Abstract
We report here the crystal structure of the RuvB motor protein from Thermus thermophilus HB8, which drives branch migration of the Holliday junction during homologous recombination. RuvB has a crescent-like architecture consisting of three consecutive domains, the first two of which are involved in ATP binding and hydrolysis. DNA is likely to interact with a large basic cleft, which encompasses the ATP-binding pocket and domain boundaries, whereas the junction-recognition protein RuvA may bind a flexible β-hairpin protruding from the N-terminal domain. The structures of two subunits, related by a noncrystallographic pseudo-2-fold axis, imply that conformational changes of motor protein coupled with ATP hydrolysis may reflect motility essential for its translocation around double-stranded DNA.
In prokaryotes, three proteins, RuvA, RuvB, and RuvC, process the universal DNA intermediate of homologous recombination, the Holliday junction (1), in a concerted manner through complex formation of these proteins with the Holliday junction (2, 3). The RuvB motor protein supplies energy generated by ATP hydrolysis and thereby promotes branch migration in cooperation with RuvA (4, 5). Sequence analyses have suggested that RuvB belongs to ATPases associated with conserved motifs (Sensors I and II) (AAA+) family (6, 7) rather than the hexameric helicase family, including Escherichia coli DnaB (8), T4 phage gp41 (9), and T7 phage gp4 helicases (10–12), with some of seven conserved motifs. Consistent with this classification, all of the helicase family members encircle single-stranded DNA, whereas RuvB and simian virus 40 large T antigen, classified as the AAA+ family, enclose double-stranded DNA (13, 14). An electron microscopic analysis has indicated that RuvB converts from a heptamer to a hexamer ring on DNA binding (15). Electron micrographs of E. coli RuvAB–Holliday junction complex showed that the two hexamer rings of RuvB lie contacting RuvA on the two opposite sides (16). Biochemical studies indicated that a heterocomplex, E. coli RuvA and Thermus thermophilus (Tth) RuvB, facilitates branch migration (17), suggesting that the RuvA–RuvB functional structure is well conserved in prokaryotes. To obtain detailed insight into its structure–function relationships, we have determined the crystal structure of Tth RuvB. Here, we primarily discuss the main-chain folding of RuvB in comparison with other ATP-binding proteins and putative interfaces of RuvB with RuvA and DNA.
Methods
Crystallization and Data Collection.
Tth RuvB was purified as described previously (17). Crystals of RuvB were obtained at 20°C in a few days by the hanging drop vapor diffusion technique. Crystallization drops containing 20 mM Tris⋅HCl (pH 8.0), 0.35 M NaCl, 10% glycerol, and 1% polyethylene glycol (PEG, Mr 4,000) were equilibrated against a reservoir with the same solution, except with 2.0 M NaCl and 10% PEG. Notably, the diffraction quality of crystals was found to sensitively depend on ratios of 5′-adenyl-imido-triphosphate (AMPPNP) to ADP. Crystals with the highest resolution limit were obtained in a crystallization solution containing 0.5 mM AMPPNP, 10 μM ADP, and 10 mM MgCl2. The crystals belonged to the space group P43212 with cell dimensions of a = b = 84.9 Å, c = 355.2 Å. Two molecules with a pseudo-noncrystallographic dyad axis were present in an asymmetric unit. All diffraction data were collected at BL24XU and BL45XU in SPring-8 (Hyogo, Japan), with R-axis IV as the detector at 100 K with nitrogen stream. All of the data were processed by denzo/scalepack (18).
Structure Determination and Refinement.
