Abstract
Background
Polymorphisms of the peptidylarginine deiminase type 4 (PADI4) gene confer susceptibility to rheumatoid arthritis (RA) in East Asian people. However, studies in European populations have produced conflicting results. This study explored the association of the PADI4 genotype with RA in a large UK Caucasian population.
Methods
The PADI4_94 (rs2240340) single nucleotide polymorphism (SNP) was directly genotyped in a cohort of unrelated UK Caucasian patients with RA (n=3732) and population controls (n=3039). Imputed data from the Wellcome Trust Case Control Consortium (WTCCC) was used to investigate the association of PADI4_94 with RA in an independent group of RA cases (n=1859) and controls (n=10 599). A further 56 SNPs spanning the PADI4 gene were investigated for association with RA using data from the WTCCC study.
Results
The PADI4_94 genotype was not associated with RA in either the present cohort or the WTCCC cohort. Combined analysis of all the cases of RA (n=5591) and controls (n=13 638) gave an overall OR of 1.01 (95% CI 0.96 to 1.05, p=0.72). No association with anti-CCP antibodies and no interaction with either shared epitope or PTPN22 was detected. No evidence for association with RA was identified for any of the PADI4 SNPs investigated. Meta-analysis of previously published studies and our data confirmed no significant association between the PADI4_94 genotype and RA in people of European descent (OR 1.06, 95% CI 0.99 to 1.13, p=0.12).
Conclusion
In the largest study performed to date, the PADI4 genotype was not a significant risk factor for RA in people of European ancestry, in contrast to Asian populations.
Introduction
Rheumatoid arthritis (RA) is a complex autoimmune disease in which genetic and environmental factors contribute to the pathogenesis. Around 60% of the risk of RA is genetic, a third of which is accounted for by HLA-DRB1.1 Numerous polymorphisms outside the HLA region have recently been confirmed as RA susceptibility loci in Caucasians, but fewer have been tested across different ethnic populations and the question of whether racial heterogeneity exists—that is, whether possession of a particular allele may confer disease susceptibility in one ethnic group but not another—is contentious.2–4 The peptidylarginine deiminase 4 (PADI4) gene is of particular interest as it has been tested in Asian, European and North American populations and its relative effect in relation to RA susceptibility across these groups remains controversial.5–13
The PADI4 gene encodes the type 4 peptidylarginine deiminase enzyme which catalyses the post-translational modification of arginine to citrulline, generating citrullinated proteins. Antibodies to these peptides are highly specific for RA and often predate the development of disease, suggesting a critical role in the pathogenesis of RA. PADI4 therefore represents an attractive RA candidate gene and was first reported to be associated with RA in a Japanese population in 2003.5 This association has been consistently replicated in East Asian populations6 11 12; however findings in cohorts of European ancestry have been inconsistent. Studies in Spanish, Swedish and UK populations reported no evidence for association of PADI4 with RA.7 9 10 Conversely, PADI4 was found to be associated with RA in North American and German populations and two published meta-analyses suggested that PADI4 polymorphisms do confer susceptibility to RA in those of European descent, albeit to a lesser degree than in Asian subjects.10 13–15 Consequently, it was hypothesised that these European studies were underpowered to detect a true but modest genetic effect. The present study was designed to address this issue by exploring the association between the PADI4 genotype and RA in a large UK population.
Materials and methods
Study design
The PADI4_94 single nucleotide polymorphism (SNP) (rs2240340) was selected for investigation as it has the strongest evidence for association with RA in Asians and Caucasians.5 12 14 15 It was genotyped in an independent UK Caucasian population of 3732 patients with RA and 3039 controls (see online supplement). In addition, imputed genotypes for the PADI4_94 SNP were compared between 1859 patients with RA and 2935 controls from the Wellcome Trust Case Control Consortium (WTCCC) study.16 Where linkage disequilibrium (LD) is high and confidence scores for imputed genotypes exceed 95%, the accuracy of imputation in predicting actual genotype counts exceeds 98.4%.17 An expanded reference group of 10 599 subjects was created by using imputed genotype data for PADI4_94 from the four non-autoimmune disease case subjects (hypertension, coronary artery disease, type 2 diabetes and bipolar disorder) genotyped as part of the WTCCC study and combining this with the genotype data from the healthy controls. The data from the present cohort and the WTCCC study were combined to provide a robust estimate of effect size for this SNP in the UK population, giving a combined sample size of 5591 cases of RA and 13 638 controls. In addition, imputed genotype data for the original WTCCC cohort (1860 cases of RA, 2938 controls) were used to investigate other SNPs spanning the PADI4 gene for evidence of association with RA.
