Abstract
Sulfite oxidizing enzymes (SOEs) are molybdenum cofactor dependent enzymes that are found in plants, animals and bacteria. Sulfite oxidase (SO) is found in animals and plants, while sulfite dehydrogenase (SDH) is found in bacteria. In animals, SO catalyzes the oxidation of toxic sulfite to sulfate as the final step in the catabolism of the sulfur-containing amino acids, methionine and cysteine. In humans, sulfite oxidase deficiency is an inherited recessive disorder that produces severe neonatal neurological problems that lead to early death. Plant SO (PSO) also plays an important role in sulfite detoxification, but in addition serves as an intermediate enzyme in the assimilatory reduction of sulfate. In vertebrates the proposed catalytic mechanism of SO involves two intramolecular one-electron transfer (IET) steps from the molybdenum cofactor to the iron of the integral b-type heme. A similar mechanism is proposed for SDH, involving its molybdenum cofactor and c-type heme. However, PSO, which lacks an integral heme cofactor, uses molecular oxygen as its electron acceptor. Here we review recent results for SOEs from kinetic measurements, computational studies, electron paramagnetic resonance (EPR) spectroscopy, electrochemical measurements, and site-directed mutagenesis on active site residues of SOEs and of the flexible polypepetide tether that connects the heme and molybdenum domains of human SO. Rapid-kinetic studies of PSO are also discussed.
In animals, sulfite oxidase (SO) is a molybdenum cofactor dependent enzyme, that catalyzes the oxidation of sulfite to sulfate, using ferricytochrome c (cyt cox) as the physiological electron acceptor (eq 1) (1-4):
| (1) |
This reaction is biologically essential, serving as the final step in the catabolism of sulfur containing amino acids, cysteine and methionine. SO also functions in detoxifying exogenously supplied sulfite and sulfur dioxide.
The SO family is comprised of plant assimilatory nitrate reductases (NR) and sulfite oxidizing enzymes (SOEs) found in animals, plants and bacteria. The SOEs include plant and animal SO and bacterial sulfite dehydrogenase (SDH). In animals, SO is located in the mitochondrial intermembrane space (5, 6). Plant SO (PSO) is localized in the peroxisomes and does not react with cyt c, but instead, PSO uses molecular oxygen as its terminal electron acceptor (7, 8). SDH is located in the periplasm (9), and the enzyme from Starkeya novella is the most thoroughly characterized example.
SO is physiologically vital in human metabolism, and severe neurological damage and early childhood death result from deficiency of SO activity due to an inborn metabolic defect. The symptoms of inherited sulfite oxidase deficiency include dislocation of the ocular lenses, attenuated growth of the brain and mental retardation (10, 11). Other symptoms that have been reported in patients with SO deficiency include poor feeding, hypoactivity, dyspnea, diffuse edema and seizures (12). The disease results either from molybdenum cofactor deficiency, which arises from a defect in its biosynthesis, or from certain point mutations within the SO enzyme (isolated SO deficiency) (13-15).
Prior to 2010 there had been no successful treatment for SO deficiency. Very recently the case of an infant female patient, who was diagnosed with molybdenum cofactor deficiency at 6 days old, has been reported (16). The patient showed a homozygous G175R (GGG-to-AGG) change in exon 10 of the MOCS1 gene, a mutation which disrupts the production of cyclic pyranopterin monophosphate (cPMP) a key step in the biosynthesis of the molybdenum cofactor (16-18). After substitution therapy with purified cyclic pyranopterin monophosphate (cPMP) (13), starting at 36 days old, all urinary metabolites of sulfite oxidase (sulfite, S-sulfocysteine, and thiosulfate) and xanthine oxidase deficiency (xanthine and uric acid) returned to almost normal readings and remained constant for more than one year. Thus, knowledge of the biosynthetic pathway of the molybdenum cofactor has made possible the first clinical treatment of SO deficiency. It is possible that an increased understanding of the catalytic mechanism of SOEs will result in additional treatment protocols for these disorders.
