Abstract
Background
Studies of several tumour types have shown that expression profiling of cellular protein extracted from surgical tissue specimens by direct mass spectrometry analysis can accurately discriminate tumour from normal tissue and in some cases can sub-classify disease. We have evaluated the potential value of this approach to classify various clinico-pathological features in colorectal cancer by employing matrix-assisted laser desorption ionisation time of-flight-mass spectrometry (MALDI-TOF MS).
Methods
Protein extracts from 31 tumour and 33 normal mucosa specimens were purified, subjected to MALDI-Tof MS and then analysed using the 'GenePattern' suite of computational tools (Broad Institute, MIT, USA). Comparative Gene Marker Selection with either a t-test or a signal-to-noise ratio (SNR) test statistic was used to identify and rank differentially expressed marker peaks. The k-nearest neighbours algorithm was used to build classification models either using separate training and test datasets or else by using an iterative, 'leave-one-out' cross-validation method.
Results
73 protein peaks in the mass range 1800-16000Da were differentially expressed in tumour verses adjacent normal mucosa tissue (P ≤ 0.01, false discovery rate ≤ 0.05). Unsupervised hierarchical cluster analysis classified most tumour and normal mucosa into distinct cluster groups. Supervised prediction correctly classified the tumour/normal mucosa status of specimens in an independent test spectra dataset with 100% sensitivity and specificity (95% confidence interval: 67.9-99.2%). Supervised prediction using 'leave-one-out' cross validation algorithms for tumour spectra correctly classified 10/13 poorly differentiated and 16/18 well/moderately differentiated tumours (P = < 0.001; receiver-operator characteristics - ROC - error, 0.171); disease recurrence was correctly predicted in 5/6 cases and disease-free survival (median follow-up time, 25 months) was correctly predicted in 22/23 cases (P = < 0.001; ROC error, 0.105). A similar analysis of normal mucosa spectra correctly predicted 11/14 patients with, and 15/19 patients without lymph node involvement (P = 0.001; ROC error, 0.212).
Conclusions
Protein expression profiling of surgically resected CRC tissue extracts by MALDI-TOF MS has potential value in studies aimed at improved molecular classification of this disease. Further studies, with longer follow-up times and larger patient cohorts, that would permit independent validation of supervised classification models, would be required to confirm the predictive value of tumour spectra for disease recurrence/patient survival.
Background
Colorectal cancer (CRC) is the second commonest malignancy and has a five-year survival rate of approximately 50% [1,2]. The majority of patients, particularly with early stage disease (Dukes' A, Stage I), are treated with surgery [3]. For more advanced disease (Dukes' C and D, Stage III or IV) surgery combined with adjuvant chemotherapy has proven survival benefits [4-6]. However, the disease outcome is very variable and prognosis and prediction of treatment response based on conventional disease staging criteria is not reliable [6,7]. There has therefore been considerable interest in the development of more robust prognostic and predictive disease markers for patient stratification with the ultimate aim of tailoring treatment to the individual patient [8,9].
Markers based on circulating carcinoembryonic antigen (CEA) levels and various tumour-associated gene mutations including microsatellite instability (MSI), loss of heterozygosity of 18q, deleted in colorectal cancer (DCC), mutations in KRAS, BRAF and PIK3CA genes have all been shown to be of some prognostic or predictive value (reviewed in [8,10]). In particular, the mutational status of KRAS, BRAF and PIK3CA genes has recently been proposed as a reliable marker for predicting responders to new targeted agents for the epidermal growth factor receptor (EGFR) [11,12]. In addition, gene expression profiling studies of both mRNA [13] and microRNA [14] have revealed tumour-associated gene expression signatures that form the basis for a molecular classification of disease sub-types that define disease course and treatment response (reviewed in [8]). These studies on gene mutations and RNA expression have been paralleled by analysis of the tumour cell proteome, most commonly employing the technique of two-dimensional difference gel electrophoresis (2D-DIGE) to identify proteins that are differentially expressed in tumour verses normal mucosa tissue (reviewed in [15]). An expanding list of candidate prognostic markers have emerged from these studies including for example, cathepsin D, S100A4 and APAF-1 [15].
As an alternative to 2D-DIGE, studies of other tumour types have also employed the technique of direct protein expression profiling of tumour/normal tissue by surface enhanced laser desorption ionisation time-of-flight mass spectrometry (SELDI-TOF) or by matrix-assisted laser desorption ionisation time of-flight-mass spectrometry (MALDI-TOF) mass spectrometry [16,17]. This approach, which is most commonly associated with the development of serum-based diagnostic markers, offers a number of advantages over 2D-DIGE. Although the technique yields no information on the actual identities of proteins, the reproducible spectral profiles that are relatively simple to generate in high throughput studies allow robust classification models of different proteome populations to be built. For example, studies of lung [18], breast [19], head and neck cancer [20] have all shown that the spectral profiles of tumour and normal tissue can be accurately discriminated and in some cases sub-classified by direct protein profiling using SELDI/MALDI-TOF mass spectrometry. Only one previous study has reported on the detection of differences between normal mucosa, adenoma and colorectal carcinoma by using SELDI-TOF MS [21].
In the present study, we have evaluated the potential value of protein expression profiling of CRC tissue by MALDI-TOF mass spectrometry. In addition to comparing tumour with adjacent normal mucosa, we have investigated whether spectral profiles of tumour tissue can be used to classify various clinico-pathological features of disease. Since previous 2D-DIGE studies have reported abnormalities of protein expression profiles in tumour-adjacent normal tissue [22], we have also extended this analysis to normal mucosa tissue.
