Abstract
The Polycomb group of proteins (PcG) is important for transcriptional repression and silencing in all higher eukaryotes. In Drosophila, PcG proteins are recruited to the DNA by Polycomb-group response elements (PREs), regulatory sequences whose activity depends on the binding of many different sequence-specific DNA-binding proteins. We previously showed that a binding site for the Sp1/KLF family of zinc-finger proteins is required for PRE activity. Here, we report that the Sp1/KLF family member Spps binds specifically to Ubx and engrailed PREs, and that Spps binds to polytene chromosomes in a pattern virtually identical to that of the PcG protein, Psc. A deletion of the Spps gene causes lethality late in development and a loss in pairing-sensitive silencing, an activity associated with PREs. Finally, the Spps mutation enhances the phenotype of pho mutants. We suggest that Spps may work with, or in parallel to, Pho to recruit PcG protein complexes to PREs.
Keywords: Sp1/Klf family members, Gene expression, Polycomb group genes
INTRODUCTION
Polycomb group genes (PcG) are an evolutionarily conserved group of proteins that were originally identified as repressors of homeotic genes in Drosophila. PcG gene products act as multi-protein complexes associating with chromatin through DNA elements called Polycomb group response elements (PREs), to repressively modify histones (for reviews, see Schwartz and Pirrotta, 2008; Müller and Verrijzer, 2009). Initially, two major PcG complexes were biochemically isolated, Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). PRC1 core is made up of the PcG proteins Polycomb (Pc), Polyhomeotic (Ph), Posterior sex combs (Psc) and dRing/Sex combs extra (Sce) (Shao et al., 1999; Francis et al., 2001). The Pc subunit has a chromodomain that binds to H3K27me3 and dRing has H2A mono-ubiquitin ligase activity (Lagarou et al., 2008). PRC2 comprises Enhancer of Zeste [E(z)], extra sex combs (Esc) or Esc-like, Suppressor of zeste 12 [Su(z)12] and Nurf 55 (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Müller et al., 2002), and mediates histone H3K27 tri-methylation via the methyl-transferase activity of E(z). More recently, additional PcG-protein complexes have been isolated. These include dRAF and Pcl-PRC2. dRaf contains PSC and dRing in a complex with dKDM2, a histone demethylase (Lagarou et al., 2008). Pcl-PRC2 is required to generate high levels of H3K27 tri-methylation at PcG target sites (Nekrasov et al., 2007). In addition, Pcl may be important for recruitment of PcG protein complexes in larvae (Savla et al., 2008). The identification of PcG protein complexes is an area of intense investigation and it is likely that additional complexes will be identified. Given that not all PcG mutants have the same phenotype (Soto et al., 1995), it is likely that not all PcG targets are regulated by the same constellation of proteins.
Pleiohomeotic (Pho) is a PcG member that has specific DNA-binding activity and is thought to be important in recruiting PcG complexes to PREs (Brown et al., 1998; Fritsch et al., 1999). Pho has been shown to bind E(z), Esc and Pc in vitro, and thus could recruit PRC1 and PRC2 through direct protein-protein interactions (Mohd-Sarip et al., 2002; Mohd-Sarip et al., 2005; Wang et al., 2004). Pho is not a stable component of either PRC1 or PRC2 but it is found in a complex (PhoRC) with the PcG protein, dSfmbt (Klymenko et al., 2006). dSfmbt selectively binds to mono- or di-methylated H3K9 or H4K20, and may contribute to PcG repression by interacting with methylated histones in the chromatin flanking the PRE. dSfmbt can also physically and genetically interact with Sex combs on middle leg (Scm), a related MBT-repeat protein that can interact with Ph (Grimm et al., 2009) and associates substoichiometrically with PRC1 (Saurin et al., 2001; Peterson et al., 2004). Thus, in principle, PhoRC could recruit PRC1 to the DNA through an interaction with Scm. Although there are a lot of models, the reality is that very little is known about how PcG protein complexes become recruited to PREs.
Functional studies show that Pho-binding sites are a major determinant of PREs. Consistent with this, genome-wide ChIP-on-ChIP studies show that Pho is present at most or all PREs (Oktaba et al., 2008; Kwong et al., 2008; Schuettengruber et al., 2009). However, Pho-binding sites alone are not sufficient for PRE activity. PREs are made up of binding sites for many different proteins. Other proteins suggested to be important for PRE activity include GAGA factor (GAF), Pipsqueak, Dsp1, Zeste and Grainyhead/NTF1 (for a review, see Müller and Kassis, 2006). It is unclear what exactly these factors do at PREs; the genome-wide ChIP-on-ChIP data suggest that GAF, Dsp1 and Zeste binding does not closely correlate with DNA fragments bound by other PcG proteins, although some overlap is present. To date, Pho is the DNA-binding protein that tracks most closely with other PcG proteins (Schuettengruber et al., 2009; Négre et al., 2006; Kwong et al., 2008; Oktaba et al., 2008).