The first mercury sites were found in a difference Patterson map from the Hg derivative, which was prepared from cocrystallization of the point mutant protein (S87C) with 0.5 mM ethylmercurithiosalicylic acid. Heavy atom sites of other four derivatives were found in cross-difference Fourier maps. Subsequently, the heavy atom parameters were refined by mlphare (19) and sharp (20). The multiple isomorphous replacement and anomalous scattering phase was improved by density modification techniques with the program dm (19), which contained solvent flattening, histogram matching, and molecular averaging (Table 1). The model was built with quanta98 (Molecular Simulations, Waltham, MA) and refined with cns 0.9 (21) by using a data set that was obtained from a single crystal with the best diffraction quality. Bulk-solvent correction, noncrystallographic symmetry (ncs) restrains, and group B factor refinements were applied. Both of the final models, corresponding to the A and B subunits in an asymmetric unit, consist of 314-aa residues of RuvB except for the disordered regions of N-terminal four and C-terminal six amino acids, and the adenine base moiety of AMPPNP or ADP bound to each subunit. The sugar and phosphate moieties of the nucleotides were eliminated from structure refinement.
Table 1.
Statistics from crystallographic analysis
| Data set: | Native | EMTS (S87C) | Se-Met | K2PtCl4 | HgCl2 (S87C) | EMTS (S157C) |
| Source: | SPring8 BL24 | SPring8 BL24 | SPring8 BL24 | SPring8 BL45 | SPring8 BL45 | Spring8 BL24 |
|---|---|---|---|---|---|---|
| Wavelength, Å | 0.885 | 0.834 | 0.834 | 1.071 | 1.005 | 0.835 |
| Resolution range, Å | 12–3.2 | 12–4.5 | 12–4 | 20–4.2 | 20–3.6 | 15–4.2 |
| Completeness, % | 99.6 | 100 | 97.7 | 99.3 | 99.8 | 97.6 |
| *Rmerge, % | 12.1 | 11.6 | 12.7 | 7.3 | 10.8 | 8.3 |
| †Riso, % | 21.0 | 22.8 | 31.1 | 20.6 | 25.8 | |
| Number of sites | 4 | 9 | 4 | 2 | 1 | |
| ‡Phasing power (acentric/centric) | 1.86/1.49 | 1.56/1.15 | 1.16/0.9 | 0.70/0.73 | 0.86/0.78 | |
| Mean overall figure of merit (acentric/centric) | 0.290/0.396 | |||||
| Reflections (work/free) | 20179/1062 | |||||
| §R/Rfree | 26.3/29.7 | |||||
| Ramachandran, % | ||||||
| Favored | 78.0 | |||||
| Generous | 22.0 | |||||
| Disallowed | 0.0 | |||||
| r.m.s. deviation from ideality | ||||||
| Bond lengths, Å | 0.008 | |||||
| Bond angles, deg | 1.50 | |||||
All observed reflections, except for those for the calculation of Rfree were used for the refinement.
Rmerge = ∑| Io − 〈I〉 |/∑ Io (18).
Riso = ∑ | Fp − 〈Fph〉 |/∑ Fp.
Isomorphous phasing power is defined as 〈Fh〉/rms (ɛ), where 〈Fh〉 is the mean calculated amplitude for the heavy-atom model and rms(ɛ) is the rms lack of closure error for the isomorphous differences (19).
Conventional R factor = ∑ | Fo − Fc |/∑ Fo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. The Rfree value was calculated using a randomly selected 5% of the data set that omitted through all stages of refinement.
Results and Discussion
Overall Structure.