Analysis of data
Allele and genotype frequencies were compared between patients with RA and controls using the χ2 test for trend implemented in PLINK. The threshold for significance was defined at p<0.05.
Meta-analysis
Meta-analysis of the results together with previous studies investigating association of PADI4_94 with RA in populations of European ancestry was performed (see online supplement). A random effects model was used and between-study heterogeneity assessed using the Cochran Q-statistic (p<0.1 considered significant).
Interaction analysis
Data were available for shared epitope (SE) and the PTPN22 R620W SNP (rs2476601) in the current cohort. The risk of RA associated with carriage of PADI4_94, PTPN22 and SE risk alleles (1 or 2 copies) alone and in combination was calculated using logistic regression. Interaction effects were quantified by calculating the attributable proportion,18 which assesses the proportion of the incidence that is due to interaction (ie, beyond the additive effects of each independent variant). There is evidence of biological interaction if the attributable proportion is not equal to 0.
Power
The present cohort had >80% power to detect the published OR of 1.13 at the 5% significance level (α=0.05) with a risk allele frequency of 0.42.15
Results
The genotyping success rate was >97% in both cases and controls. In this independent cohort of 3732 cases of RA and 3039 controls, no significant difference in PADI_94 allele or genotype frequencies was detected (table 1).
Table 1.
PADI4_94 (rs2240340) genotypes in current and WTCCC cohorts
| RA cases* | Controls* | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | MAF | 11 | 12 | 22 | MAF | Allele 2 vs allele 1 OR (95% CI) | p Trend | 12 vs 11 OR (95% CI) | 22 vs 11 OR (95% CI) | |
| Current cohort | 1238 (33.2) | 1841 (49.3) | 653 (17.5) | 0.42 | 1018 (33.5) | 1508 (49.6) | 513 (16.9) | 0.42 | 1.02 (0.95 to 1.09) | 0.58 | 1.00 (0.90 to 1.12) | 1.05 (0.91 to 1.20) |
| WTCCC | 665 (35.8) | 860 (48.3) | 334 (18.0) | 0.41 | 1031 (35.1) | 1434 (48.9) | 470 (16.0) | 0.40 | 1.03 (0.95 to 1.12) | 0.54 | 0.93 (0.82 to 1.06) | 1.10 (0.93 to 1.30) |
| WTCCC plus non-autoimmune controls | 665 (35.8) | 860 (48.3) | 334 (18.0) | 0.41 | 3614 (34.1) | 5154 (48.6) | 1831 (17.3) | 0.42 | 0.98 (0.91 to 1.05) | 0.59 | 0.91 (0.81 to 1.01) | 0.99 (0.86 to 1.14) |
| Combined analysis | 1903 (34.0) | 2701 (48.3) | 987 (17.7) | 0.42 | 4632 (34.0) | 6662 (48.8) | 2344 (17.2) | 0.42 | 1.01 (0.96 to 1.05) | 0.72 | 0.99 (0.92 to 1.06) | 1.02 (0.94 to 1.12) |
Data shown as number (%).
1, major (common) allele; 2, minor (rare) allele; MAF, minor allele frequency; WTCCC, Wellcome Trust Case Control Consortium.
WTCCC cohort
Imputed minor allele frequencies for PADI_94 in the WTCCC cohort were similar to those previously reported in European populations and to those observed in the current study. No association between PADI4_94 genotype and RA was observed (table 1).
Combined analysis
Combined analysis of the current and WTCCC cohorts showed no evidence for association between PADI4_94 genotype and RA. No significant heterogeneity between these two cohorts was detected (phet=0.43, I2=0%) and meta-analysis under a random effects model yielded a similar overall OR of 1.00 (95% CI 0.95 to 1.05). There was no significant difference in genotype distribution between the WTCCC patients with non-autoimmune disease, the original WTCCC controls and the current controls (p=0.20, χ2=6.0). Excluding the WTCCC patients with non-autoimmune disease from the analysis did not affect the outcome (OR 1.02 (95% CI 0.97 to 1.08)).