Methods
Clinical specimens
Tissue samples were collected from a total of 36 patients with confirmed CRC at the time of surgical resection at Colchester General Hospital, Essex UK. All specimens were obtained following informed consent in accordance with local UK NHS Ethics Committee approval (protocol reference: MH 528). Surgically excised specimens were washed extensively in ice-cold 150 mM NaCl and samples of normal colonic mucosa (>10 cm from tumour margin) and tumour tissues were excised using a scalpel and then snap frozen and transferred to a - 80°C freezer. The total time from surgical resection to snap freezing of specimens was <30 mins.
Protein extraction and purification
Frozen tissue samples (approximately 250 mg) were ground using a mortar and pestle and then lysed for 30 mins at 4°C in 1.0 ml of 10 mM Tris-HCl pH 7.5, 200 mM NaCl containing Protease inhibitor cocktail (Roche Pharmaceuticals) and 1% N-octyl-β-D-glucopyranoside (Sigma Aldrich). The cell lysate was then centrifuged at 12,000 × g for 30 mins and the supernatant representing the solubilised fraction was removed. Protein was further purified by reversed phase hydrophobic interaction chromatography using a commercially available super-paramagnetic microparticle kit (MB-HIC-C8, Bruker Daltonics). Briefly, 10 μl of 30-35 mg/ml protein solution was adsorbed to 10 μl of beads after addition of 20 μl kit binding buffer. After three washes with 200 μl 0.1% trifluoroacetic acid, protein was eluted in 20 μl of 50% (v/v) acetonitrile (Fisher Scientific) Eluted protein was stored at 4°C for no more than 1 hr prior to matrix co-crystallisation.
MALDI-TOF mass spectrometry
To facilitate reproducible co-crystallisation of protein with matrix solution, a modification of the slow crystallisation method [23] was used. Briefly, 20 ul of purified protein was mixed with 20 μl of acetonitrile containing 0.1% trifluoroacetic acid, saturated with sinapic acid (Sigma Aldrich). A 20 μl aqueous solution containing diammonium citrate (200 mM) and nitrotetracetic acid (0.1%) was added and crystal formation was allowed to proceed for 2-3 hrs. Crystallised matrix-protein samples were spotted onto a stainless steel MALDI target plate and spectra were acquired using a MALDI-TOF mass spectrometer (Reflex IV; Bruker Daltonics) with the following instrument settings: ion source 1, 20 kV; ion source 2, 16.65 kV; lens voltage, 9.5 kV; pulsed ion extraction, 200 ns. Ionisation was achieved by irradiation with a nitrogen laser (e = 337 nm) operating at 25 Hz and 20% laser power. For matrix suppression, we used a high gating factor with signal suppression up to 1500 Da. Mass spectra were detected in linear positive mode. Detector gain was set at 1600 V, sample rate at 1.0 and electronic gain at 100 mV with real-time smoothing. Spectra were acquired in duplicate from 500 laser shots delivered as 5 × 100 pulses and were internally calibrated using 'FlexAnalysis' spectral processing software (Version 2.0; Bruker Daltonics) with reference marker peaks at 2426.9Da, 6109.5 Da and 12471.6 Da. External calibration used the following reference standards: bombesin (1620.86 Da), somatostatin (3149.57 Da), insulin (5734.51 Da), ubiquitin I (8565.76 Da), cytochrome c (12,360.97 Da) and myoglobin (16,952.30 Da).
Spectral processing and analysis
Calibrated spectra were exported as ASCII files and were digitally processed by smoothing, de-noising, baseline subtraction and normalisation (by total ion current) using the 'SpecAlign' suite of spectral computational tools [24,25]. Validation of the reproducibility of the resulting mass spectrometry profiles and elimination of 'outliers' was accomplished as described elsewhere [26]. Duplicate spectra with a cross-correlation function of < 0.950 were discarded. From the initial cohort of specimens, representing matched tumour and adjacent normal mucosa from 36 patients, a total of 64 spectra representing 31 tumours and 33 normal mucosa were obtained (see Table 1). Of the 5 tumour and 3 mucosa specimens that were excluded from analysis, 2 tumour and one mucosa failed to yield reproducible spectra on repeated protein preparations. The remaining 3 tumour and 2 mucosa specimens consistently gave spectra of poor quality (outliers), presumably as a result of specimen deterioration. Matching peaks were aligned across spectra by using the combined Fast Fourier Transform/Peak matching method [25] and modelled peak areas for the entire set of spectra were exported as a single csv file.
Table 1.