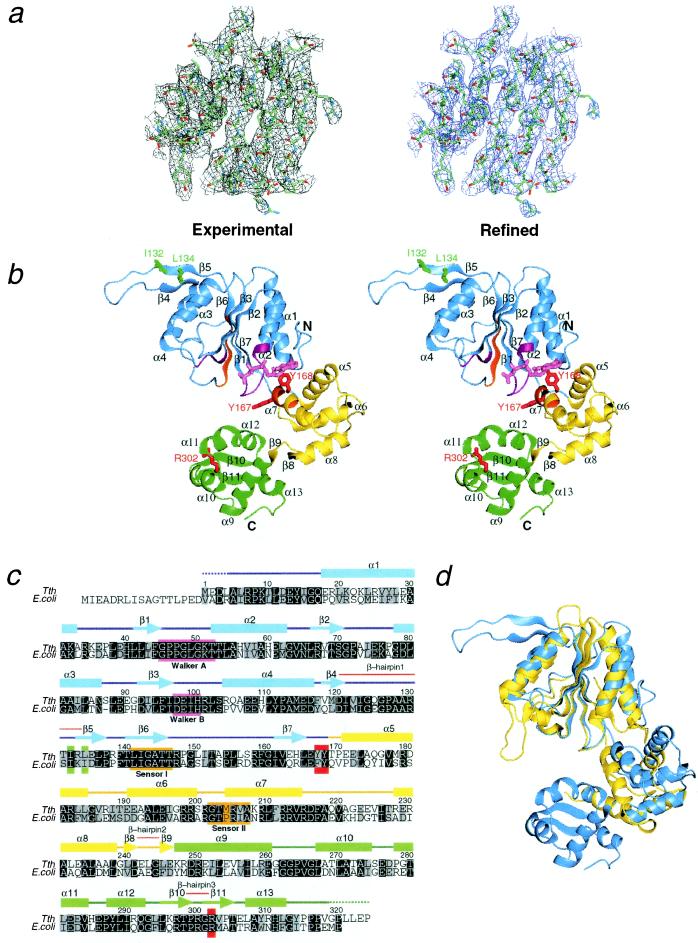
The crystal contains two RuvB subunits related by a noncrystallographic pseudo-2-fold axis in an asymmetric unit. These two molecules, designated as A and B, show similar overall architectures (Fig. 1a), each of which binds nucleotide at the same position. The RuvB monomer, with approximate dimensions of 60 Å × 50 Å × 30 Å, is divided into three consecutive domains (domains N, M, and C), which form a crescent-like configuration (Fig. 1 a and b). Domain N is a triangle-shaped nucleotide-binding domain with the Rossman fold, composed of five parallel β-strands and the surrounding four α-helices. Domain M is composed of four α-helices and the connecting loops, whereas domain C contains five α-helices and one β-hairpin. The spatial configurations among these three domains are topologically similar to those of E. coli DNA polymerase III δ′ subunit (22), another AAA+ family member.
Figure 1.
Structure and topology of RuvB from Tth. (a) Experimental (Left) and final 2Fo − Fc (Right) electron density maps determined for the central β-sheet region of domain N. (b) Stereoview of the overall fold bound to a AMPPNP. The monomer is viewed from the nucleotide-binding side. Domains N, M, and C are colored in blue, yellow, and green, respectively. Walker motifs and sensor motifs are colored in magenta and orange, respectively. Residues involved in RuvB activities are labeled by green (RuvA binding) and red (DNA binding or nucleotide binding). The bound nucleotide is colored pink. (c) Multiple sequence alignments with the same color code as defined above. Amino acid conservation with identical and similar (R/K, D/E, S/T, L/I/V/F/Y/W/M) residues is indicated by black and gray backgrounds, respectively. Secondary structure elements are schematically displayed over the sequences: helices and strands are shown by boxes and arrows, respectively. (d) Ribbon diagram representing superposition of the ATPase domains between RuvB (blue) and NSF (yellow), which have been classified into the AAA+ and AAA families, respectively.
Domain N architecture closely resembles the nucleotide-binding folds of δ′-subunit, NSF-D2 domain (23, 24), and HslU (25), all of which are members of ATPases associated with diverse cellular activities (AAA) or AAA+ family: the central β-sheets yield high scores of equivalence with 0.8 Å of rms deviation of 150 Cα atoms between RuvB and NSF (Fig. 1c). A notable feature is a unique β-hairpin (β4 to β5, designated as β-hairpin 1) that protrudes from the central core β-sheets of domain N to the solvent. In contrast to the highly conserved architecture of domain N, domain M adopts a unique fold, consisting of four α-helices and one β-hairpin (β8 to β9, designated as β-hairpin 2) preceding domain C, although the spatial configurations of domains N and M are topologically similar to those of the AAA+ family members. Domain C consists of five α-helices and one β-hairpin (β10 to β11, designated as β-hairpin 3). According to three-dimensional search in dali-server (26), the spatial arrangement of the four helices (α9 to α12) following β-hairpin 3 agrees well with the putative DNA-binding domain of metallothionein repressor SmtB, whose topology is related to “winged helix” DNA-binding proteins (27).
