HBV replication and expression [13,14]. However, in most previous studies, the activity of RNAi against HBV is often evaluated with only one HBV strain [15-18]. Nine HBV genotypes (including a newly identified genotype "I"), designated as the letters A through I, have been recognized with an accompanying sequence divergence of >8% over the entire genome [19-21]. The influence of genotypes on HBV replication efficacy and antigen expression level had been proved to be various and that may further associate with clinical outcomes and antiviral treatments responses [22]. Hence, RNAi designed for one genotype may not necessarily be effective against another genotype. Given the high heterogeneity of HBV strains and the sensitivity of siRNA to the sequence changes, designing siRNA targets against the conservative site on HBV genome is essential to ensure activity across all genotypes [23].
In shRNA expression systems, two different promoters are predominantly used: U6 and H1, both driven by human polymerase III (poly III). Compared to Pol II promoters, Pol III promoters generally possess a greater capacity to synthesize RNA transcripts of a higher yield and rarely induce interferon responses [17,24]. However, a previous study noted that U6 Pol III-expressed shRNAs may cause serious toxicity in vivo by saturating the endogenous miR pathway [25]. In this report, we constructed 40 shRNA plasmids (Table 1) with various targets, using a human H1 Pol III promoter. The target sequences of the final four selected shRNA plasmids demonstrated high sequence conservation among A to I genotypes and significant inhibition activity, in both Huh7 cells and mice, against the expressions of HBV RNA, DNA and antigens in genotypes A, B, C, D and I. The inhibitory efficacy of these shRNAs (B245, B376, B1581 and B1789) however, varies significantly against the various genotypes for different viral markers in different models (Figure 3, 4, 5 and 6). Such differences in efficiency may be due to differences in the mRNA's secondary structure or the target site accessibility [26]. B245 was the most effective of the four candidates.
It should be noted that both the cell-transfection model and hydrodynamic injection model more closely resemble an acute model of a HBV infection. This is a potential limitation in this study, as most individuals who need anti-HBV therapy are chronically infected. Compared to the HBV transgenic mouse models and stably transfected cell lines, the former are more flexible and convenient in evaluating the efficacy of shRNAs as a way to inhibit various HBV strains. Nevertheless, the effective shRNA candidates should be studied further in different models.
Because HBV contains overlapping open reading frames (ORFs) and all four HBV transcripts overlap in their 3' terminals, a single siRNA targeting multiple areas could be designed to maximize inhibitory potency [23]. The siRNAs targeting C ORF, such as B2389~B2397, presented in Table 1, show activity only against the 3.5 kb pregenomic RNA, but are unlikely to show any activity against the other three transcripts (Figure 1). Meanwhile, all four siRNAs demonstrated more silencing activity with regards to HBsAg expression than HBeAg expression for various genotypes in the cell cultures and mice. The targets on both however were the same in the HBV transcripts for the two proteins (Figure 4 and Figure 5), which was also observed in a previous study [23]. HBcAg, a viral capsid correlated with viral replication [27,28], was silenced as effectively as HBsAg, but HBeAg was not (Figure 4).
The registered agents currently available for the treatment of HBV infections, such as interferon and nucleoside analogues, can dramatically decrease HBV DNA levels and induce particular HBeAg loss, but will rarely cause HBsAg loss in chronic hepatitis B patients [29-32]. RNA interference, on the other hand, can theoretically be directed to cleave any target RNA, providing a novel methodology for anti-HBV therapy [33]. In the present study, and supported by other studies [13,34,35], using RNAi as an inhibitor for HBV effectively reduces viral antigen levels, including HBsAg. It can be speculated that RNAi-treatments may offer complementary effects for current anti-HBV therapy. However, the final application of RNAi-based anti-HBV drugs depends on the development of effective and safe RNAi delivery systems.
Conclusions
In summary, four candidate shRNA plasmids significantly inhibited HBV genotypes A, B, C, D and I in vitro and in vivo. A potential avenue of investigation would be a combination strategy of various siRNA in a single transcript to improve efficacy and also prevent or at least delay the rise of viral escape mutants.
Methods
HBV Plasmids
Five HBV 1.35-fold genome plasmids - N10 (genotype Ae, AY707087), C4371 (genotype Ba, GU357842), Y1021 (genotype C1, GU357845), Y10 (genotype D1, GU357846) and W29 (genotype I1, GU357844) were used for transfection and hydrodynamic injection. The constructions and molecular and phenotypic characteristics are described in our previous report [36].