Clinico-pathological features of patient specimens
| Tumour | 1NM | Age | Gender | Dukes' stage | TNM stage | Differentiation | Vascular invasion |
2LNs harvested | LNs pos | Patient status | 3Follow-up time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| - | 001NM | 78 | F | B | pT3, pN0, pR0 | Poor | Absent | 15 | 0 | Well & symptom free | 48 |
| 002T | 002NM | 91 | M | B | pT3, pN0, pR0 | Moderate | Absent | 9 | 0 | Deceased (recurrence) | 35 |
| 003T | 003NM | 75 | M | C1 | pT3, pN1, pR0 | Poor | Absent | 10 | 3 | Well & symptom free) | 36 |
| 004T | 004NM | 74 | F | C1 | pT4, pN1, pR2 | Poor | Present | 11 | 3 | Deceased (recurrence) | <1 |
| 005T | 005NM | 76 | M | B | pT3, pN0, pR0 | Poor | Absent | 6 | 0 | Well & symptom free | 49 |
| - | 006NM | 69 | F | A | pT2, pN0, pR0 | Well | Absent | 11 | 0 | Well & symptom free | 40 |
| - | 007NM | 52 | M | C1 | pT3, pN1, pR0 | Poor | Absent | 23 | 3 | Well & symptom free | 48 |
| 008T | 008NM | 63 | F | C1 | pT4, pN0, pR0 | Poor | Absent | 10 | 0 | Deceased (recurrence) | 40 |
| 009T | 009NM | 68 | M | B | pT3, pN0, pR0 | Poor | Absent | 8 | 8 | Well & symptom free | 36 |
| 011T | 011NM | 77 | M | C1 | pT4,p N1, pR0 | Poor | Absent | 15 | 3 | Well & symptom free | 40 |
| 016T | 016NM | 61 | M | C2 | pT2, pN2, pR0 | Moderate | Present | 14 | 5 | Well & symptom free | 43 |
| 017T | 017NM | 65 | F | B | pT3, pN0, pR0 | Moderate | Absent | 14 | 0 | Well & symptom free | 39 |
| 020T | 020NM | 65 | F | B | pT3, pN0, pR0 | Poor | Absent | 12 | 0 | Well & symptom free | 36 |
| 021T | 021NM | 72 | M | B | pT4, pN1, pR0 | Moderate | Present | 5 | 1 | Well & symptom free | 28 |
| 023T | 023NM | 59 | M | B | pT3, pN0, pR0 | Moderate | Absent | 10 | 0 | Well & symptom free | 20 |
| 024T | 024NM | 41 | F | C2 | pT4, pN1, pRx | Well | Absent | 15 | 2 | Deceased (recurrence) | 30 |
| 025T | - | 82 | M | B | pT4, pN0, pMx, pRx | Poor | Absent | 7 | 0 | Deceased (recurrence) | 13 |
| 026T | 026NM | 76 | F | A | pT2, pN0, pR0 | Moderate | Absent | 5 | 0 | Deceased (recurrence) | 36 |
| 028T | 028NM | 86 | F | C1 | pT3, pN1, pR0 | Moderate | Absent | 12 | 0 | Well & symptom free | 36 |
| 029T | 029NM | 71 | F | B | pT3, pN0, pR0 | Well | Absent | 32 | 0 | Well & symptom free | 36 |
| 031T | 031NM | 82 | M | C2 | pT3, pN2, pR0 | Poor | Present | 11 | 3 | Well & symptom free | 36 |
| 032T | 032NM | 69 | F | B | pT4, pN0, pR0 | Moderate | Absent | 11 | 0 | Well & symptom free | 23 |
| 033T | 033NM | 72 | M | C1 | pT4, pN1, pR0 | Moderate | Absent | 8 | 1 | Well & symptom free | 22 |
| 034T | 034NM | 58 | M | C1 | pT4, pN1, pR0 | Moderate | Absent | 5 | 3 | Well & symptom free | 25 |
| - | 035NM | 77 | F | B | pT3, pN0, pR0 | Poor | Absent | 7 | 0 | Well & symptom free | 25 |
| 036T | - | 81 | F | B | pT3, pN0, pR0 | Moderate | Absent | 13 | 0 | Well & symptom free | 21 |
| 037T | 037NM | 77 | F | B | pT3, pN0, pR0 | Well | Absent | 7 | 0 | Well & symptom free | 19 |
| 038T | 038NM | 76 | F | A | pT2, pN1, pR0 | Poor | Absent | 5 | 1 | Well & symptom free | 20 |
| 039T | 039NM | 75 | F | B | pT3, pN0, pR0 | Moderate | Absent | 16 | 0 | Well & symptom free | 23 |
| 2012T | 2012NM | 62 | M | C1 | pT3, pN1, pR0 | Poor | Present | 18 | 3 | Well & symptom free | 20 |
| 2018T | 2018NM | 83 | F | A | pT1, pN0, pR0 | Moderate | Absent | 6 | 0 | Deceased (unrelated) | 2 |
| 2022T | 2022NM | 56 | M | B | pT3, pN0, pR0 | Well | Present | 20 | 0 | Well & symptom free | 20 |
| 2044T | 2044NM | 82 | M | A | pT2, pN0, pR0 | Moderate | Absent | 10 | 0 | Well & symptom free | 21 |
| - | 2080NM | 72 | F | A | pT2, pN0, pR0 | Moderate | Absent | 5 | 0 | Well & symptom free | 21 |
| 2084T | - | 38 | M | B | ypT3, ypN0, ypR0 | Poor | Absent | 10 | 0 | Well & symptom free | 20 |
| 2085T | 2085NM | 78 | F | C1 | pT3, pN1, pR0 | Moderate | Absent | 11 | 1 | Deceased (unrelated) | <1 |
1NM = normal mucosa; 2LN = lymph node; 3follow-up time in months
Subsequent spectral analysis was implemented in the 'GenePattern' suite of software tools (Broad Institute, MIT, USA) [27]. Hierarchical clustering used Euclidean correlation as the column distance measure with pair-wise average linkage as the clustering method. Comparative Gene Marker Selection [28,29] with either a t-test or a signal-to-noise ratio (SNR) test statistic was used to identify and rank differentially expressed marker peaks and to assign Bonferroni-corrected P and false discovery rate (FDR) values [28-30]. The k-nearest neighbours (kNN) algorithm [29] was used to build a classification model for tumour vs normal using separate training and test datasets. For this purpose, two thirds of the spectra, comprised of a representative proportion of tumour and normal spectra, were randomly assigned to a training dataset, with the remaining third being used as an independent test dataset. Spectra were randomly assigned using the GenePattern 'SplitDatasetTrainTest' module [27]. Alternatively the kNN algorithm was used in an iterative, 'leave-one-out' cross-validation mode. Other statistical analysis used the SPSS software.
Results
Spectral profiles in tumour and normal mucosa tissues
Table 1 summarises the clinico-pathological data for the 36 CRC patients from whom specimens were obtained. In most cases, spectra of adequate quality from matching pairs of tumour and adjacent normal mucosa were obtained. However, some tissue protein preparations consistently yielded spectra of poor quality or that were poorly reproducible (see Methods section); these were excluded from the analysis. The resulting 64 spectra, representing 31 tumour and 33 normal mucosa specimens, generated a total of 265 protein peaks in the mass range 1800-16000Da. Illustrative examples of raw MALDI-TOF spectral profiles are shown in additional file 1. Although the overall intensity profile of individual protein peaks was very heterogeneous across different specimens, unsupervised hierarchical cluster analysis classified most tumour and normal mucosa into distinct cluster groups (Figure 1) consistent with major differences in the tumour verses normal protein expression profiles.