Nucleotide Binding.
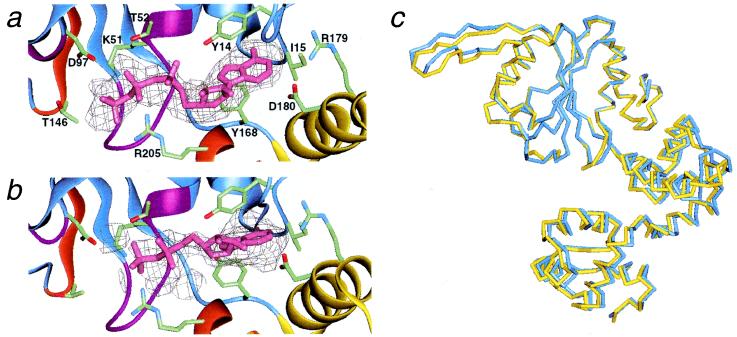
In both of the “A” and “B” molecules, the adenine nucleotides are located near the boundary between domains N and M (Fig. 1a). They are bound to the common structural motifs among Walker-type ATPases, which consist of the carboxyl sides of the five parallel β-strands, the P-loop, and the N terminus of the following α2-helix. The simulated annealed Fo − Fc omit map revealed similar electron densities between the two molecules, except for a region nearest to the P-loop (Fig. 2 a and b). The adenine base is bound to the bottom of a hydrophobic cleft, which consists of Y14, I15, and Y168. The aromatic side chains of conserved Y14 and Y168 interact with the adenine ring. The two polar residues, R179 and D180, on the α5-helix may also be involved in binding to the adenine.
Figure 2.
Electron density maps and ribbon models of nucleotide-binding sites in the two ncs subunits. Possible residues that interact with nucleotides are depicted: Y14, I15, Y168, R179, and D180 are in contact with the adenine bases; K51 and T52 (Walker A), D97 (Walker B), T146 (Sensor I), and R205 (Sensor II) may interact with the phosphate groups. The stick models of (a) AMPPNP and (b) ADP were represented with corresponding simulated annealed Fo − Fc omit maps at a 1.5σ contour. The nucleotide atoms were omitted from the map calculation. Ribbons corresponding to the two sensor motifs and the two Walker motifs are indicated by the same color as in Fig. 1c. (c) Structural differences between the “A” (blue) and “B” (yellow) forms. Here, only the Cα backbones of domain N (ATPase domain) were superimposed between the two ncs molecules.
On the other hand, electron densities corresponding to the ribose and phosphates are not equivalent between the “A” and “B” molecules: density in the “A” molecule appears to fit to a triphosphate (Fig. 2a), whereas that in the “B” molecule lacks a γ-phosphate (Fig. 2b). Thus, provided that this apparent density represents the entire nucleotide conformations, each RuvB structure in the asymmetric unit may separately reflect the ATP- or ADP-bound state. However, we cannot exclude the possibility that structural disorder obscured the density of the γ-phosphate moiety of AMPPNP in the “B” molecule, because the ribose and phosphate moieties were not included in refinement calculations. The Walker A motif (P-loop) is positioned to interact with phosphate group, and the Walker B motif lies at an adequate position to interact with Mg2+. In addition, T146 in Sensor I (6, 7) and R205 in Sensor II (6, 7) are positioned to possibly make polar interactions with the β- or γ-phosphate groups. The environments around the nucleotides are similar to those of other AAA+ family members. The structural features of these motifs are in good agreement with the mutational analysis of E. coli RuvB protein: mutations of these motifs affect ATPase and abolish branch migration activities (ref. 28 and unpublished data for Sensor I, II).