Our interest has been to understand how PcG complexes are recruited to PREs, what constitutes a PRE and, in particular, how PREs work in the context of regulation of the engrailed gene. To this end, we have focused our analysis on an 181 bp PRE (–576 to –395 bp) upstream of the engrailed gene. Our approach has been to identify DNA-binding sites within this PRE that are important for PRE function and subsequently identify the proteins that bind to them. Importantly, this approach should allow us to identify proteins with redundant activities that may be missed by screening for mutations that cause PcG phenotypes. We previously reported that a Sp1/KLF binding site is required for the activity of the 181-bp engrailed PRE and that Sp1/KLF binding sites are present in all known PREs (Brown et al., 2005). Here, we describe the identification of a protein that binds this site: Spps. In polytene chromosomes, Spps is bound to the same chromosome bands as the PcG protein Psc. Furthermore, Spps is bound to the en and Ubx PREs. Finally, mutation of Spps suppresses pairing-sensitive silencing, an activity associated with PREs, and enhances the phenotype of pho mutants. These data show that Spps is important for PRE activity. We suggest it may play a role in recruiting PcG proteins to PREs.
MATERIALS AND METHODS
Antibodies
Rabbit polyclonal antibodies were raised against a gel purified HIS-Tag fusion protein that expresses amino acids 267 to 968 of Spps. Squashes and immunofluorescent staining of polytene chromosomes were performed as described previously (Brown et al., 2003) with the following changes. Primary antibodies were used at: αPho, 1:200; αPsc, 1:25; αSpps, 1:200; α-mouse Alexafluor 488 (1:400) and α-rabbit Alexafluor 555 (1:500) were used as secondary antibodies.
Chromatin immunoprecipitation
The ChIP in S2 cells was performed using methods and Ubx primer sets described previously (Wang et al., 2004). The primer sequences for PCR over the en region are available on request. For ChIP from larval tissues, imaginal disks, brains and some cuticle were harvested from 40 larvae (2×20 larvae) for each ChIP experiment. The 3rd instar larvae were dissected in PBS and then stored on ice in S2 culture media (GIBCO). The media was removed and the disks were fixed in 2% Ultrapure formaldehyde in 50 mM HEPES (pH 7.6), 100 mM NaCl, 0.1 mM EDTA, 0.5 mM EGTA with protease inhibitors for 15 minutes at room temperature. The disks were washed for 5 minutes with stop solution (1×PBS, 0.01% Triton X-100, 125 mM glycine) followed by two 5 minute washes in 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100. At this stage, the disks could be frozen at –80°C [in 10 mM Tris-HCl (pH 8), 1 mM EDTA, 0.5 mM EGTA] or used immediately by replacing the wash buffer with 300 μl reaction buffer [50 mM Tris-HCl (pH 7.4), 1 mM EDTA]. The disks were homogenized then sonicated with a Biorupter sonicator according to the manufacturers' instructions. The debris was spun out, the 2×300 μl samples were pooled and an input control sample was removed. The remaining volume was divided into three equal aliquots and ChIP was carried out using the Millipore ChIP kit following the protocol supplied by the manufacturer. Pho and Spps antibodies were used at 1:200 dilutions. qPCR was performed on a Roche 480 Lightcycler with the 480 SYBR Master mix using standard protocols.
Oligos for larval qPCR
Oligos used were as follows: En PRE, GCTTATGAAAAGTGTCTGTG and GGGGCTTGTTAGGCAGCAAT; En gene control, CGCCTTAAGGTGAGATTCAGTT and GGCGGTGTCAATATTTTGGT; PRED, CGAAATGCTACTGCTCTCTA and GCGTAGTCTTATCT GTATCT; Ubx non PRE, CCAGCATAAAACCGAAAGGA and CGCCAAACATTCAGAGGATAG.
Targeted knockout of Spps
We used the ends out recombination strategy of Gong and Golic (Gong and Golic, 2003) to replace the Spps genomic region with GFP under the control of the Armadillo promoter (ARM). Flanking DNA (2 kb) from either side of the Spps-coding region (CG13609 and cav) was amplified by PCR and cloned into the zero blunt TOPO vector (Invitrogen). CG13609 was flanked with EcoRI and KpnI sites and cav was flanked with SpeI sites. The two flanking sequences and intervening ARM/GFP was sub-cloned into pW30 as a NotI-KpnI fragment (Gong et al., 2005). The exact nature of the Spps1 targeted knockout mutation was confirmed by PCR analysis and sequencing.
Generation of germline clones and other genetic crosses
To generate germline clones, w; P{neoFRT}82B Spps1/TM6B, Tb1 virgin females were crossed to yw hsFLP; P{neoFRT}82B P{ovoD1-18}3R/TM3, Sb1 males. After 1 day, parents were transferred to a new vial and progeny were aged for 1 day and then heat-shocked at 37°C for 1 hour on 2 consecutive days (Chou and Perrimon, 1996). Virgin females of the genotype yw hsFLP; P{neoFRT}82B Spps1/P{neoFRT}82B P{ovoD1-18}3R were crossed to w; Spps1/TM6B, Tb1 males. Progeny were collected for embryo staining, polytene chromosome squashes and to assess the lethal phase.