Bioinformatics Analysis
To define the conservative sites on HBV genomes amongst the various genotypes, all available complete genome sequences of HBV, as of April 2009, were downloaded from GenBank. Multiple alignment was done with ClustalX2 under default settings (Gap Opening:10, Gap Extension: 0.2, Delay Divergent Sequences(%): 30, DNA Transition Weight: 0.5, Use Negative Matrix: Off). The most representative and informative sequence in terms of phylogeny were collected as a dataset and the most similar sequences were removed using all pairwise distance scan. A total of 327 HBV genomes including A-I genotypes and nearly all reported subtypes were remained in the final dataset. The genotypes and subtypes of six HBV genomes isolated in the study were submitted to phylogenetic analysis using MEGA 4.0 software (data not shown). Forty sites with conservative sequences were selected and the shRNA plasmids were constructed (Table 1). The designed siRNA were evaluated for potential off-target effects by the online SOS program http://rnai.cs.unm.edu/offTarget. The sequences and positions of the forty designed shRNA targets are shown in Table 1.
ShRNA Plasmids
ShRNA plasmids were cloned downstream of the human H1 promoter in the vector pSUPER [37]. The target sites for siRNA were chosen based on conservative sites among the major HBV genotypes and subtypes. An shRNA plasmid targeting the firefly luciferase gene was used as a control (L1254: TGG CTA CAT TCT GGA GAC ATA).
Cell Culture and In Vitro Transfection
The plasmids used for in vitro transfection were purified with PlasmidSelect Xtra Starter Kit (GE Health, Sweden) and the concentrations were determined by the UV-spectrophotometric method. To determine the ability of siRNA to inhibit HBV gene expression in cell cultures, Huh7 cells were co-transfected with 4 μg of HBV plasmids, 1 μg of shRNA plasmids and 0.4 μg of a pcDNA3.1-SEAP plasmid using Lipofectamine 2000 (Invitrogen, Shanghai, China) following the manufacturer's instructions. They were then harvested four days later. The pcDNA3.1-SEAP plasmid is a reporter plasmid expressing secreted alkaline phosphatase and used for transfection efficiency standardization by estimating SEAP enzymatic activity (Pierce; Kunming, China) in the culture supernatant.
Evaluation for Potential Adverse Effects of siRNA
Possible adverse effects of shRNA on cells were evaluated using morphology criterion, growth rate assessment, and by noting the cytotoxicity profile of transfected cells. Cytotoxicity was determined through a WST-8 assay (Cell Counting Kit-8, Beyotime, Shanghai, China) [38,39]. The number of viable cells was then determined by absorbance measured at 450 nm on an automated plate reader. The potential off-target effects of siRNA were evaluated by monitoring the IFN response. Huh7 cells were transfected with 1 μg of shRNA plasmids. Non-transfected cells treated or untreated with 500 IU of IFNα-2a (Anfulong, Huadali Company, China) for 24 h served as a positive control [40]. Expression profile of four major interferon-stimulated (STAT1, OAS1, GBP1 and MX1) were analyzed by a quantitative RT-realtime PCR using the previously reported primers while the GAPDH level served as a control[41].
Mice Experiments
To evaluate the anti-viral effects of siRNA in vivo, an HBV hydrodynamic injection was conducted in BALB/c mice. Briefly, 50 μg of purified HBV plasmid and 10 μg shRNA plasmids were diluted to 2 mL with physiological saline and then injected into the tail vein within 5-10 s. Mice sera were assayed every day for HBsAg and HBeAg from Day 0 to Day 9. For each group, five mice aging from 4-6 weeks were used [42]. All animals received humane care and the study protocol complied with the institution's ethics guidelines.
Measurement of HBV RNA and DNA
For detection of the cytoplasmic HBV RNA, total RNA was extracted from cells using Tripure Isolation Reagent (Roche Applied Science, Switzerland) according to the manufacturer's instructions. Potential residual DNA contamination of RNA preparations were excluded by DNase I digestion. Ten nanograms of RNA were analysed by AccessQuick realtime RT-PCR System (Promega, USA) on a CFX96 instrument (Bio-Rad, USA). The HBV pg/pc (pregenomic/preCore) RNA level was detected by primers PGP (-CACCTCTGCCTAATCATC, nt1826-nt1843) and BC1 (GGAAAGAAGTCAGAAGGCAA, nt1974-nt1955) [43] using probe CP2 (HEX-ATGTTCATGTCCTACTGTTCAAGCC-BHQ2). The transcript copy number was normalized to those of GAPDH.