The individual backbones of the three domains superimpose well between the two ncs subunits: rms deviation values of Cα atoms are 0.76 Å for domain N, excluding the β-hairpin 1, which shows the largest conformational change, 0.15 Å for domain M, excluding the flexible loop between α7- and α8-helices and 0.26 Å for domain C. On the other hand, the two subunits exhibit three-degree pivots of the domain M and C against the domain N (Fig. 2b). Their domain movements, coupled with variations in the backbone geometry of the linker (E166 to T169), may be associated with the occupancy of ADP in the “B” molecule. Notably, this linker contains Y168 interacting with the adenine ring, and thus its complete dissociation from the hydrophobic adenine pocket may induce an even more drastic conformational change in the RuvB protein.
Interfaces with DNA and RuvA.
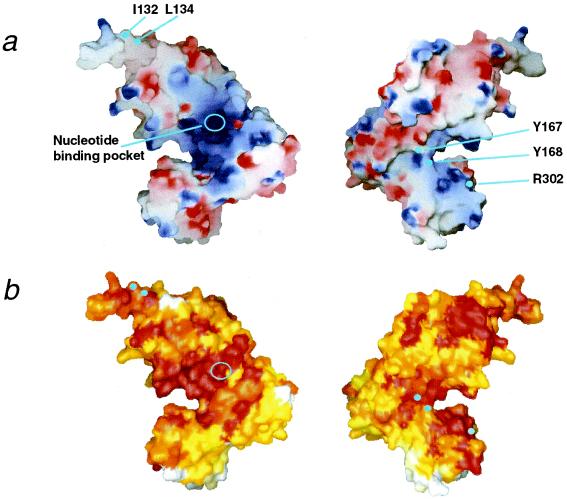
The surface charge distribution of RuvB is substantially different from those found in δ′-subunit, NSF-D2, and HslU. In particular, one side of domain M and the linker region, followed by domain N with the nucleotide, are remarkably positive. This positive area, extending to a part of the domain C surface (Fig. 3a), mostly overlaps with the region conserved among 18 RuvB orthologs (Fig. 3b). Notably, the two conserved Y167 and Y168 residues are located at the edge of this positively charged area. The substitutions of Ala for the corresponding F183 and Y184 in E. coli RuvB result in a total loss of their repair function (7), implying that these two residues may play important roles in driving branch migration. In the Tth RuvB structure, the aromatic ring of Y167 points to the solvent and interacts with R291 in a conserved region of domain C. This region contains the β-hairpin 3, and the mutation that altered the corresponding arginine (R318) in E. coli RuvB abolished stimulation of ATPase activity by DNA. Notably, the crystallographic studies of the Rep and PcrA helicase complexes with DNA highlighted the important role of the aromatic and positive side chains in DNA translocation (29–31). Taken together, these findings imply that the conserved basic area around the aromatic residues in RuvB may constitute a part of the DNA-binding interface.
Figure 3.
Characteristics of the RuvB molecular surface. (a) Electrostatic potential calculated with grasp (33) and (b) amino acid conservation, calculated with clustalw (34) and Toh's modified program (7) and based on 18 RuvB homologues, were mapped onto the grasp molecular surfaces by gradation coloring. Blue and red in a represent positively and negatively charged regions on a scale from −10.5 to +10.5, respectively, and white and red in b indicate low and high values of conservation, respectively. Perspectives are the same as in Fig. 1a on the Left and are from the rear on the Right.
The β-hairpin 1 in domain N is an unusually hydrophobic block pointing to the solvent. E. coli RuvB mutants (I148T and I150T), which changed the hydrophobic residues corresponding to I132 and L134 located on β5 in Tth RuvB structure, abolished physical and functional interactions with RuvA, whereas they retained an intrinsic ATPase activity (unpublished data). Therefore, β-hairpin 1 may constitute the interface with a hydrophobic region of RuvA. Intriguingly, mutational and biochemical analyses of RuvA also implied that a hydrophobic patch of domain III participates in RuvB binding (32). The hydrophobic interaction between β-hairpin 1 and domain III may be crucial for ATP-dependent branch migration.
Possible Subunit Organization.