Studies in Asian populations have shown that PADI4_94 exerts an allele dose-dependent effect in RA susceptibility, with the greatest effect being seen when comparing minor allele homozygotes (2/2) with major allele homozygotes (1/1).5 12 In contrast, we found no evidence for association in any of the genotypic models tested (table 1).
Stratification
Stratification analysis creates more homogenous subsets of patients and thus may increase the power to detect association despite loss of sample size. Phenotype data were available for a proportion of patients within the current cohort. Stratification by autoantibody status, gender and SE revealed no evidence for association in any of the subgroups tested (table 2).
Table 2.
PADI4_94 genotype in current cohort stratified by autoantibody status, carriage of SE, presence of erosions and gender
| Cases* | Controls* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | Total | 11 | 12 | 22 | Total | Allele 2 vs 1 OR (95% CI) | p Trend | |
| CCP +ve vs all controls | 498 (33.2) | 722 (48.2) | 279 (18.6) | 1499 | 1018 (33.5) | 1508 (49.6) | 513 (16.9) | 3039 | 1.04 (0.95 to 1.14) | 0.36 |
| CCP −ve vs all controls | 241 (34.4) | 347 (49.5) | 113 (16.1) | 701 | 1018 (33.5) | 1508 (49.6) | 513 (16.9) | 3039 | 0.97 (0.86 to 1.09) | 0.57 |
| CCP +ve vs CCP −ve | 498 (33.2) | 722 (48.2) | 279 (18.6) | 1499 | 241 (34.4) | 347 (49.5) | 113 (16.1) | 701 | 1.08 (0.95 to 1.23) | 0.25 |
| RF +ve vs all controls | 849 (33.4) | 1250 (49.1) | 444 (17.5) | 2543 | 1018 (33.5) | 1508 (49.6) | 513 (16.9) | 3039 | 1.01 (0.94 to 1.09) | 0.71 |
| RF −ve vs all controls | 328 (34.2) | 467 (48.7) | 165 (17.2) | 960 | 1018 (33.5) | 1508 (49.6) | 513 (16.9) | 3039 | 0.99 (0.89 to 1.10) | 0.89 |
| Erosions | 394 (33.8) | 576 (49.4) | 195 (16.8) | 1165 | 1018 (33.5) | 1508 (49.6) | 513 (16.9) | 3039 | 0.99 (0.9 to 1.09) | 0.85 |
| No erosions | 183 (33.0) | 270 (48.8) | 101 (18.2) | 554 | 1018 (33.5) | 1508 (49.6) | 513 (16.9) | 3039 | 1.04 (0.91 to 1.18) | 0.57 |
| SE +ve (1 or 2 copies) | 747 (33.3) | 1093 (48.6) | 407 (18.1) | 2247 | 214 (36.2) | 276 (46.6) | 102 (17.2) | 592 | 1.08 (0.95 to 1.23) | 0.24 |
| SE −ve (0 copies) | 244 (32.7) | 373 (49.9) | 130 (17.4) | 747 | 231 (32.6) | 359 (50.6) | 119 (16.8) | 709 | 1.01 (0.87 to 1.17) | 0.88 |
| Men | 334 (32.3) | 524 (50.7) | 176 (17.0) | 1034 | 304 (32.6) | 471 (50.5) | 158 (16.9) | 933 | 1.01 (0.89 to 1.15) | 0.91 |
| Women | 896 (33.6) | 1294 (48.6) | 474 (17.8) | 2664 | 657 (33.9) | 964 (49.8) | 315 (16.3) | 1936 | 1.04 (0.95 to 1.13) | 0.38 |
Data shown as number (%).
1, major (common) allele; 2, minor (rare) allele; RF, rheumatoid factor; SE, shared epitope.