Figure 1.
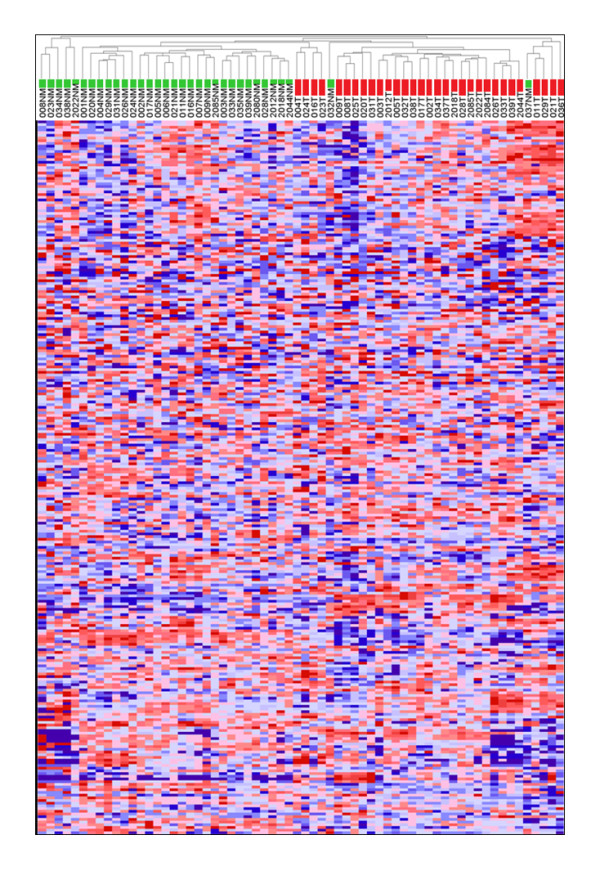
Unsupervised hierarchical cluster analysis of tumour and normal mucosa spectra. The dendrogram and heatmap show the clustering of Tumour (T) and normal mucosa (NM) spectra using Euclidean correlation as the column distance measure with pair-wise average linkage as the clustering method. Row clustering (not shown) used Spearman's rank correlation as distance measure with pair-wise complete linkage as the clustering method. Specimens are colour-coded as green (NM) and red (T).
To quantitatively evaluate the differences between the protein expression profiles of tumour verses normal tissue, the Comparative Gene Marker Selection algorithm [28] was applied to the spectral data-set to determine the level of significance of difference between tumour and normal for each protein peak. Figure 2 shows the frequency distribution (occurrences) of protein peak P values (Feature P) that were binned in increments of 0.05. Above P = 0.05, the representation of protein peaks was fairly evenly distributed. However, nearly 100 peaks gave a P value < 0.05, indicating that a sizable fraction of proteins detected by MALDI-TOF mass spectrometry discriminate between tumour and normal colonic tissue. Applying a threshold of P ≤ 0.01, FDR ≤ 0.05, the expression profile of a total of 73 protein peaks was significantly different between tumour and normal tissue with 57 being up-regulated in normal tissue and 16 being up-regulated in tumour tissue. Figure 3 shows a heat-map profile of these 'marker peaks' and additional file 2 summarises their statistical features.
Figure 2.
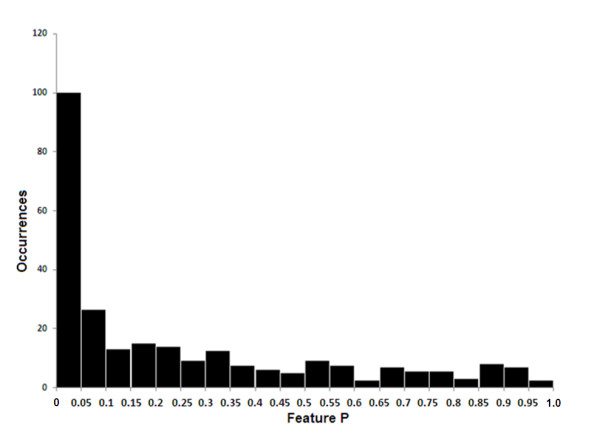
Probability distribution of marker peaks distinguishing tumour from normal mucosa. Spectra from all 64 tumour and normal tissue samples were analysed by Comparative Gene Marker Selection [28] using the SNR test statistic to identify peaks (features) that discriminate tumour from normal tissue. The feature P histogram shows the number of peaks (occurrences) that fall within binned P values.
Figure 3.
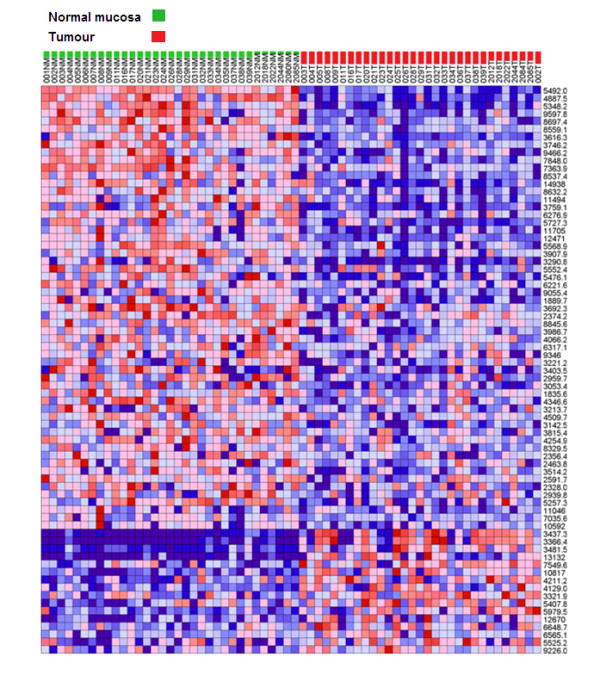
Heat map profile of marker peaks discriminating tumour from normal mucosa. The expression profiles and m/z values of the top 73 ranked peaks identified by Comparative Gene Marker Selection [28] (P = ≤ 0.01, FDR = ≤ 0.05) are depicted for all 64 tissue specimens.