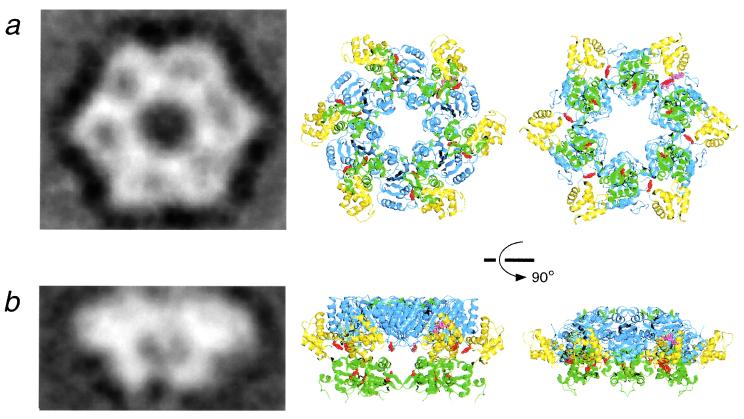
The two tentative models have revealed that the RuvB hexameric ring structures can be constructed without serious steric hindrance by superimposing each domain N onto the ATPase domains of the HslU (25) and NSF hexamers (23), respectively (Fig. 4 Center and Right). Actually, each top view of the models agrees well with an averaged image created from electron micrographs of negatively stained RuvB in complex with duplex DNA (15). The HslU model shows about a 120-Å diameter with a 23-Å hole, and likewise the NSF model exhibits a 130-Å diameter and a 30-Å hole. These hole sizes are sufficient to accommodate a single DNA duplex. In addition, the aromatic and positive residues possibly involved in DNA binding, Y167 and R302, are positioned near the hole. The overall side views are also similar in shape and size, except for the bottom region, which corresponds to domain C. The altered orientation of the entire subunit or domain C caused by DNA binding may account for the discrepancy between the side views. Indeed, this versatility of subunit interfaces in ring structures has been highlighted by a recent crystallographic study, which revealed that ATP- or ADP-dependent rotation between adjacent subunits occurs in the T7 gene 4 helicase (12).
Figure 4.
Comparison of the hypothetical hexamer model of RuvB with the electron microscopic image. (a) Projection image (Left) of negative stained RuvB complexed with a 30-bp DNA, obtained by averaging 140 top views in our previous work (15). The resolution of the averaged image was 30.0 Å. The top views of the hexamer model (Center and Right) were constructed by superimposing each ATPase domain of RuvB (AMPPNP form) (blue region) onto the corresponding regions of HslU crystal structure (25) and the NSF crystal structure (23), respectively. The domains N, M, C, labeled residues, and the bound nucleotides are represented with the same color code as defined in Fig. 1. (b) Projection image (Left) of RuvB–DNA obtained by averaging 266 side views. This image of the single ring was taken from one-half of the double ring, which encircles duplex DNA. The resolution of the averaged image was 34.3 Å. Side view of the hexamer model (Right). [Reproduced with permission from ref. 15 (Copyright 2000, Academic Press).]
Another notable feature of the model is that the six putative RuvA-binding β-hairpins (β-hairpin 1) are located on the top of the hexameric ring (Fig. 4b). This location is consistent with the images of the double hexamer ring, where the interfaces of RuvB with RuvA are oriented toward each other in the absence of RuvA (13, 15). Even in our crystal, interestingly, the same β-hairpin and the adjacent α3-helix form a major interface between the two ncs subunits, and this 135-Å-long ncs pair can be conformed to a unit of the double-ring structure without serious collision, implying that the two RuvB rings are linked through this β-hairpin (15).
The oligomeric states of RuvB vary from monomer to heptamer, sensitively depending on protein and salt concentration, whereas the protomer of E. coli RuvB was reported to be a dimer (35). However, it remains unclear which functional significance resides in different oligomeric states, and how conversion takes place between different RuvB protomers. It appear that the present crystal structure in the absence of DNA hardly exhibits true functional interfaces between RuvB subunits, and hence answers to the above questions would require the atomic resolution structure of the functional hexameric ring complexed with DNA.