Meta-analysis
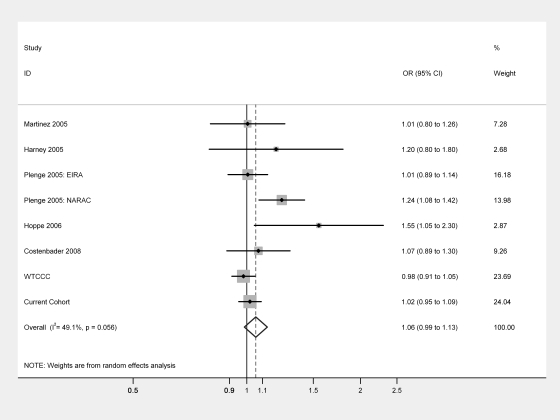
Five eligible studies investigating the PADI4_94 SNP for association with RA in Caucasian populations were identified.8–10 13 19 Eight separate comparisons were available for the allelic model (minor allele 2 vs common allele 1) and seven for the genotypic model (1/2 vs 1/1 and 2/2 vs 1/1). No significant association between PADI4_94 and RA was detected for any of the models tested (figure 1). The pooled OR for allele 2 vs allele 1 was 1.06 (95% CI 0.99 to 1.13, p=0.12). The OR for 1/2 vs 1/1 was 1.04 (95% CI 0.92 to 1.17, p=0.53) and for 2/2 vs 1/1 was 1.07 (95% CI0.96 to1.19, p=0.20). Significant between-study heterogeneity was noted (phet=0.06, I2=49.1%). Analysis restricted to European cohorts (ie, excluding North American cohorts whose genetic background may be more ethnically diverse) gave an OR of 1.01 (95% CI 0.96 to 1.07) with no significant heterogeneity (phet=0.31, I2=16.8%).
Figure 1.
Meta-analysis of PADI4_94 in populations of European descent. Odds ratio (OR), minor allele (2) vs common allele (1). Weight expressed as percentage.
Interaction analysis
Significant interaction between SE and PTPN22 was detected in anti-CCP positive RA, consistent with previous reports (tables 3 and 4).20 In contrast, there was no significant interaction between PADI4_94 and either PTPN22 or SE. In a model containing all three factors, the highest ORs were seen with possession of SE alleles in conjunction with the PTPN22 risk allele, regardless of the presence or absence of the PADI4_94 putative risk allele (data not shown).
Table 3.
Details of studies included in meta-analysis and subsequent analysis of SE, PTN22 and PADI4_94 risk allele combinations
| Sample size | PADI4_94 allele 2 vs 1 | ||||
|---|---|---|---|---|---|
| Study | Population | RA cases | Controls | OR (95% CI) | p Value |
| Martinez (2005)9 | Spain | 248 | 394 | 1.01 (0.80 to 1.26) | 1.00 |
| Harney (2005)8 | UK | 100 | 94 | 1.20 (0.80 to 1.80) | 0.41 |
| Plenge (2005): EIRA10 | Sweden | 1498 | 858 | 1.01 (0.89 to 1.14) | 0.93 |
| Plenge (2005): NARAC10 | North America | 895 | 748 | 1.24 (1.06 to 1.42) | 0.003 |
| Hoppe (2006)13 | German | 102 | 102 | 1.55 (1.05 to 2.30) | 0.04 |
| Costenbader (2008)19 | North America | 430 | 426 | 1.07 (0.89 to 1.30) | 0.49 |
| WTCCC | UK | 1859 | 10599 | 0.98 (0.91 to 1.05) | 0.58 |
| Current cohort | UK | 3732 | 3039 | 1.02 (0.95 to 1.09) | 0.72 |
EIRA, Epidemiological Investigation of Rheumatoid Arthritis; NARAC, North American Rheumatoid Arthritis Consortium; SE, shared epitope; WTCCC, Wellcome Trust Case Control Consortium.
Table 4.