To assess the genetic interaction of pho and Spps, a w; Spps1/TM6B, Tb1; pho1/ciD stock was made and progeny were analyzed. pho1 homozygotes normally die as pharate adults with a PcG phenotype (sex combs on the second and third legs visible through the pupal cases in pharate adults). Spps1/TM6B, Tb1; pho1/pho1 pharate adults with a PcG phenotype were among the progeny of w; Spps1/TM6b, Tb1; pho1/ciD. However, we noticed that all Spps1/Spps1 pharate adults lack the PcG phenotype. This suggested that either Spps1/Spps1; pho1/pho1 do not survive to the pharate stage, or that Spps suppresses the pho phenotype. In order to determine which was the case, we tested 40 Spps1/Spps1 pharate adults for the presence of the homozygous pho1 mutation by PCR (Brown et al., 1998). No homozygous pho1 mutants were detected, showing that Spps1/Spps1; pho1/pho1 animals do not survive to the pharate adult stage.
To test whether the pharate adult lethal phase of Spps1 homozygotes was due to the mutation of Spps and not another mutation on the chromosomes, Spps1/TM6B, Tb1 flies were crossed to two different deficiency stocks, Df(3R)crb87-4 st e/TM6C and w1118; Df(3R)Exel6198, P{XP-U}Exel6198/TM6B, Tb1. In both cases, Spps1/Df flies died as pharate adults with no obvious transformations. To test whether the suppression of mini-white silencing also mapped to the Spps mutation, w; PRE-mw-1/PRE-mw-1; Spps1/TM3, Ser virgins were crossed to w; PRE-mw-1/+; Df(3R)crb87-4 st e/+ males. Like PRE-mw-1/+ and PRE-mw-1/PRE-mw-1; Spps1/Spps1 pharate adults, PRE-mw-1/+ and PRE-mw-1/PRE-mw-1; Spps1/ Df(3R)crb87-4 st e pharate adults had red eyes, showing that the suppression of mini-white silencing was due to the Spps1 mutation and not to another mutation on the chromosome.
To express Spps under the control of enGAL4, we used line P{Mae-UAS.6.11}CG5669[DP01353], an EP-line (Staudt et al., 2005) with the EP-element inserted just upstream of the Spps transcription start site, and crossed it to enGAL4.
RESULTS
We have previously shown that a Sp1/KLF-binding site is required for activity of a 181 bp PRE from the Drosophila en gene in two assays: pairing-sensitive silencing and maintenance of repression of a reporter gene in embryos (Americo et al., 2002; Brown et al., 2005). There are nine members of the Sp1/KLF family in Drosophila. Of the nine members, six of the proteins showed strong in vitro binding to the Sp1 site in the 181 bp PRE (Brown et al., 2005). These included: Bteb2, D-Sp1, btd, CG5669, CG12029 and CG9895. Of these genes, D-Sp1 and btd are involved in the development of the head, mechanosensory bristles and ventral imaginal disks (Schock et al., 1999; Wimmer et al., 1996; Estella et al., 2003), and it is unlikely that they play a role in Polycomb recruitment in general. Bteb2 and CG12029 are not expressed ubiquitously, a property of most PcG genes. We have not been able to detect the expression of CG9895 in embryos. Thus, we have concentrated our efforts on CG5669. Because of the phenotype of the CG5669 mutant (see below), we have named this gene Spps (Sp1-like factor for Pairing Sensitive-silencing). Spps protein localization and mutant phenotype suggest it is important for PRE activity.
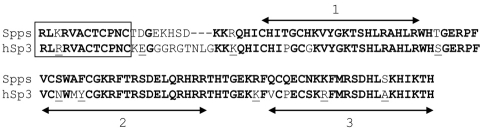
Of the over 20 mammalian Sp1/KLF family members, Spps shares the greatest sequence similarity to Sp1, Sp3 and Sp4. The homology spans the zinc-finger domains and a small region N-terminal to the zinc fingers that matches the consensus for a buttonhead (btd) box, a conserved region of unknown function also present in the btd protein. Over a 106 amino acid stretch of Spps there is 79%, 81% and 83% identity with the human Sp1, Sp3 and Sp4 factors, respectively. The sequence comparison of Spps with human Sp3 is shown in Fig. 1. The sequence similarity of Spps to the human factors is greater than the similarity to any of the other Drosophila Sp1/KLF family members.
Fig. 1.
Amino acid sequence comparison between Drosophila Spps and human Sp3. The boxed amino acids are the btd box, a domain present in Sp-subfamily members (Suske et al., 2005). The double-headed arrows indicate the extent of the three zinc-finger domains. Amino acids in bold are conserved between the two proteins. Underlined amino acids have similar properties.
Spps RNA is maternally deposited and ubiquitously expressed throughout embryogenesis (data not shown), an expression pattern that is typical of most PcG proteins. We made an antibody against the bacterially expressed C-terminal two-thirds of the Spps protein and affinity purified it for use in the studies described here. Spps is a nuclear protein, expressed ubiquitously throughout embryonic and larval development (Fig. 2 and data not shown). In Fig. 2A, the Spps is seen in all nuclei of the anterior half of a stage 13 embryo. The nuclear localization of Spps may be more easily seen in Fig. 2B. Here, Spps expression is under the control of an enGAL4 driver, causing its accumulation in a striped pattern and in particular cells of the nervous system. This confirms that our antibody is able to detect the Spps protein. In addition, we wanted to know whether our antibody was specific for the Spps protein, especially as the antigen we used contains the C-terminus, which includes the zinc-finger region that contains sequences similar to eight other Sp1/KLF family members. Importantly, in maternal-zygotic Spps mutants, that contain a deletion of the Spps-coding region, we detected no nuclear staining with the affinity-purified Spps antibody (Fig. 2C,D) (generation of this mutant is described below). Thus, our antibody is specific for Spps.