The development of recombinant human sulfite oxidase (HSO) has made it possible to prepare a library of mutations to study isolated SO deficiency, especially the effects of point mutations on the kinetic and spectroscopic properties of purified HSO (19-23). In a previous review Feng and coworkers have discussed the effects of several mutations on the intramolecular electron transfer (IET) kinetics of SO (24). For example, three mutations that have been associated with SO deficiency in patients (HSOR160Q (21), HSOG473D and HSOA208D (20) are near the active site of SO. Laser flash photolysis measurements of their IET rates showed that the ket values were decreased by three orders of magnitude relative to wild type. Thus, IET of these variants became rate-limiting and equal to the overall turnover rate (kcat) determined from steady-state kinetic studies (24). Feng also discussed several future research directions concerning SO that still needed to be addressed. These topics included: 1) studies of electron transfer, using SDH as a model; 2) the importance of the flexible tether connecting the heme and molybdenum domains in docking the two domains for electron transfer; 3) pulsed EPR experiments to elucidate the nature of the sulfite/sulfate-bound and chloride-bound forms of SO; 4) computational modeling and simulations to provide further understanding of chloride-bound SO and to promote greater understanding of the role of specific amino acids in IET and SO catalysis. This review addresses these questions using molecular biology, rapid kinetics (laser flash photolysis and stopped-flow), advanced EPR spectroscopy, protein crystallography, and computational modeling.
Structural Motif of Sulfite Oxidizing Enzymes
Crystal structures have been solved for SO from Arabidopsis thaliana (25), chicken liver (26) and SDH from S. novella (27) (Figure 1). In addition, structures of recombinant wild type chicken SO proteins with and without sulfate-bound and recombinant R138Q chicken SO have been reported (28). Recently, structures of mutant forms of SDH have been solved and will be discussed in this review (29-31).
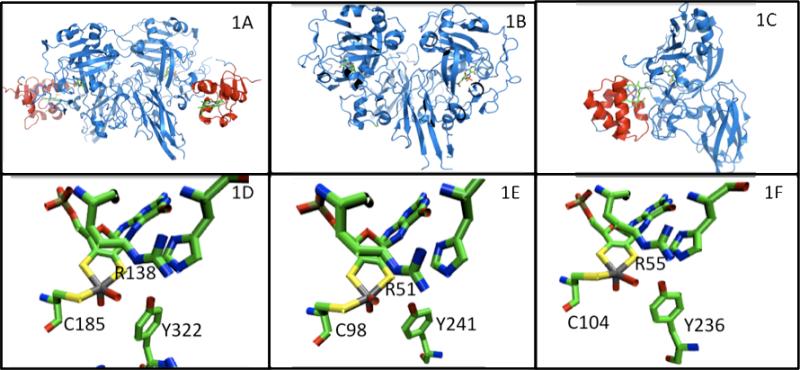
Figure 1.
Top row: Ribbon diagrams of the crystal structures of SOEs: 1A CSO (26), 1B PSO (32), 1C SDH (29); blue represents the molybdenum and C-terminal domain; red represents the heme domain. Bottom row: Selected amino acid residues near the molybdenum active site of the corresponding SOEs of the top row.
Active Site
The molybdenum active site is buried within a positively charged pocket, and has tyrosine, histidine and arginine residues that are conserved throughout SOEs and plant assimilatory nitrate reductases and have direct interactions with sulfate in CSO (26). The molybdenum active sites exhibit similar square pyramidal geometry around the molybdenum atom (Figure 1). There is an axial terminal oxo group, and the equatorial plane has three sulfur atoms (two from the dithiolene moity of the pyranopterin cofactor and one from a cysteine residue) and a water/hydroxo ligand.
Chicken Sulfite Oxidase (CSO)
CSO is the only intact eukaryotic crystal structure solved to date and possesses both a heme and a molybdenum domain. It is a 110 kDa homodimer and each subunit has three domains, an N-terminal b5-type heme domain (Figure 1A, colored red), a central molybdenum binding domain, and a C-terminal interface domain (Figure 1A, colored blue) (26). The distance between the heme and molybdenum centers, within this structure is ~ 32 Å. However, the b5-type heme and the molybdenum domains are connected by a flexible polypeptide tether that varies in length, depending upon the SO species. CSO has a tether that contains 13 amino acids, whereas the tether in HSO is 14 amino acids in length (based upon sequence alignment, Figure 2). The sequence of the tether of CSO is very different from that of HSO.
Figure 2.
Sequence alignment of the flexible tether regions of CSO and HSO. Amino acids highlighted in red are conserved between the two species, while those in blue are similar. Mutations discussed in this work are indicated in the HSO sequence: proline residues mutated to alanines are in bold type, and deleted residues are underlined.
The crystal structures reported for two forms of recombinant CSO (wt rCSO and rCSOR138Q) contain only the C-terminal dimerization domain and the catalytic core domain, which houses the molybdenum cofactor center (28). Neither of these structures exhibit a heme domain. Superposition of the structures of wt rCSO and native CSO show that they have nearly identical structures. Both the wt rCSO and rCSOR138Q SOEs were crystallized in the resting state (without sulfate) and with sulfate bound. wt rCSO contains two bound sulfates per monomer, whereas rCSOR138Q contains only one bound sulfate per monomer. The active site of SO has three positively charged residues, R138, R190 and R450, which are presumed to help guide the negatively charged substrate into the active site of the enzyme. The active site pocket of the sulfate-bound and resting state of the wt rCSO and rCSOR138Q forms, showed significant changes in the position of R450, indicating that this residue undergoes a conformational change upon substrate binding.