Odds ratios for developing rheumatoid arthritis (RA) according to presence or absence of SE, PTPN22 R620W and PADI4_94 risk alleles: allele 2 (minor allele) vs allele 1 (common allele)
| Presence of risk allele | All cases | CCP positive cases | CCP negative cases | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PADI4_94 | PTPN22 | Case:control | OR (95% CI) | AP (CI) | Case:control | OR (95% CI) | AP (CI) | Case:control | OR (95% CI) | AP (CI) |
| None | None | 876:810 | Referent | 0.0 (–0.4 to 0.4) | 340:810 | Referent | 0.0 (–0.3 to 0.3) | 182:810 | Referent | 0.0 (–0.4 to 0.4) |
| Present | None | 1771:1618 | 1.0 (0.9 to 1.1) | 680:1618 | 1.0 (0.9 to 1.2) | 349:1618 | 1.0 (0.8 to 1.2) | |||
| None | Present | 334:191 | 1.6 (1.3 to 2.0) | 146:191 | 1.8 (1.4 to 2.3) | 51:191 | 1.2 (0.8 to 1.7) | |||
| Present | Present | 679:386 | 1.6 (1.4 to 1.9) | 300:386 | 1.9 (1.5 to 2.3) | 102:386 | 1.2 (0.9 to 1.5) | |||
| PADI4_94 | SE | |||||||||
| None | None | 244:231 | Referent | 0.1 (–0.1 to 0.3) | 62:231 | Referent | 0.1 (–0.1 to 0.3) | 94:231 | Referent | 0.2 (–0.1 to 0.5) |
| Present | None | 503:478 | 1.0 (0.8 to 1.2) | 154:478 | 1.2 (0.9 to 1.7) | 162:478 | 0.8 (0.6 to 1.1) | |||
| None | Present | 747:214 | 3.3 (2.6 to 4.2) | 382:214 | 6.7 (4.8 to 9.2) | 118:214 | 1.4 (1.0 to 1.9) | |||
| Present | Present | 1500:378 | 3.8 (3.0 to 4.6) | 747:378 | 7.4 (5.4 to 10.0) | 240:378 | 1.6 (1.2 to 2.1) | |||
| PTPN22 | SE | |||||||||
| None | None | 553:563 | Referent | 0.3 (0.1 to 0.5) | 145:563 | Referent | 0.4 (0.1 to 0.6) | 204:563 | Referent | 0.2 (–0.1 to 0.5) |
| Present | None | 200:141 | 1.4 (1.1 to 1.8) | 71:141 | 2.0 (0.7 to 5.7) | 52:141 | 1.0 (0.7 to 1.5) | |||
| None | Present | 1610:474 | 3.5 (3.0 to 4.0) | 779:474 | 6.4 (5.1 to 7.9) | 270:474 | 1.6 (1.3 to 2.0) | |||
| Present | Present | 626:116 | 5.5 (4.4 to 6.9) | 347:116 | 11.6 (8.8 to 15.3) | 87:116 | 2.1 (1.5 to 2.9) | |||
Present, 1 or 2 copies of SE, PTPN22 R620W risk allele or PADI4_94 putative risk allele. Significant results in bold.
None, no copies.
AP, attributable proportion; CCP, cyclic citrullinated peptide; SE, shared epitope.
Further PADI4 SNPs
Imputed genotype data were available for 1860 cases of RA and 2938 controls from the WTCCC study. A further 56 SNPs spanning the PADI4 gene with imputation confidence scores of over 99% were identified. No evidence for association with RA was detected for any of these SNPs (see table 1 in online supplement). Importantly, PADI4_89 and PADI4_90, which have previously been reported to be associated with RA in Caucasians, were not associated in this UK cohort (see table 2 in online supplement).13 21
Discussion
In the largest study performed to date, we found no evidence for association between the PADI4_94 SNP and RA in a combined sample of over 19 000 UK subjects. The results were consistent across two large independent populations, lending weight to these findings.
This contrasts with the convincing evidence that PADI4 is an RA susceptibility gene in East Asian subjects.5 6 11 12 The strongest association is seen with PADI4_94, with an estimated OR of 1.31, making it the major genetic risk factor outside the HLA region in this group.12 14 15 Consistent with our findings, several other groups have failed to find an association between PADI4_94 and RA in populations of European ancestry. However, meta-analyses of published data in Caucasians demonstrated evidence for association with a summary OR of 1.13.14 15 For a study to have 80% power to detect an OR of 1.13 at p<0.05, more than 4200 subjects would be required. All previous studies have been underpowered to detect an OR of this level, conceivably accounting for the frequent failure to replicate the association. However, these meta-analyses were based on comparatively limited data (pooled total of 2950 RA cases and 2300 controls) and findings may have been influenced by publication bias and heterogeneity between studies. The present study circumvents these problems by using two independent cohorts, each with more than 80% power to detect the published OR, thus minimising the chance of a type II error. Furthermore, there was no significant difference when comparing minor allele homozygotes with major allele homozygotes (OR 1.02), which is where the greatest effect is seen in Asian populations (OR 1.73).12 Our findings suggest that previous reported associations between PADI4_94 and RA in European populations are false-positive results.