Fig. 2.
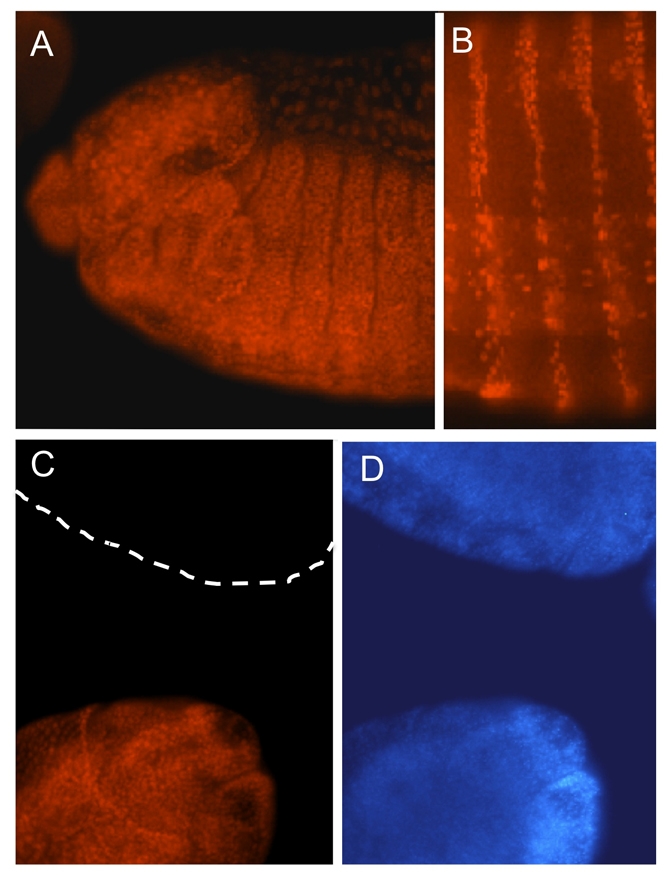
Spps is a ubiquitous nuclear protein. (A) Anterior half of a stage 13 embryo (anterior leftwards, dorsal upwards), showing ubiquitous staining of Spps antigen. (B) Ventral view of abdominal segments of a stage 16 embryo expressing Spps via an enGal4 driver that causes Spps to be expressed in stripes and in specific cells of the CNS. This shows that the Spps antibody can detect Spps. (C,D) Embryos derived from Spps germline clones, stained with Spps antibody (C) and DAPI (D). The lower embryo received a wild-type Spps gene from the father, whereas the upper embryo received a mutant Spps1 chromosome from the father and thus has no Spps antigen. The absence of Spps staining in the upper embryo (broken line in C) shows the specificity of the Spps antibody for Spps. Both embryos are stage 15. The anterior end of the lower embryo and the lateral posterior end of the upper embryo are shown.
Does Spps associate with Polycomb targets?
We analyzed the pattern of binding of Spps to the polytene chromosomes of Drosophila in comparison with the binding pattern of the PcG protein Psc. It is striking that the patterns of binding of Psc and Spps are virtually identical, both in location and in intensity (Fig. 3A). This suggests that Spps functions at the same targets as Psc. This complete correlation in localization and intensity of polytene bands with Psc is somewhat different from that seen with polytene chromosomes double labeled with antibodies against Pho and Psc (Brown et al., 2003) (Fig. 3B). Although most of the bands of Pho and Psc are coincident, there are often differences in the intensity of staining relative to other bands in the spread. Some examples of this are highlighted in the last panel in Fig. 3B. In addition, there are polytene bands that have only Pho or Psc bound. Thus, Spps shows an even more specific association with Psc than does Pho, a protein known to be important for PRE activity.
Fig. 3.
Immunostaining of polytene chromosomes. (A) Green and red panels show the binding of antibodies against Psc and Spps respectively. The third panel shows the merged images. Virtually all of the bands are coincident. Boxed regions are magnified in the merged figure. (B) Green and red panels show the binding of antibodies against Psc and PHO proteins respectively. The third panel shows the merged images. The green asterisks represent some of the positions where Psc is bound at a higher incidence than PHO based on the comparable intensities of other bands nearby. The red asterisks represent positions where PHO is bound at a higher incidence than Psc when compared with the intensities of nearby bands.