Plant Sulfite Oxidase
The crystal structure of PSO reveals that this enzyme is a homodimer, with an overall fold similar to CSO. PSO has two domains: an N-terminal Moco and a C-terminal dimerization domain (32), with no additional heme domain (Figure 1B). The active site is positively charged and has the conserved tyr, his, arg motif found in CSO (Figure 1E). The purpose of SO in plants is to detoxify excess sulfite in the cell, preventing sulfitolysis and also to serve as an intermediate enzyme in the assimilatory reduction of sulfate (33).
Sulfite Dehydrogenase
Sulfite dehydrogenase is a heterodimer, containing both a molybdenum-binding subunit (SorA) and a smaller heme c containing subunit (SorB) (29). The domains of the SorA subunit have the same overall fold as the Moco and dimerization domains of both CSO and PSO (Figure 1C). Unlike the eukaryotic CSO, SDH does not possess a flexible polypeptide tether. Also, the heme and molybdenum centers in bacterial SDH are within a closer proximity (~ 16 Å), than the distance found in CSO. The SorB subunit and the heme b domain of chicken liver SO have a similar overall shape. The crystal structures of the SDHY236F (30), SDHR55M and SDHH57A (31) mutants of SDH have also been solved. Combining these results with those for wt SDH has enabled potential electron transfer pathways between the heme and molybdenum redox centers to be postulated and evaluated by steady-state kinetics (31) and laser flash photolysis measurements (34).
The structure of SDHR55M
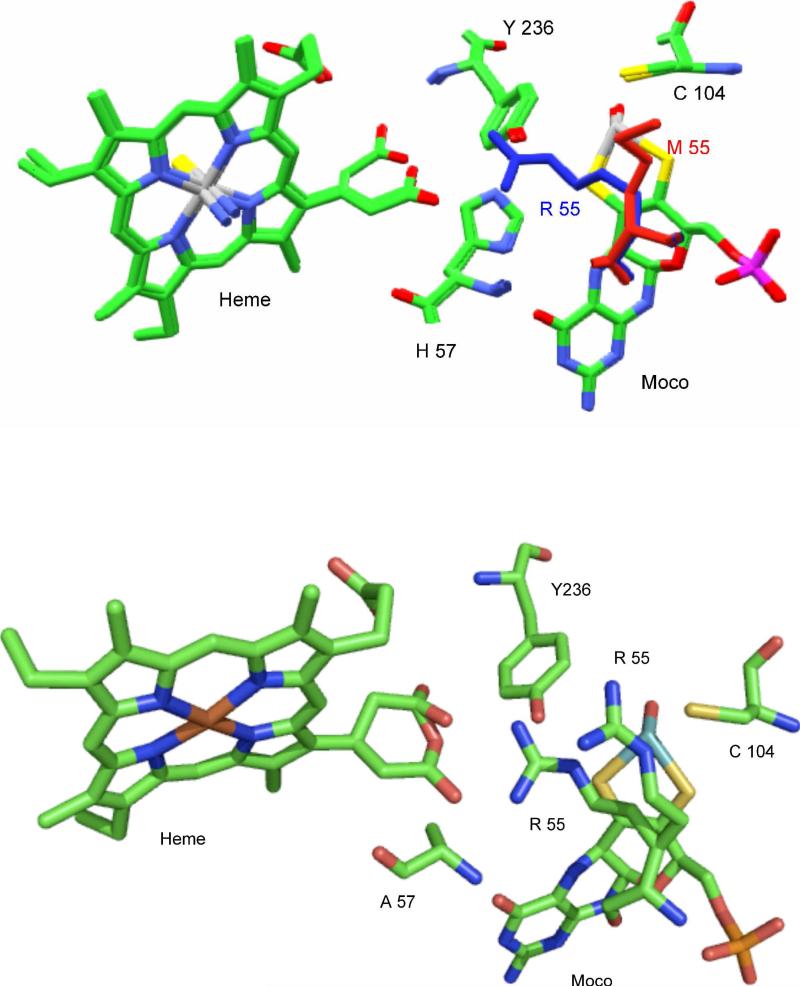
Within SDHwt, the side chain of R55 occupies a space that is close to the substrate binding site, forming hydrogen bonds with the equatorial oxo ligand of the molybdenum cofactor, Gln33 OE1 and a water molecule. It also forms a salt bridge with the propionate-6 of the heme moiety of the cytochrome subunit, which effectively locks the propionate group into the appropriate position for one of the proposed electron transfer pathways (29). However, within the SDHR55M crystal structure, the side chain of M55 does not occupy the same space as does R55 in the wt structure. M55 bends away, packing into a small cavity between the side chains of L121 and Q33. The space previously occupied by R55 in SDHwt is empty (Figure 3A) (31).