The differential effect of the PADI4_94 SNP in populations of Asian and European descent is unusual as, although the frequency of complex disease-associated polymorphisms may vary across ethnic groups, their genetic effects are usually consistent.2 There are several plausible explanations for this discrepancy. First, linkage disequilibrium varies between races, so PADI4_94 may be in linkage disequilibrium with the true disease-associated allele in Asian but not Caucasian populations. However, PADI4 SNP and haplotype frequencies are similar in the two populations.7 Second, it is possible that different PADI4 polymorphisms may be associated with RA in Caucasians. However, this would be unlikely given that we investigated numerous SNPs spanning the PADI4 gene within the WTCCC cohort and found no evidence for association across the locus as a whole. Third, the biological effect of PADI4_94 variants may be modulated by environmental exposures such as smoking or interactions with other genes which may differ between races. The PTPN22 R620W variant is a potential candidate for introducing variation via gene–gene interaction as it is a major risk factor for RA in Caucasians but has not been detected in Japanese populations. However, we found no interaction between PADI4_94 and PTPN22 in this study. Alternatively, it may be that PADI4 is associated only with a subgroup of patients with RA. Hoppe et al recently suggested that the association between PADI4 genotype and RA may be restricted to patients with more severe disease.22 It is unlikely, however, that this would account for the differences observed as the cohorts tested in the WTCCC and current studies were comparable with the Japanese cohort in terms of disease characteristics. Moreover, PADI4 genotype was not associated with the presence of cyclic citrullinated peptide (CCP) antibody, rheumatoid factor or erosions in this study, and we have previously found no relationship between PADI4 genotype and disease severity in UK patients with early inflammatory arthritis followed prospectively.23
Finally, the disparity in genetic effect may reflect genuine differences in pathogenic processes between European and Asian populations. The mechanism by which the PADI4 genotype may influence RA susceptibility has not yet been elucidated. Suzuki et al showed that the PADI4 susceptibility haplotype had significantly increased mRNA stability compared with the non-susceptibility haplotype. In theory this could result in increased PAD4 enzyme, with consequently increased protein citrullination which may break immune tolerance leading to production of anti-citrullinated peptide antibodies (ACPA) and disease.5 They went on to demonstrate an association between homozygosity for the PADI4 susceptibility haplotype and the presence of ACPA. However, we and many others have failed to find any link between PADI4 genotype and presence of anti-CCP antibodies, drawing this hypothesis into question.8 10 23–25 Furthermore, given that individuals carrying SE alleles may be predisposed to mount an immune response to citrullinated peptides, the absence of a significant interaction between PADI4_94 and SE in this study is of relevance.26 We did confirm the previously reported interaction between PTPN22 and SE, supporting the validity of our data.20 In a Korean cohort, PADI4 and SE had additive effects with regard to the risk of RA, although no significant interaction was reported.11 It would be interesting to investigate this further in Asian populations as distribution of HLA subtypes varies across races and it has been suggested that this could lead to differential modulation of the genetic effects of PADI4.8 11
In conclusion, in contrast to Asian populations, the PADI4 genotype is not a significant risk factor for RA in people of European descent. Identification of biological mechanisms responsible for this disparity may yield novel insights into the pathogenic processes underpinning rheumatoid disease.
Acknowledgments
The authors thank the Arthritis Research Campaign for their support (arc grant reference number 17552), the Wellcome Trust Case Control Consortium (WTCCC) for providing the genotype data included and acknowledge support from the Manchester Biomedical Research Centre.
Footnotes
Competing interests: None.
Ethics approval: This study was conducted with the approval of the North West Research ethics committee (MREC 99/9/84).
Provenance and peer review: Not commissioned; externally peer reviewed.
Membership of the BIRAC Consortium and YEAR Consortium is shown in the online supplement.