Sp1/KLF-binding sites are present in all known PREs; therefore, we investigated whether we could show a specific association of Spps with PREs by chromatin immunoprecipitation in S2 cells and in larvae. First, we looked at the en 181 bp PRE and the 2 kb upstream of this that also exhibits PRE activity (see Fig. 4) (Kassis, 1994). The pattern of ChIP for Spps corresponds to the pattern of ChIP that we see with Pho, Ph and Pc at this PRE. In addition, we tested whether Spps binds to another well studied PRE, PRED from Ubx (Fritsch et al., 1999). Ubx PRED also contains a consensus-binding site for Sp1/KLF factors (Brown et al., 2005). As in the en PRE, there is an excellent correspondence between Spps, Pho, Ph and Pc binding to Ubx PRED. We also used ChIP followed by qPCR on larval samples and found that, like Pho, Spps binds to the en and Ubx PREs in larvae (Fig. 4E,F). Spps is therefore a very good candidate for a DNA-binding protein involved in the PcG response.
Fig. 4.
Spps colocalizes with Pho, Ph and Pc on en and Ubx PREs in S2 cells and larvae. (A) Schematic of the en transcription start and the upstream 2.4 kb containing the identified PREs and the fragments amplified in the PCR after ChIP. The diagram is not to scale. (B) PCR of fragments after ChIP with antibody shown on the left. The numbers above the lanes indicate the fragment that was amplified in the PCR as shown in A. RpII140 primers were included as a negative control, as PcG proteins do not bind to this DNA. A DNA control and a sample incubated with pre-bleed from the rabbit used to generate the Spps antibody are also shown. (C) Schematic of Ubx DNA fragments used to analyze ChIP products over PRED. (D) PCR of fragments after ChIP with antibody shown on the left and the same controls as in B. (E,F) qPCR showing that Pho and Spps bind to the en and Ubx PREs in larvae.
Generation of a Spps mutant
We generated a deletion of the Spps-coding region using the ends-out recombination system developed by Gong and Golic (Gong and Golic, 2003). We replaced endogenous Spps with GFP under the control of the Armadillo promoter. The nature of the mutation was confirmed by PCR analysis, showing that the Arm-GFP is inserted at the expected location and that Spps is absent (data not shown). The genes on either side of Spps were sequenced in the mutant to confirm that the open reading frame of these two genes (CG13609 and caravaggio, cav) were still intact.
Mutation of Spps interferes with PRE function
Most homozygous Spps1 mutants die as pharate adults, though some hatch and die shortly thereafter. Flies that are Spps1 heterozygous with a deficiency that deletes Spps die at a similar stage. Aside from some wing defects (most flies do not open the wings at all), there is no other morphological defect, i.e. they do not show the homeotic phenotypes generally seen in PcG mutants. Spps has a large maternal component. In order to assess the role of the maternal contribution of Spps, we made germline clones. Surprisingly, the phenotype of maternal-zygotic mutants was exactly the same as that of the zygotic mutants, death at the pharate adult stage with no homeotic transformations. Consistent with this, in polytene chromosomes from Spps maternal-zygotic mutant larvae, we see no Spps antigen, but Psc is still bound to the DNA (data not shown). Furthermore, Ubx is not misexpressed in imaginal disks of Spps mutants (data not shown). This leads us to one of two conclusions. Either Spps plays no role in embryogenesis or larval development and instead plays an exclusively pupal/adult role or there is another protein that has some functional redundancy with Spps. Partial functional redundancy is seen with a number of members of the PcG genes [pho and pho-like (Brown et al., 2003), Esc and esc-like (Ohno et al., 2008; Kurzhais et al., 2008; Wang et al., 2006), Psc and Su(z)2 (Lo et al., 2009)] and with the Drosophila [Btd and Sp1 (Wimmer et al., 1996; Estella et al., 2003)] and mammalian Sp1/KLF factors [Sp1 and Sp3 (Li et al., 2004)]. Because the Sp1/KLF-binding site present in the en 181-bp PRE is important for PRE activity in an embryonic reporter assay (Brown et al., 2005), we believe that there must be a protein that acts through this site in embryos and provides an activity redundant to that of Spps. Currently, we do not know what the redundant protein is. It could be another member of the Sp1/KLF family, or possibly a different protein able to bind the same sequence.
Spps is necessary for PRE-mediated mini-white silencing
When PREs are cloned in reporter constructs with the eye color marker gene mini-white, they commonly silence its expression. This silencing activity is often stronger in flies that are homozygous for the reporter construct and is known as pairing-sensitive silencing (PSS). The Sp1/KLF-binding site in the 181-bp en PRE is required for PSS (Americo et al., 2002). In flies homozygous for the Spps1 mutation, PRE-mediated mini-white silencing is suppressed, both in flies heterozygous for the PRE reporter gene, and in those homozygous for the insert. Fig. 5 shows the eyes of flies homozygous for a reporter construct with the 181-bp en PRE and mini-white at two different chromosomal insertion sites, in a fly with one wild-type copy of Spps and in a Spps1 homozygous mutant. In flies with one wild-type copy of Spps, the PRE suppresses mini-white expression, and the eyes are white. By contrast, in Spps1 homozygous mutants the eye color is red in one line, and orange in the other, showing that loss of Spps reduces or eliminates PSS in these flies. Spps also suppresses mini-white silencing mediated by Ubx PRED (Fig. 5G,H). Importantly, a mutation in Spps does not change the eye color of a line containing a mini-white reporter without a PRE (Fig. 5A,B). Thus, a mutation in Spps was able to abrogate the mini-white silencing activity of both the 181 bp en PRE and Ubx PRED, suggesting a general role in PRE-mediated silencing late in development.