Figure 3.
Top: superposition of the active site structures of SDHwt and SDHR55M, showing that the M55 side chain (red) in SDHR55M does not occupy the same space as R55 (blue) in SDHwt. M55 bends away, packing into a small cavity between the side chains of L121 and Q33, and the space previously occupied by R55 in SDHwt is empty (31). Bottom: structure of the active site of SDHH57A, showing the two disordered conformations of the R55 side chain (31).
The structure of SDHH57A
Kappler and co-workers found that the SDHH57A structure confirms that the substitution of H57 with an alanine identifies an additional water molecule that sits within close proximity to the imidazole ring in SDHwt (31). The water molecule is able to form hydrogen bonds with the N5 and O4 of the molybdopterin cofactor. Like SDHY236F, SDHH57A appears to have increased the mobility of the R55 side chain and the interacting propionate-6 side chain of the heme (30, 31). Kappler and co-workers found that both Y236 and H57 are necessary to stabilize R55 in a position for optimal hydrogen bonding to the heme 6–propionate (Figure 3B).
Mechanism of the Sulfite Oxidizing Enzymes
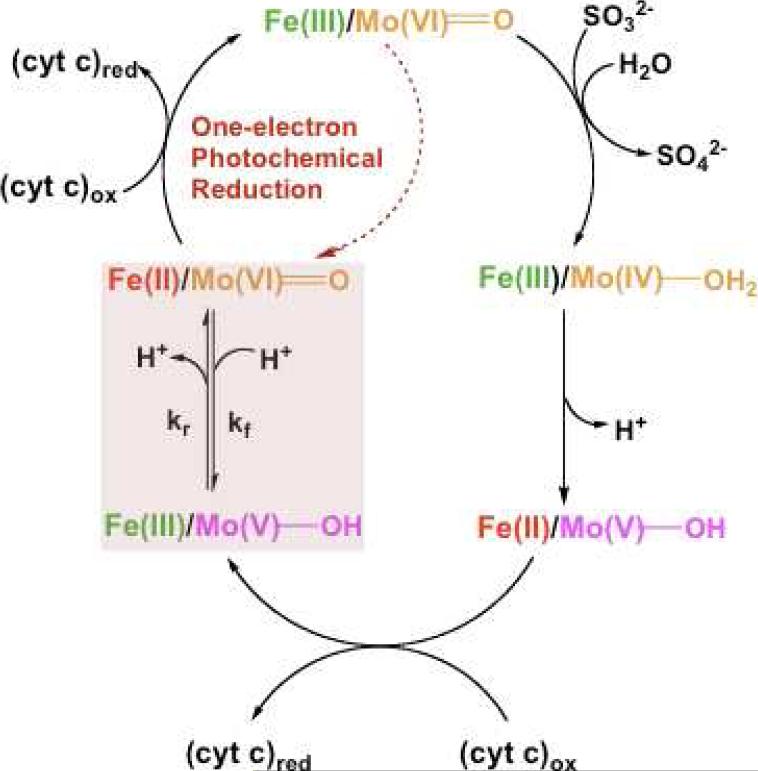
The catalytic mechanism of animal sulfite oxidase, originally proposed by Hille (Figure 4), (35-38) has become widely accepted. The proposed mechanism of animal SO starts with the fully oxidized resting Mo(VI)/Fe(III) state. Two-electron oxidation of sulfite to sulfate occurs at the molybdenum center, which is reduced to Mo(IV). The first intramolecular electron transfer (IET) step occurs when the reduced Mo(IV) transfers one electron to the oxidized Fe(III) b5 heme. The resultant Mo(V)/Fe(II) species contains a diamagnetic Fe(II) center and a paramagnetic 4d1 Mo(V) center which is detectable by electron paramagnetic resonance (EPR) spectroscopy (39-42) (see following section). The Mo(V)Fe(II) species transfers an electron to the physiological electron acceptor (cyt c)ox, producing the Mo(V)/Fe(III) state. A second IET step generates the Mo(VI)/Fe(II) state, and reduction of a second molecule of cyt c regenerates the fully oxidized Mo(VI)/Fe(III) state of the enzyme. The portion of the catalytic mechanism, in which the oxidized enzyme is reduced by two electrons and sulfate is produced, is referred to as the reductive half reaction. The oxidative half reaction thus involves two one-electron transfers of electrons to cyt c to yield oxidized SO and two molecules of reduced cyt c.