To rigorously demonstrate that tumour and normal mucosa tissue could be distinguished using their protein spectral profiles, the 64 spectra were randomly split into separate training and test datasets. The training spectra dataset was used to optimise a kNN algorithm [29] for predicting tumour or normal status. As summarised in additional file 3, the model correctly predicted the status of specimens in the independent test spectra dataset with 100% sensitivity and specificity (95% confidence interval: 67.9-99.2%).
Classification of clinico-pathological characteristics from tumour spectra
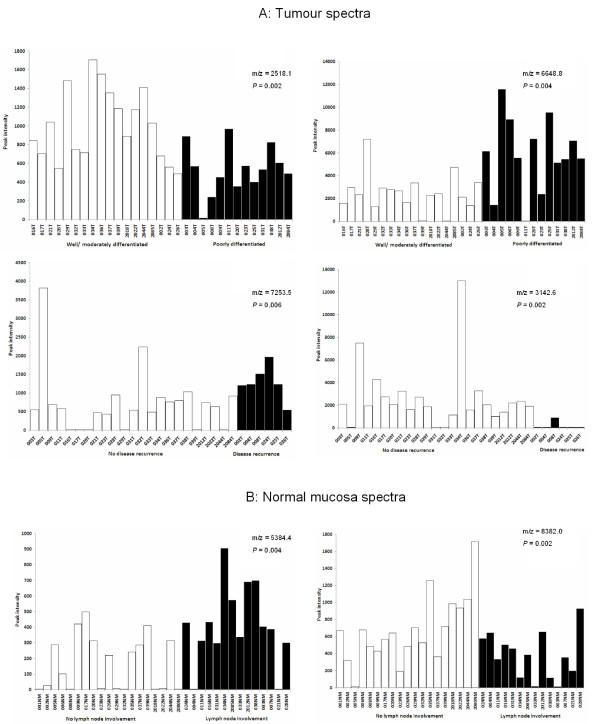
To determine whether the protein expression profiles of tumour tissue could be used to predict individual clinico-pathological characteristics of patients (Table 1), the kNN algorithm was used to optimise a series of classification models. Since the limited numbers of datasets precluded analysis by using independent train and test spectra, the kNN algorithm was used in an iterative, 'leave-one-out' cross-validation mode. Table 2 summarises the results of this analysis. The predictive model for distinguishing poorly differentiated from well/moderately differentiated tumours gave a receiver-operator characteristics (ROC) error of 0.171, correctly classifying 10/13 poorly differentiated and 16/18 well/moderately differentiated tumours (P = < 0.001). Additional file 4 summarises the kNN algorithm results and Figure 4A shows the expression profiles of the top two ranked discriminating peaks. The kNN model for disease recurrence also gave a low ROC error (0.105 - see Table 2A). As summarised in additional file 5, the model correctly predicted 5/6 patients with recurrent disease and 22/23 who are disease-free (P = < 0.001). Figure 4A shows the expression profiles of the top two ranked marker peaks for classifying disease outcome.
Table 2.
Performance of predictive models for classification of clinico-pathological characteristics in tumour tissue
| CHARACTERISTICS |
1Advanced Dukes' stage |
Poorly differentiated | Lymph node involvement |
Invasiveness | 2Disease recurrence |
|---|---|---|---|---|---|
| Number of features | 5 | 2 | 4 | 9 | 10 |
| Positive prediction rate | 6/12 | 10/13 | 5/13 | 3/7 | 5/6 |
| Sensitivity | 0.500 | 0.769 | 0.385 | 0.429 | 0.833 |
| 3CI | 0.223-0.777 | 0.460-0.938 | 0.151-0.677 | 0.118-0.798 | 0.364-0.991 |
| Positive predictive value | 0.750 | 0.833 | 0.625 | 0.750 | 0.833 |
| CI | 0.356-0.955 | 0.509-0.971 | 0.259-0.898 | 0.219-0.986 | 0.364-0.991 |
| Negative prediction rate | 17/19 | 16/18 | 15/18 | 23/24 | 22/23 |
| Specificity | 0.894 | 0.889 | 0.833 | 0.958 | 0.957 |
| CI | 0.654-0.981 | 0.639-0.981 | 0.577-0.956 | 0.768-0.998 | 0.760-0.998 |
| Negative predictive value | 0.739 | 0.842 | 0.652 | 0.852 | 0.957 |
| CI | 0.513-0.889 | 0.585-0.958 | 0.428-0.828 | 0.654-0.951 | 0.760-0.998 |
| Absolute error | 0.258 | 0.161 | 0.355 | 0.161 | 0.069 |
| 4ROC error | 0.302 | 0.171 | 0.391 | 0.307 | 0.105 |
| Fisher's exact test | P = 0.020 | P = < 0.001 | P = 0.133 | P = 0.027 | P = < 0.001 |
1Includes Dukes' C1 and C2; 2 Median follow-up time for recurrent disease patients: 33 months; median follow-up time for disease-free patient: 27 months (analysis excludes patients who died through surgical complications - see Table 1); 3CI = 95% confidence interval; 4ROC = receiver-operator characteristics
The KNN algorithm [29] was used in 'leave-one-out' cross-validation prediction with the number of features (marker peaks) specified. Marker peaks were selected using a t-test statistic except for lymph node involvement and invasiveness characteristics of tumour tissue where the SNR test statistic was used.