Fig. 5.

Pairing-sensitive silencing is alleviated in Spps1 homozygotes. (A,B) Eye from a fly homozygous for a mini-white construct without a PRE in either a heterozygous or homozygous Spps1 background, showing that Spps1 does not alter the eye color in the absence of a PRE. (C,D) Eye from a fly homozygous for a PRE-mini-white construct in either a heterozygous or homozygous Spps1 background, showing that Spps alleviates pairing-sensitive silencing. This PRE-mini-white construct contains en-181 bp PRE in pCaSpeR [insert 8-10C on chromosome 2R (Kassis, 1994)]. (E,F) Eyes from flies with the insertion of another en-181 bp PRE pCaSpeR construct (4-8-3-1B, inserted on chromosome 2L, unpublished line). (G,H) Eyes from a line of flies with an insertion of a construct that has Ubx-PRED cloned in pCaSpeR (line K2-1, inserted on the X chromosome).
Spps enhances the pho mutant phenotype
In order to further investigate the role of Spps in PcG-mediated repression, we looked for genetic interactions between Spps1 and mutations in other PcG genes. We first looked at whether a mutation in Spps affected the extra sex comb phenotypes seen in flies heterozygous for Psc1. We did not see any enhancement of the extra sex comb phenotype in Spps1/Spps1; Psc1/+. Similarly, Spps does not enhance the extra sex comb phenotype seen in Pc4 heterozygotes. We next examined whether there was a genetic interaction between pho and Spps. There were two reasons for testing this. First, both Pho and Spps are PRE DNA-binding proteins; thus, they may work together or in parallel pathways to recruit PcG protein complexes to the DNA. Second, YY1 and Sp1, mammalian proteins that contain zinc-finger regions highly similar to Pho and Spps, are known to interact through their zinc-finger domains (Lee et al., 1993). Therefore, we examined the phenotype of Spps1; pho1 double mutants. Although both pho1 and Spps1 mutants die as pharate adults, Spps1; pho1 double mutants die earlier in development, as third instar larvae or often before. Although theoretically one of every three Spps1 homozygous third instar larvae should be a pho1 homozygote in a Spps1/TM6b; pho1/CiD stock, the frequency was much less, about one in 15 (4/62) and double mutant larvae were often small. We examined the expression pattern of Ubx and En in pho1 and Spps1 single mutants, and in Spps1; pho1 double mutants in imaginal disks. As expected from the lack of homeotic phenotype, we did not see any mis-expression of Ubx or En in Spps1 mutant larvae. The phenotype of Ubx and En expression in pho1;Spps1 double mutants was extremely variable and sometimes the disks were misshapen and it was difficult to discern which disk was which. Perhaps because the larvae are sick, in some double mutants there was a lot of mis-expression, in others there was much less (data not shown). Nevertheless, our data show that Spps; pho mutants die at a much earlier stage than either single mutant, and it is reasonable to propose that they may act either together or in parallel pathways to recruit PcG protein complexes to DNA.
DISCUSSION
Spps, a member of the Sp1/KLF family of proteins binds polytene chromosomes in a pattern comparable with the PcG protein Psc and, as shown by chromatin immunoprecipitation, is bound to both the en and Ubx PREs in S2 cells and in larvae. Furthermore, a mutation in Spps abrogates PRE activity in a mini-white assay, and enhances the phenotypes seen in a pho mutant. We suggest that Spps acts either with or in parallel to Pho to recruit PcG protein complexes to the DNA. This result is particularly interesting in lieu of the recent report that the PcG protein Scm is recruited to the DNA independently of Pho (Wang et al., 2010). Those authors speculate that Scm is in a complex with another PRE-DNA-binding protein, and show that, like Pho, Scm plays a role in recruitment of PRC1 and PRC2 to the PRE. It will be interesting to explore whether Spps or another Sp1/KLF family member recruits Scm to the DNA.
The mammalian homologues of Pho and Spps, YY1 and Sp1 are extremely versatile proteins. Their activities can be changed from repressor to activator or vice versa depending on the cellular and binding site context (for reviews, see Gordon et al., 2006; Lomberk and Urrutia, 2005; Li et al., 2004). The activity of both these proteins is sensitive to the influence of many different co-repressors and co-activators. Both factors have been shown to bend DNA (Natesan and Gilman, 1993; Sjottem et al., 1997). Finally, YY1 and Sp1 have also been shown to interact directly by a number of groups (Lee et al., 1993; Li et al., 2008). We find it very intriguing that such proteins bind to the PREs of Drosophila genes. Given that PREs may mediate the action of both the Polycomb and Trithorax group proteins, DNA-binding/recruitment proteins with such versatility and adaptability could be one way to facilitate the change from repression to activation. In fact, there is a report that Pho and Phol, in addition to their association with PREs, are bound to regions of chromatin with active histone modifications (Schuettengruber et al., 2009). Finally, a single Pho-binding site in a PRE in the even-skipped gene has been shown to be important for both activation and repression, dependent on the context (Fujioka et al., 2008). It will be interesting to explore whether Spps also has a dual role in gene regulation.