References
- 1.MacGregor AJ, Snieder H, Rigby AS, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum 2000;43:30–7 [DOI] [PubMed] [Google Scholar]
- 2.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet 2004;36:1312–18 [DOI] [PubMed] [Google Scholar]
- 3.Barton A, Thomson W, Ke X, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet 2008;40:1156–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raychaudhuri S, Remmers EF, Lee AT, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet 2008;40:1216–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki A, Yamada R, Chang X, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet 2003;34:395–402 [DOI] [PubMed] [Google Scholar]
- 6.Ikari K, Kuwahara M, Nakamura T, et al. Association between PADI4 and rheumatoid arthritis: a replication study. Arthritis Rheum 2005;52:3054–7 [DOI] [PubMed] [Google Scholar]
- 7.Barton A, Bowes J, Eyre S, et al. A functional haplotype of the PADI4 gene associated with rheumatoid arthritis in a Japanese population is not associated in a United Kingdom population. Arthritis Rheum 2004;50:1117–21 [DOI] [PubMed] [Google Scholar]
- 8.Harney SM, Meisel C, Sims AM, et al. Genetic and genomic studies of PADI4 in rheumatoid arthritis. Rheumatology (Oxford) 2005;44:869–72 [DOI] [PubMed] [Google Scholar]
- 9.Martinez A, Valdivia A, Pascual-Salcedo D, et al. PADI4 polymorphisms are not associated with rheumatoid arthritis in the Spanish population. Rheumatology (Oxford) 2005;44:1263–6 [DOI] [PubMed] [Google Scholar]
- 10.Plenge RM, Padyukov L, Remmers EF, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet 2005;77:1044–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang CP, Lee HS, Ju H, et al. A functional haplotype of the PADI4 gene associated with increased rheumatoid arthritis susceptibility in Koreans. Arthritis Rheum 2006;54:90–6 [DOI] [PubMed] [Google Scholar]
- 12.Takata Y, Inoue H, Sato A, et al. Replication of reported genetic associations of PADI4, FCRL3, SLC22A4 and RUNX1 genes with rheumatoid arthritis: results of an independent Japanese population and evidence from meta-analysis of East Asian studies. J Hum Genet 2008;53:163–73 [DOI] [PubMed] [Google Scholar]
- 13.Hoppe B, Häupl T, Gruber R, et al. Detailed analysis of the variability of peptidylarginine deiminase type 4 in German patients with rheumatoid arthritis: a case-control study. Arthritis Res Ther 2006;8:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwamoto T, Ikari K, Nakamura T, et al. Association between PADI4 and rheumatoid arthritis: a meta-analysis. Rheumatology (Oxford) 2006;45:804–7 [DOI] [PubMed] [Google Scholar]
- 15.Lee YH, Rho YH, Choi SJ, et al. PADI4 polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Rheumatol Int 2007;27:827–33 [DOI] [PubMed] [Google Scholar]
- 16.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13 [DOI] [PubMed] [Google Scholar]
- 18.Andersson T, Alfredsson L, Källberg H, et al. Calculating measures of biological interaction. Eur J Epidemiol 2005;20:575–9 [DOI] [PubMed] [Google Scholar]
- 19.Costenbader KH, Chang SC, De Vivo I, et al. Genetic polymorphisms in PTPN22, PADI-4, and CTLA-4 and risk for rheumatoid arthritis in two longitudinal cohort studies: evidence of gene-environment interactions with heavy cigarette smoking. Arthritis Res Ther 2008;10:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallberg H, Padyukov L, Plenge RM, et al. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet 2007;80:867–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandjbakhch F, Fajardy I, Ferre B, et al. A functional haplotype of PADI4 gene in rheumatoid arthritis: positive correlation in a French population. J Rheumatol 2009;36:881–6 [DOI] [PubMed] [Google Scholar]
- 22.Hoppe B, Häupl T, Egerer K, et al. Influence of peptidylarginine deiminase type 4 genotype and shared epitope on clinical characteristics and autoantibody profile of rheumatoid arthritis. Ann Rheum Dis 2009;68:898–903 [DOI] [PubMed] [Google Scholar]
- 23.Barton A, Bowes J, Eyre S, et al. Investigation of polymorphisms in the PADI4 gene in determining severity of inflammatory polyarthritis. Ann Rheum Dis 2005;64:1311–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poór G, Nagy ZB, Schmidt Z, et al. Genetic background of anticyclic citrullinated peptide autoantibody production in Hungarian patients with rheumatoid arthritis. Ann N Y Acad Sci 2007;1110:23–32 [DOI] [PubMed] [Google Scholar]
- 25.Harris ML, Darrah E, Lam GK, et al. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum 2008;58:1958–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill JA, Southwood S, Sette A, et al. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol 2003;171:538–41 [DOI] [PubMed] [Google Scholar]