For the HBV DNA assay, 100 μL of supernatant was pre-heated at 50°C for 20 minutes and then treated with 1 U DNase I for 2 hours to eliminate residual plasmids. The reaction was terminated by EDTA at a final concentration of 10 mM. The mixture was then incubated at 70°C for 10 min and the HBV DNA was extracted using QIAamp DNA blood kits (QIAGEN, Hilden, Germany). HBV DNA quantification assays were performed using a commercial real-time PCR kit (Kehua, Shanghai, China).
Determination of HBV Antigens
HBsAg, HBeAg and HBcAg levels were determined by chemiluminescence using commercial assay kits (Wantai, Beijing, China). The relative level of each antigen was expressed as an S/CO (signal/cutoff) value, on a linear range from 1 to 1000 for all three assays. The lower detection limit was 10 pg/mL for the HBsAg and HBeAg assays, and 50 pg/ml for the HBcAg assay. In regards to the intracelluar HBV antigen assay, the transfected cells were treated with a suitable lysis buffer (20 mM HEPES, 1 mM EGTA, 100 mM NaCl, 5 mM Mg2Cl, 0.4% n-Dodecyl β-D- maltoside, n-Dodecyl β-D-maltoside, and 10% Glycerol) at room temperature for 30 minutes and the supernatants were separated through centrifugation and used for immunoassay.
Statistical Evaluation
Statistical analyses were performed using independent Student t test or Mann-Whitney U test (GraphPad Software, San Diego California USA,). Differences were considered to be statistically significant for p values ≤ 0.05.
Abbreviations
SIRNA: small interfering RNA; SHRNA: short hairpin RNA; OFF-TARGET EFFECT: non-specific effects resulting from the introduction of siRNA; STAT1: signal transducers and activators of transcription1; OAS1: 2'-5'-oligoadenylate synthetase 1; MX1: interferon-induced GTP-binding protein; GBP1: guanylate binding protein 1; HBV: hepatitis B virus; HBSAG: hepatitis B surface antigen; HBEAG: hepatitis B e antigen; HBCAG: hepatitis B core antigen.
Authors' contributions
YLZ, TC, JZ and NSX conceived the study, participated in its design and coordination and drafted the manuscript. YLZ and QY carried out the molecular genetic studies, analyzed the aligned sequences, found conserved targets, participated in the study design and were involved in the shRNA design. YZL and YJC constructed all shRNA plasmids. YZL, YJC, CL, TZ, DZX, RYL, LWY and YBW performed all cell and mice experiments (including all transfections, hydrodynamic injections, WST-8 assays, RT-PCR and chemiluminescence immunoassays). YLZ, YJC, TC and QY conducted the data analysis and interpretation. AEY, JWS, QY, JZ and NSX helped to draft the manuscript and critically revised its final version. TC, JZ and NSX obtained funding. All authors read and approved the final manuscript.
Contributor Information
Ya-Li Zhang, Email: zhangyali@xmu.edu.cn.
Tong Cheng, Email: tcheng@xmu.edu.cn.
Yi-Jun Cai, Email: caiyijun@xmu.edu.cn.
Quan Yuan, Email: yuanquan@xmu.edu.cn.
Che Liu, Email: liuche@xmu.edu.cn.
Tao Zhang, Email: zhangtnidvd@xmu.edu.cn.
De-Zhen Xia, Email: xiadz@xmu.edu.cn.
Rui-Yin Li, Email: liruiyin@xmu.edu.cn.
Lian-Wei Yang, Email: ylianwei@xmu.edu.cn.
Ying-Bin Wang, Email: ybwang@xmu.edu.cn.
Anthony ET Yeo, Email: ayeo@xmu.edu.cn.
James Wai-Kuo Shih, Email: jwshih@xmu.edu.cn.
Jun Zhang, Email: zhangj@xmu.edu.cn.
Ning-shao Xia, Email: nsxia@xmu.edu.cn.
Acknowledgements
This work was supported by a grant from Key Special Subjects of Infectious Diseases (2008ZX10002-011 and 2008ZX10002-012) and from the Excellent Youth Foundation of Fujian Scientific Committee (2009J06020). We gratefully acknowledge Lucy Zhu from McGill University for editorial assistance in writing this paper. We also thank Dr. Hai Yu and Dr. Chenghao Huang (NIDVD, Xiamen) for their technical help with this work.