Acknowledgments
We thank Jim Kennison for helpful discussions, Monica Cooper and Jim Kennison for the PRED-mw line, and Yuzhong Cheng and Melissa Cunningham for comments on this paper. We also thank Kris Langlais, Karl Pfeifer and Cameron Johnson for help with the larval ChIP and qPCR, and the Bloomington stock center for fly stocks. This research was supported by the Intramural Research Program of the NIH, NICHD. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Americo J., Whiteley M., Brown J. L., Fujioka M., Jaynes J. B., Kassis J. A. (2002). A complex array of DNA binding proteins required for pairing-sensitive silencing by a Polycomb group response element from the Drosophila engrailed gene. Genetics 160, 1561-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Mucci D., Whiteley M., Dirksen M. L., Kassis J. A. (1998). The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1, 1057-1057 [DOI] [PubMed] [Google Scholar]
- Brown J. L., Fritsch C., Müller J., Kassis J. A. (2003). The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130, 285-285 [DOI] [PubMed] [Google Scholar]
- Brown J. L., Grau D. J., DeVido S. K., Kassis J. A. (2005). An Sp1/KLF binding site is important for the activity of a Polycomb group response element from the Drosophila engrailed gene. Nucleic Acids Res. 33, 5181-5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002). Role of histone H3 lysine K27 methylation in Polycomb-group silencing. Science 298, 1039-1039 [DOI] [PubMed] [Google Scholar]
- Chou T. B., Perrimon N. (1996). The autosomal Flp-Dfs technique for generating germline mosaics in Drosophila melanogaster. Genetics 144, 1673-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemin B., Melfi R., McCabe D., Steitz V., Imhof A., Pirrotta V. (2002). Drosophila Enhancer of Zeste/Esc complexes have a histone methyltransferase activity that marks chromosomal Polycomb sites. Cell 11, 185-185 [DOI] [PubMed] [Google Scholar]
- Estella C., Rieckhof G., Calleja M., Morata G. (2003). The role of buttonhead and Sp1 in the development of the ventral imaginal disks of Drosophila. Development 130, 5929-5929 [DOI] [PubMed] [Google Scholar]
- Francis N. J., Saurin A. J., Shao Z., Kingston R. E. (2001). Reconstitution of a functional core Polycomb repressive complex. Mol. Cell 8, 546-546 [DOI] [PubMed] [Google Scholar]
- Fritsch C., Brown J. L., Müller J., Kassis J. A. (1999). The DNA binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126, 3905-3905 [DOI] [PubMed] [Google Scholar]
- Fujioka M., Yusibova G. L., Zhou J., Jaynes J. B. (2008). The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development 135, 4131-4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M., Bi X., Rong Y. S. (2005). Targeted mutagenesis of Drosophila atm and mre11 genes. Drosoph. Inf. Serv. 88, 79-79 [Google Scholar]
- Gong W. J., Golic K. G. (2003). Ends out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100, 2556-2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S., Akopyan G., Garban H., Bonavida B. (2006). Transcription factor YY1: structure, function and therapeutic implications in cancer biology. Oncogene 25, 1125-1125 [DOI] [PubMed] [Google Scholar]
- Grimm C., Matos R., Ly-Hartig N., Steuerwald U., Lindner D., Rybin V., Müller J., Muller C. W. (2009). Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 28, 1965-1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. (1994). Unusual properties of regulatory DNA from the Drosophila engrailed gene: three ``pairing-sensitive'' sites within a 1.6kb region. Genetics 136, 1025-1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T., Papp B., Fischle W., Kocher T., Schelder M., Fristch C., Wild B., Wilm M., Müller J. (2006). A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2, 1110-1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzhais R. L., Tie F., Stratton C. A., Harte P. J. (2008). Drosophila ESC-like can substitute for ESC and becomes required for Polycomb silencing if ESC is absent. Dev. Biol. 313, 293-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002). Histone methyltransferase activity associated with a human multi-protein complex containing Enhancer of Zeste protein. Genes Dev. 16, 2893-2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C., Adryan B., Bell I., Meadows L., Russell Manak R., White R. (2008). Stability and dynamics of polycomb target sites in Drosophila development. PLoS Biol. 4, 1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A., Mohd-Sarip A., Moshkin Y. M., Chalkley G. E., Bezstarosti K., Demmers J. A. A., Verrijzer C. P. (2008). dKDM2 couples histone H2A ubiquitination to histone H3 demethylation during Polycomb group silencing. Genes Dev. 22, 2799-2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-S., Galvin K. M., Shi Y. (1993). Evidence for physical interaction between the zinc finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. USA 90, 6145-6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu H., Wang Z., Liu X., Guo L., Huang L., Gao L., McNutt M. A., Li G. (2008). The role of transcription factors Sp1 and YY1 in proximal promoter region in initiation of transcription of the Mu opioid receptor gene in human lymphocytes. J. Cell. Biochem. 104, 237-237 [DOI] [PubMed] [Google Scholar]
- Li L., He S., Sun J.-M., Davie J. R. (2004). Gene Regulation by Sp1 and Sp3. Biochem. Cell. Biol. 82, 460-460 [DOI] [PubMed] [Google Scholar]