Figure 4.
Relative ion intensity profiles of marker peaks used in predictive algorithms of tumour/mucosa clinico-pathological features. The peak intensity profiles of the top two-ranked scoring peaks are shown for tumour spectra (A) for classifying differentiation and disease recurrence and for normal mucosa spectra (B) for classifying lymph node involvement (see Table 2). The performance of predictive models for these clinico-pathological features are shown in additional file 4 (differentiation), in additional file 5 (disease recurrence) and in additional file 6 (lymph node involvement). The t-test P value is given for each marker peak.
Classification of clinico-pathological characteristics from normal mucosa spectra
In a similar analysis of normal mucosa spectra (Table 3), only the characteristic of lymph node involvement gave a low ROC error (0.212). As shown in Table 3 and in additional file 6, the kNN algorithm correctly predicted 11/14 patients with, and 15/19 patients without lymph node involvement (P = 0.001). Figure 4B shows the expression profiles of the top two ranked marker peaks for classifying the characteristic of lymph node involvement.
Table 3.
Performance of predictive models for classification of clinico-pathological characteristics in normal mucosa tissue.
| CHARACTERISTICS |
1Advanced Dukes' stage |
Poorly differentiated | Lymph node involvement |
Invasiveness | 2Disease recurrence |
|---|---|---|---|---|---|
| Number of features | 7 | 5 | 3 | 6 | 7 |
| Positive prediction rate | 8/13 | 8/14 | 11/14 | 3/7 | 0/5 |
| Sensitivity | 0.615 | 0.571 | 0.786 | 0.429 | 0.000 |
| 3CI | 0.322-0.849 | 0.296-0.812 | 0.488-0.943 | 0.116-0.798 | 0.000-0.537 |
| Positive predictive value | 0.500 | 0.444 | 0.733 | 0.500 | 0.000 |
| CI | 0.255-0.749 | 0.224-0.686 | 0.448-0.911 | 0.139-0.860 | 0.000-0.945 |
| Negative prediction rate | 12/20 | 9/19 | 15/19 | 23/26 | 25/26 |
| Specificity | 0.600 | 0.474 | 0.789 | 0.885 | 0.962 |
| CI | 0.364-0.800 | 0.252-0.705 | 0.539-0.930 | 0.687-0.970 | 0.784-0.998 |
| Negative predictive value | 0.706 | 0.600 | 0.833 | 0.852 | 0.833 |
| CI | 0.440-0.886 | 0.329-0.825 | 0.577-0.956 | 0.654-0.951 | 0.645-0.937 |
| Absolute error | 0.394 | 0.485 | 0.212 | 0.212 | 0.194 |
| 4ROC error | 0.392 | 0.477 | 0.212 | 0.343 | 0.519 |
| Fisher's exact test | P = 0.139 | P = 0.267 | P = 0.001 | P = 0.082 | P = 0.839 |
1Includes Dukes' C1 and C2; 2 Median follow-up time for recurrent disease patients: 33 months; median follow-up time for disease-free patient: 27 months (analysis excludes patients who died through surgical complications - see Table 1); 3CI = 95% confidence interval; 4ROC = receiver-operator characteristics
The KNN algorithm [29] was used in 'leave-one-out' cross-validation prediction with the number of features (marker peaks) specified. Marker peaks were selected using a t-test statistic except for lymph node involvement and invasiveness characteristics of tumour tissue where the SNR test statistic was used.
Discussion
Although previous studies employing 2D-DIGE analysis of CRC tissues have documented a number of proteins that are either up- or down-regulated in tumour verses normal mucosa [15], the extent to which protein expression profile differences can be detected by direct MALDI-TOF analysis in CRC was not previously known. Analysis of complex protein mixtures by MALDI-TOF MS is inherently limited by the resolution afforded by this type of instrument. Also, only a minor fraction of protein species are efficiently ionisable and therefore detectable. However, our results show that, in common with similar studies in some other solid tumour types [18-20], MALDI-TOF MS readily detects a sizable fraction of protein marker peaks whose expression level is significantly different between tumour and normal mucosa. By using an optimised kNN training model, the classification of tumour and normal tissue was correctly predicted with 100% sensitivity and specificity (95% confidence interval: 0.679-0.992) in an independent test dataset. This performance compares favourably with other studies, for example in head and neck squamous cell carcinoma, in which supervised prediction using SELDI-TOF spectral data correctly classified healthy mucosa and tumour tissue with an accuracy of 94.5% and 92.9% respectively [20].
In further evaluating the potential value of spectra generated from tumour tissue for classifying various clinic-pathological characteristics of disease, we observed low ROC errors with the kNN predictive models for differentiation (0.171) and disease recurrence (0.105). Since histological differentiation stage is a characteristic that is intrinsic to the tumour tissue (and would most closely reflect the actual tumour cell proteome), the ability of the spectra to discriminate well/moderately differentiated from poorly differentiated histologies is perhaps unsurprising. The good performance of the predictive model for disease recurrence is consistent with data from several microarray expression profiling studies that have clearly demonstrated associations between patterns of tumour-associated gene expression and prognosis/treatment response [8,13,14]. However, given that in our study, only six patients had succumbed to recurrent disease at the time of data analysis (median follow-up time for recurrent disease patients: 33 months; median follow-up time for disease-free patient: 27 months), our results should be interpreted with caution. It is also important to emphasise that because of the relatively small number of tumour specimens, rigorous validation of correlations with disease recurrence and histological differentiation stage in an independent 'test' datsaset was not possible in our study.