References
- Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337(24):1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2(8256):1129–1133. doi: 10.1016/S0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- Mast EE, Alter MJ, Margolis HS. Strategies to prevent and control hepatitis B and C virus infections: a global perspective. Vaccine. 1999;17(13-14):1730–1733. doi: 10.1016/S0264-410X(98)00415-0. [DOI] [PubMed] [Google Scholar]
- Vial T, Descotes J. Clinical toxicity of the interferons. Drug Saf. 1994;10(2):115–150. doi: 10.2165/00002018-199410020-00003. [DOI] [PubMed] [Google Scholar]
- Fattovich G, Giustina G, Favarato S, Ruol A. A survey of adverse events in 11,241 patients with chronic viral hepatitis treated with alfa interferon. J Hepatol. 1996;24(1):38–47. doi: 10.1016/S0168-8278(96)80184-X. [DOI] [PubMed] [Google Scholar]
- Liaw YF. Antiviral therapy of chronic hepatitis B: opportunities and challenges in Asia. J Hepatol. 2009;51(2):403–410. doi: 10.1016/j.jhep.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP. RNA interference inhibitors of hepatitis B virus. Ann N Y Acad Sci. 2009;1175:15–23. doi: 10.1111/j.1749-6632.2009.04974.x. [DOI] [PubMed] [Google Scholar]
- Deng L, Li G, Xi L, Yin A, Gao Y, You W, Wang X, Sun B. Hepatitis B Virus Inhibition in Mice by Lentiviral Vector Mediated Small Interference RNA. BMC Gastroenterol. 2009;9(1):73. doi: 10.1186/1471-230X-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PL, Althage A, Chung J, Chisari FV. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci USA. 2002;99(21):13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21(6):639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37(4):764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- Ying RS, Zhu C, Fan XG, Li N, Tian XF, Liu HB, Zhang BX. Hepatitis B virus is inhibited by RNA interference in cell culture and in mice. Antiviral Res. 2007;73(1):24–30. doi: 10.1016/j.antiviral.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8(5):769–776. doi: 10.1016/S1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Chen Y, Cheng G, Mahato RI. RNAi for treating hepatitis B viral infection. Pharm Res. 2008;25(1):72–86. doi: 10.1007/s11095-007-9504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely A, Naidoo T, Mufamadi S, Crowther C, Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol Ther. 2008;16(6):1105–1112. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- Olinger CM, Jutavijittum P, Hubschen JM, Yousukh A, Samountry B, Thammavong T, Toriyama K, Muller CP. Possible new hepatitis B virus genotype, southeast Asia. Emerg Infect Dis. 2008;14(11):1777–1780. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Trinh TN, Abe K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J Virol. 2008;82(11):5657–5663. doi: 10.1128/JVI.02556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Roquelaure B, Tamalet C. Detection of a newly identified hepatitis B virus genotype in southeastern France. J Clin Virol. 2009;45(2):165–167. doi: 10.1016/j.jcv.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, Kramvis A, Shimada T, Izumi N. et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology. 2006;44(4):915–924. doi: 10.1002/hep.21345. [DOI] [PubMed] [Google Scholar]
- Wu HL, Huang LR, Huang CC, Lai HL, Liu CJ, Huang YT, Hsu YW, Lu CY, Chen DS, Chen PJ. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128(3):708–716. doi: 10.1053/j.gastro.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MF, Joshi S. RNA-polymerase III-driven expression cassettes in human gene therapy. Curr Opin Mol Ther. 1999;1(5):580–594. [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Keck K, Volper EM, Spengler RM, Long DD, Chan CY, Ding Y, McCaffrey AP. Rational design leads to more potent RNA interference against hepatitis B virus: factors effecting silencing efficiency. Mol Ther. 2009;17(3):538–547. doi: 10.1038/mt.