- Lo S. M., Ahuja N. K., Francis N. J. (2009). Polycomb group protein suppressor of zeste(2) is a functional homolog of Posterior Sex combs. Mol. Cell. Biol. 29, 515-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G., Urrutia R. (2005). He family feud: turning off Sp1 by Sp1-KLF proteins. Biochem. J. 392, 1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., Venturini F., Chalkley G. E., Verrijzer C. P. (2002). Pleiohomeotic can link Polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22, 7473-7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., Cleard F., Mishra R. K., Karch F., Verrijzer C. P. (2005). Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 19, 1755-1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Kassis J. (2006). Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16, 476-476 [DOI] [PubMed] [Google Scholar]
- Müller J., Verrijzer P. (2009). Biochemical mechanisms of gene regulation by Polycomb group protein complexes. Curr. Opin. Genet. Dev. 19, 150-150 [DOI] [PubMed] [Google Scholar]
- Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., Simon J. A. (2002). Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197-197 [DOI] [PubMed] [Google Scholar]
- Natesan S., Gilman M. Z. (1993). DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 7, 2497-2497 [DOI] [PubMed] [Google Scholar]
- Négre N., Hennetin J., Sun L. V., Lavrov S., Bellis M., White K. P., Cavalli G. (2006). Chromosomal Distribution of PcG Proteins during Drosophila Development. PLoS Biol. 4, 0917-0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M., Klymenko T., Fraterman S., Papp B., Oktaba K., Kocher T., Cohen A., Stunnenberg H. G., Wilm M., Müller J. (2007). Pcl-PCR2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 26, 4078-4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K., McCabe D., Czermin B., Imhof A., Pirrotta V. (2008). ESC, ESCL and their roles in Polycomb group mechanism. Mech. Dev. 125, 527-527 [DOI] [PubMed] [Google Scholar]
- Oktaba K., Gutierrez L., Gagneur J., Girardot C., Sengupta A., Furlong E. E. M., Müller J. (2008). Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15, 877-877 [DOI] [PubMed] [Google Scholar]
- Peterson A. J., Mallin D. R., Francis N. J., Ketel C. S., Stamm J., Voeller R. K., Kingston R. E., Simon J. A. (2004). Requirement for Sex comb on middle leg protein interactions in Drosophila Polycomb group repression. Genetics 167, 1225-1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savla U., Benes J., Zhang J., Jones R. S. (2008). Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development 135, 813-813 [DOI] [PubMed] [Google Scholar]
- Saurin A. J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R. E. (2001). A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412, 655-655 [DOI] [PubMed] [Google Scholar]
- Schock P., Purnell B. A., Wimmer E. A., J©ckle H. (1999). Common and diverged functions of the Drosophila gene pair D-Sp1 and buttonhead. Mech. Dev. 89, 125-125 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Ganapathi M., Leblanc B., Portoso M., Jaschek R., Tolhuis B., van Hohuizen M., Tanay A., Cavalli G. (2009). Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLOS Biology 7, 0146-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Pirrotta V. (2008). Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20, 266-266 [DOI] [PubMed] [Google Scholar]
- Shao Z., Raible F., Mollaaghabaha R., Guyon J. R., Wu C. T., Bender W., Kingston R. E. (1999). Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98, 37-37 [DOI] [PubMed] [Google Scholar]
- Sjottem E., Andersen C., Johansen T. (1997). Structural and functional analyses of DNA bending induced by Sp1 family transcription factors. J. Mol. Biol. 267, 490-490 [DOI] [PubMed] [Google Scholar]
- Soto M. C., Chou T.-B., Bender W. (1995). Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140, 231-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt N., Molitor A., Somogyi K., Mata J., Curado S., Eulenberg K., Meise M., Siegmund T., H©der T., Hilfiker A., et al. (2005). Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS Genet. 1, e55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G., Bruford E., Philipsen S. (2005). Mammalian SP/KLF transcription factors: bring in the family. Genomics 85, 551-551 [DOI] [PubMed] [Google Scholar]
- Wang L., Brown J. L., Cao R., Zhang Y., Kassis J. A., Jones R. S. (2004). Hierarchical recruitment of Polycomb group silencing complexes. Mol. Cell 14, 637-637 [DOI] [PubMed] [Google Scholar]
- Wang L., Jahren N., Vargus M. L., Anderson E. F., Benes J., Zhang J., Miller E. L., Jones R. S., Simon J. A. (2006). Alternative ESC and ESC-like subunits of Polycomb group histone methyltransferase complex are differentially employed during Drosophila development. Mol. Cell. Biol. 26, 2637-2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Jahren N., Miller E. L., Ketel C. S., Mallin D. R., Simon J. A. (2010). Comparative analysis of chromatin binding by Sex Comb on Midleg (SCM) and other Polycomb group repressors at a Drosophila Hox gene. Mol. Cell. Biol. 30, 2584-2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer E. A., Frommer G., Purnell B. A., J©ckle H. (1996). Buttonhead and D-Sp1: a novel Drosophila gene pair. Mech. Dev. 59, 53-53 [DOI] [PubMed] [Google Scholar]