Several lines of evidence indicate that the normal mucosa from surgically resected CRC tumour specimens display abnormalities in gene and protein expression. These abnormalities have been attributed to precancerous 'field effect' changes in tumour-adjacent mucosa and have been reported to affect protein expression [22], CpG island gene methylation [31] and gene microarray expression profiles [32]. Indeed one study has reported that gene expression profiling of non-neoplastic mucosa may predict clinical outcome of CRC patients [32]. These findings are reminiscent of reports from studies of other solid tumour types, most strikingly in hepatocellular carcimoma in which gene expression patterns of non-neoplastic liver tissue were predictive of patient survival, whereas tumour tissue gene expression signatures were of no prognostic value [33]. It was therefore of interest in our study to determine whether the protein expression profiles of normal mucosa could be used to classify any clinico-patholgical characteristics. Although we found no evidence for predictive value for disease relapse (ROC error, 0.519), the kNN model of normal mucosa spectra for lymph node involvement did give a low ROC error (0.212); the corresponding kNN model for tumour spectra did not show predictive value (0.391). One plausible scenario to explain the predictive value of normal mucosa spectra for lymph node involvement is that paracrine/inflammatory mechanisms, involving proximal affected lymph nodes, may induce changes to the microenvironment of tumour-adjacent mucosa.
As an essential pre-requisite for marker validation, it would be highly desirable in future studies to determine the identities of candidate marker peaks in tumour tissue that discriminate different histological differentiation stages and predict disease recurrence. Our findings also indicate that similar studies using the alternative approach of liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) in CRC are warranted.
Conclusions
In summary, our study has shown that direct protein expression profiling of surgically resected CRC tissue by MALDI-TOF mass spectrometry has potential value in studies aimed at improved molecular classification of this disease. Further studies, with longer follow-up times and larger patient cohorts, that would permit independent validation of predictive models, would be required to confirm the predictive value of tumour spectra for disease recurrence/patient survival.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CCLL collected specimens, processed all samples and collated and analysed data. NW collected specimens and collated data. SM and TA contributed to the study design and in arrangements for specimen collection. JDN contributed to the study design, mass spectrometry and data analysis and wrote the manuscript. All authors have read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Examples of raw MALDI-TOF spectral profiles. Illustrative examples shown for 2012NM and 020T
Summary of marker peaks discriminating tumour from normal mucosa. Compilation of m/z values, ranking and statistics for 73 marker peaks.
Performance of predictive model for discriminating tumour and normal mucosa. Summary of results of optimised k-NN algorithm on an independent test dataset.
Performance of model for predicting poor differentiation based on tumour spectra. Summary of results of 'leave-one-out' cross-validation k-NN algorithm.
Performance of model for predicting disease recurrence based on tumour spectra. Summary of results of 'leave-one-out' cross-validation k-NN algorithm.
Performance of model for predicting lymph node involvement based on mucosa spectra. Summary of results of 'leave-one-out' cross-validation k-NN algorithm.
Contributor Information
Christopher CL Liao, Email: cliao@essex.ac.uk.
Nicholas Ward, Email: Nicholas.Ward@colchesterhospital.nhs.uk.
Simon Marsh, Email: Simon.Marsh@colchesterhospital.nhs.uk.
Tan Arulampalam, Email: Tan.Arulampalam@colchesterhospital.nhs.uk.
John D Norton, Email: jnorton@essex.ac.uk.
Acknowledgements
The authors acknowledge the financial support of the Royal College of Surgeons of England for this work and the contribution of Mr James Wright in preliminary experiments and of Mr Nikhil Pawa for assistance in collating patient follow-up data.
References
- Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- Jemel A, Seigel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics 2007. CA Cancer J Clin. 2007;57:43–46. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Hind R, Rew DR, Johnson CD. Surgical excision alone is adequate for treatment for primary colorectal cancer. Annals of the Royal College of Surgeons of England. 1992;74:63–67. [PMC free article] [PubMed] [Google Scholar]
- Andre T, Boni C, Mounedji-Boudiaf J, Navarro M, Tabernero J, Topham C, Zaninelli M, Clingan P, Bridgewater J, Tabah-Fisch I, Gramont A. Oxaliplatin, fluorouracil and leucovorin as adjuvant treatment for colon cancer. New England Journal of Medicine. 2004;350:2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- Goldberg RM, Rothenberg ML, Can Cutsem E, Benson AB, Blanke CD, Diasio RB, Grothey A, Ramanathan RK, Wickham R, Armstrong D, Viele C. The continuum of care: a paradigm for the management of metastatic colorectal cancer. Oncologist. 2007;12:38–50. doi: 10.1634/theoncologist.12-1-38. [DOI] [PubMed] [Google Scholar]
- Doillard JY, Bennouna J. Adjuvant chemotherapy for colon cancer: a confusing arena! Annals of Oncology. 2005;16:1853–1854. doi: 10.1093/annonc/mdi413. [DOI] [PubMed] [Google Scholar]
- Compton CC, Greene FL. The staging of colorectal cancer: 2004 and beyond. CA Cancer J Clin. 2004;54:295–308. doi: 10.3322/canjclin.54.6.295. [DOI] [PubMed] [Google Scholar]
- Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;7:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- Duffy MJ, Van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Peltomaki P, Sturgeon C, Topolcan O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348–1360. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Shankaran V, Wisinski KB, Mulcahy MF, Benson AB. The role of molecular markers in predicting response to therapy in patients with colorectal cancer. Mol Diag Ther. 2008;12:87–98. doi: 10.1007/BF03256274. [DOI] [PubMed] [Google Scholar]
- Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA. Common mutations linked to adverse colorectal cancer outcomes. Br J Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, Molinari F, De Dosso S, Saletti P, Martini M, Cipani T, Marrapese G, Mazzucchelli L, Lamba S, Veronese S, Frattini M, Bardelli A, Siena S. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLoS One. 2009;4:e7287. doi: 10.1371/journal.pone.0007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G. Gene expression profling in colorectal cancer using microarray technologies: Results and perspectives. Cancer Treatment Reviews. 2009;35:201–209. doi: 10.1016/j.ctrv.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DLW, Au GKH, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijks-Engwegen JYMN, Cats A, Smits ME, Schellens JHM, Neijnen JH. Improving colorectal cancer management: the potential of proteomics. Biomarkers in Medicine. 2008;2:253–289. doi: 10.2217/17520363.2.3.253. [DOI] [PubMed] [Google Scholar]
- Kuramitsu Y, Nakamura K. Proteomic analysis of cancer tissues: shedding light on carcinogenesis and possible biomarkers. Proteomics. 2006;6:5650–5661. doi: 10.1002/pmic.200600218. [DOI] [PubMed] [Google Scholar]
- Palmblad M, Tiss A, Cramer R. Mass spectrometry in clinical proteomics - from the present to the future. Proteomics Clin Appl. 2009;3:6–17. doi: 10.1002/prca.200800090. [DOI] [PubMed] [Google Scholar]
- Au JSK, Cho WCS, Yip T-T, Law SCK. Proteomic approach to biomarker discovery in cancer tissue from lung adenocarcinoma among non-smoking Chinese women in Hong Kong. Cancer Invest. 2008;26:128–135. doi: 10.1080/07357900701788031. [DOI] [PubMed] [Google Scholar]
- Sanders ME, Dias EC, Xu BJ, Mobley JA, Billheimer D, Roder H, Grigorieva J, Dowset M, Arteaga CL, Caprioli RM. Differentiating proteomic biomarkers in breast cancer by laser capture microdissection and MALDI MS. J Proteome Research. 2008;7:1500–1507. doi: 10.1021/pr7008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch-Ely M, Nees M, Karsai S, Ruess A, Bogumil R, Warnken U, Schnolzer M, Dietz A, Plinkert PK, Hofele C, Bosch FX. Proteomic analysis reveals successive aberrations in protein expression from healthy mucosa to invasive head and neck cancer. Oncogene. 2007;26:54–64. doi: 10.1038/sj.onc.1209770. [DOI] [PubMed] [Google Scholar]
- Melle C, Ernst G, Schimmel B, Bleul A, Mothes H, Kaufmann R, Settmacher U, Von Eggeling F. Different expression of calgizzarin (S100A11) in normal colonic epithelium, adenoma and colorectal carcinoma. Int J Oncol. 2006;28:195–200. [PubMed] [Google Scholar]
- Polley ACJ, Mulholland F, Pin C, Williams EA, Bradburn DM, Mills SJ, Mathers JC, Johnson IT. Proteomic analysis reveals field-wide changes in protein expression in the morphologically normal mucosa of patients with colorectal cancer. Cancer Res. 2006;66:6553–6562. doi: 10.1158/0008-5472.CAN-06-0534. [DOI] [PubMed] [Google Scholar]
- Xiang F, Beavis RC. Growing protein-doped sinapic acid crystals for laser desorption: an alternative preparation method for difficult samples. Organic Mass Spectrom. 1993;28:1424–1429. doi: 10.1002/oms.1210281211. [DOI] [Google Scholar]
- Wong JWH, Cagney G, Cartwright HM. SpecAlign - processing and alignment of mass spectra datasets. Bioinformatics. 2005;21:2088–2090. doi: 10.1093/bioinformatics/bti300. [DOI] [PubMed] [Google Scholar]
- Wong JWH, Durante C, Cartwright HM. Application of fast Fourier Transform Cross-Correlation for the alignment of large chromatographic and spectral datasets. Anal Chem. 2005;77:5655–5661. doi: 10.1021/ac050619p. [DOI] [PubMed] [Google Scholar]
- Whistler T, Rollin D, Vernon SD. A method for improving SELDI-TOF mass spectrometry data quality. Proteome Science. 2007;5:14. doi: 10.1186/1477-5956-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–1925. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Calgiuri MA, Bloomfield CD, Lander ES. Molecular classification of Science: class discovery and class prediction by gene expression profiling. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Belshaw NJ, Elliot GO, Foxall RJ, Dainty JR, Pal N, Coupe A, Garg D, Bradburn DM, Mathers JC, Johnson IT. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer. 2008;99:136–142. doi: 10.1038/sj.bjc.6604432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrier A, Boelle P-Y, Lemoine A, Tse C, Brault D, Chiappini F, Lacaine F, Houry S, Huguier M, Flahault A, Dudoit S. Gene expression profiling of nonneoplastic mucosa may predict clinical outcome of colon cancer patients. Dis Colon Rectum. 2005;48:2238–2248. doi: 10.1007/s10350-005-0175-9. [DOI] [PubMed] [Google Scholar]
- Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, Reich M, Chan JA, Glickman JN, Ikeda K, Hashimoto M, Watanabe G, Daidone MG. et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. New Eng J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of raw MALDI-TOF spectral profiles. Illustrative examples shown for 2012NM and 020T
Summary of marker peaks discriminating tumour from normal mucosa. Compilation of m/z values, ranking and statistics for 73 marker peaks.
Performance of predictive model for discriminating tumour and normal mucosa. Summary of results of optimised k-NN algorithm on an independent test dataset.
Performance of model for predicting poor differentiation based on tumour spectra. Summary of results of 'leave-one-out' cross-validation k-NN algorithm.
Performance of model for predicting disease recurrence based on tumour spectra. Summary of results of 'leave-one-out' cross-validation k-NN algorithm.
Performance of model for predicting lymph node involvement based on mucosa spectra. Summary of results of 'leave-one-out' cross-validation k-NN algorithm.