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredehorst R, von Wulffen H, Granato C. Quantitation of hepatitis B virus (HBV) core antigen in serum in the presence of antibodies to HBV core antigen: comparison with assays of serum HBV DNA, DNA polymerase, and HBV e antigen. J Clin Microbiol. 1985;21(4):593–598. doi: 10.1128/jcm.21.4.593-598.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Rokuhara A, Matsumoto A, Yagi S, Tanaka E, Kiyosawa K, Maki N. New enzyme immunoassay for detection of hepatitis B virus core antigen (HBcAg) and relation between levels of HBcAg and HBV DNA. J Clin Microbiol. 2003;41(5):1901–1906. doi: 10.1128/JCM.41.5.1901-1906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA. et al. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365(9454):123–129. doi: 10.1016/S0140-6736(05)17701-0. [DOI] [PubMed] [Google Scholar]
- EASL. [EASL clinical practice guidelines. Management of chronic hepatitis B] Gastroenterol Clin Biol. 2009;33(6-7):539–554. doi: 10.1016/j.gcb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Papatheodoridis GV, Manolakopoulos S. EASL clinical practice guidelines on the management of chronic hepatitis B: the need for liver biopsy. J Hepatol. 2009;51(1):226–227. doi: 10.1016/j.jhep.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Stroffolini T, Gaeta GB, Mele A. AASLD Practice Guidelines on chronic hepatitis B and HBV infection in Italy. Hepatology. 2007;46(2):608–609. doi: 10.1002/hep.21841. author reply 609. [DOI] [PubMed] [Google Scholar]
- Arbuthnot P, Longshaw V, Naidoo T, Weinberg MS. Opportunities for treating chronic hepatitis B and C virus infection using RNA interference. J Viral Hepat. 2007;14(7):447–459. doi: 10.1111/j.1365-2893.2006.00818.x. [DOI] [PubMed] [Google Scholar]
- Moore MD, McGarvey MJ, Russell RA, Cullen BR, McClure MO. Stable inhibition of hepatitis B virus proteins by small interfering RNA expressed from viral vectors. J Gene Med. 2005;7(7):918–925. doi: 10.1002/jgm.739. [DOI] [PubMed] [Google Scholar]
- Hamasaki K, Nakao K, Matsumoto K, Ichikawa T, Ishikawa H, Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543(1-3):51–54. doi: 10.1016/S0014-5793(03)00400-9. [DOI] [PubMed] [Google Scholar]
- Yu H, Yuan Q, Ge SX, Wang HY, Zhang YL, Chen QR, Zhang J, Chen PJ, Xia NS. Molecular and phylogenetic analyses suggest an additional hepatitis B virus genotype "i". PLoS One. 2010;5(2):e9297.. doi: 10.1371/journal.pone.0009297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Shiokawa T, Hattori Y, Kawano K, Ohguchi Y, Kawakami H, Toma K, Maitani Y. Effect of polyethylene glycol linker chain length of folate-linked microemulsions loading aclacinomycin A on targeting ability and antitumor effect in vitro and in vivo. Clin Cancer Res. 2005;11(5):2018–2025. doi: 10.1158/1078-0432.CCR-04-1129. [DOI] [PubMed] [Google Scholar]
- Ishiyama M, Tominaga H, Shiga M, Sasamoto K, Ohkura Y, Ueno K. A combined assay of cell viability and in vitro cytotoxicity with a highly water-soluble tetrazolium salt, neutral red and crystal violet. Biol Pharm Bull. 1996;19(11):1518–1520. doi: 10.1248/bpb.19.1518. [DOI] [PubMed] [Google Scholar]
- Sun D, Rosler C, Kidd-Ljunggren K, Nassal M. Quantitative assessment of the antiviral potencies of 21 shRNA vectors targeting conserved, including structured, hepatitis B virus sites. J Hepatol. 2010;52(6):817–826. doi: 10.1016/j.jhep.2009.10.038. [DOI] [PubMed] [Google Scholar]
- Liang YG, Liu HY, Liu BX, Bai Y, Wu H, Zhou QH, Chen J. Detection of IFN Response of Non-Specific Effects on RNAi. Chin J Lung Cancer. 2009;12(1):16–22. doi: 10.3779/j.issn.1009-3419.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Wang SH, Yeh SH, Lin WH, Wang HY, Chen DS, Chen PJ. Identification of androgen response elements in the enhancer I of hepatitis B virus: A mechanism for sex disparity in chronic hepatitis B. Hepatology. 2009. [DOI] [PubMed]
- Jammeh S, Thomas HC, Karayiannis P. Replicative competence of the T131I, K141E, and G145R surface variants of hepatitis B Virus. J Infect Dis. 2007;196(7):1010–1013. doi: 10.1086/521198. [DOI] [PubMed] [Google Scholar]