Acknowledgments
We gratefully acknowledge M. Ariyoshi, D. Tsuchiya, T. Oyama, M. Shimizu, N. Ito, Y. Katsuya, M. Yamamoto, and T. Kumasaka for expert advice during the course of this project, and H. Toh for analysis of RuvB sequence alignment. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan to H.S.
Abbreviations
- Tth
Thermus thermophilus HB8
- AAA
ATPases associated with diverse cellular activities
- AAA+
AAA with some other conserved motifs (Sensor I, II)
- ncs
noncrystallographic symmetry
- AMPPNP
5′-adenyl-imido-triphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1HQC).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031470598.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031470598
References
- 1.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 2.Shinagawa H, Iwasaki H. Trends Biochem Sci. 1996;21:107–111. [PubMed] [Google Scholar]
- 3.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 4.Tsaneva I R, Müller B, West S C. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 6.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 7.Iwasaki H, Han Y W, Okamoto T, Ohnishi T, Yoshikawa M, Yamada K, Toh H, Daiyasu H, Ogura T, Shinagawa H. Mol Microbiol. 2000;36:528–538. doi: 10.1046/j.1365-2958.2000.01842.x. [DOI] [PubMed] [Google Scholar]
- 8.San Martin M C, Stamford N P, Dammerova N, Dixon N E, Carazo J M. J Struct Biol. 1995;114:167–176. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 9.Dong F, Gogol E P, von Hippel P H. J Biol Chem. 1995;270:7462–7473. doi: 10.1074/jbc.270.13.7462. [DOI] [PubMed] [Google Scholar]
- 10.Egelman H H, Yu X, Wild R, Hingorani M M, Patel S S. Proc Natl Acad Sci USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawaya M R, Guo S, Tabor S, Richardson C C, Ellenberger T. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 12.Singleton M R, Sawaya M R, Ellenberger T, Wigley D B. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 13.Stasiak A, Tsaneva I R, West S C, Benson C J B, Yu X, Egelman E H. Proc Natl Acad Sci USA. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean F B, Borowiec J A, Eki T, Hurwitz J. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 15.Miyata T, Yamada K, Iwasaki H, Shinagawa H, Morikawa K, Mayanagi K. J Struct Biol. 2000;131:83–89. doi: 10.1006/jsbi.2000.4290. [DOI] [PubMed] [Google Scholar]
- 16.Yu X, West S C, Egelman E H. J Mol Biol. 1997;266:217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- 17.Yamada K, Fukuoh A, Iwasaki H, Shinagawa H. Mol Gen Genet. 1999;261:1001–1011. doi: 10.1007/s004380051049. [DOI] [PubMed] [Google Scholar]
- 18.Otwinoski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.de la Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 21.Brünger A T, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Guenther B, Onrust R, Sali A, O'Donnell M, Kuriyan J. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 23.Lenzen C U, Steinmann D, Whiteheart S W, Weis W I. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 24.Yu R C, Hanson P I, Jahn R, Brünger A T. Nat Struct Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 25.Bochtler M, Hartmann C, Song H K, Bourenkov G P, Bartunik H D, Huber R. Nature (London) 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 27.William J C, Sambit R K, Kenneth B T, Leo M H. J Mol Biol. 1998;275:337–346. [Google Scholar]
- 28.Hishida T, Iwasaki H, Yagi T, Shinagawa H. J Biol Chem. 1999;274:25335–25342. doi: 10.1074/jbc.274.36.25335. [DOI] [PubMed] [Google Scholar]
- 29.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 30.Velankar S S, Soultanas P, Dillingham M S, Subramanya H S, Wigley D B. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 31.Dillingham M S, Soultanas P, Wigley D B. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishino T, Iwasaki H, Kataoka M, Ariyoshi M, Fujita T, Shinagawa H, Morikawa K. J Mol Biol. 2000;298:407–416. doi: 10.1006/jmbi.2000.3675. [DOI] [PubMed] [Google Scholar]
- 33.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 34.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiba T, Iwasaki H, Nakata A, Shinagawa H. Mol Gen Genet. 1993;237:395–399. doi: 10.1007/BF00279443. [DOI] [PubMed] [Google Scholar]