Figure 4.
Proposed oxidation state changes occurring at the Mo and Fe centers of animal SO during the catalytic oxidation of sulfite and the concomitant reduction of (cyt c)ox.
EPR Spectroscopy
The utility of EPR spectroscopy for investigating the Mo(V) sites of SOEs formed in the reductive half reaction of Figure 4 has been recognized since the early 1980s, with the identification of three distinct CW EPR forms of vertebrate SO: low pH (lpH); high pH (hpH); and phosphate inhibited (Pi) (43-45). The coordination structures of the Mo(V) sites of SOEs obtained by pulsed EPR spectroscopy have been discussed previously (46), and the applications of pulsed EPR methods to SOEs have also been extensively reviewed (47). Therefore, only a few more recent EPR structural results for SOEs are presented here.
Bray and co-workers originally proposed that chloride and phosphate were directly coordinated to Mo(V) in the lpH and Pi forms, respectively, but no 35,37Cl (I = 3/2) or 31P (I = 1/2) hyperfine interactions (hfi) were directly detected for these nuclei by CW EPR (42-45, 48). In 1996, electron spin echo envelope modulation (ESEEM) studies between 8-18 GHz detected 31P hfi for the Pi form of vertebrate SO and demonstrated that it has an equatorial monodentate phosphate group (42). However, until recently, the role of Cl− in lpH SO remained elusive. A multifrequency CW EPR experiment using HSO and 35Cl−- and 37Cl−-enriched solutions showed very small spectral differences that were interpreted as indicating the presence of an axial Cl− ligand trans to the terminal oxo group, but the limited resolution of the CW EPR spectra precluded reliable determination of the hfi and nuclear quadrupole interaction (nqi) parameters required to define the structure of lpH SO (49). Very recently, the magnetic resonance parameters of 35,37Cl (I = 3/2) in lpH SO were finally measured by ESEEM methods, and density functional theory (DFT) calculations favored a structure in which the Cl− anion is hydrogen bonded to the equatorial OH ligand and surrounded by several proton donors from nearby water molecules and charged amino acid side chains (50).
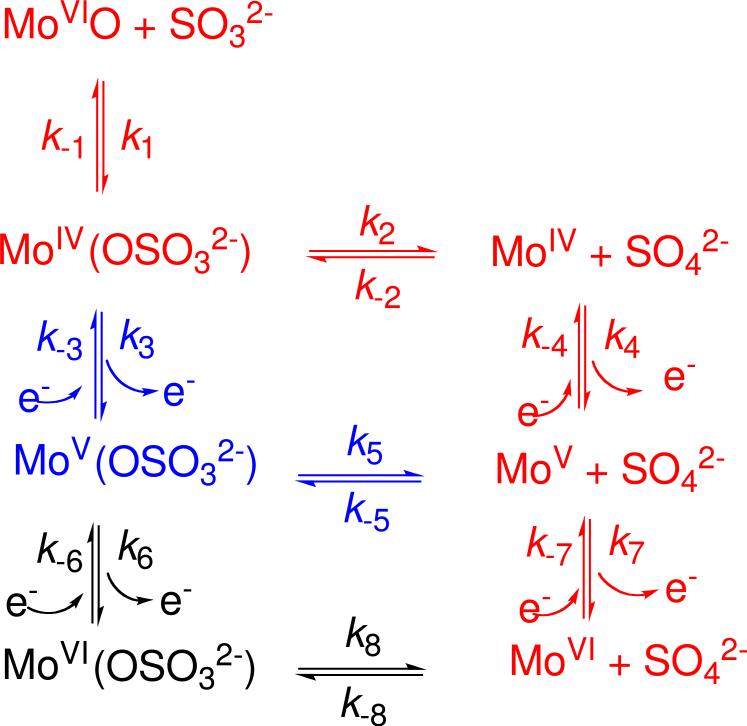
At some stage of the catalytic cycle of Figure 4, the product, SO42−, has to be released from the active site. This release might occur from either the Mo(IV) or Mo(V) states of the reductive half reaction (Scheme 1). In 1982 Bray et al. reported that the presence of excess sulfite leads to another EPR-active Mo(V) species at low pH, which they attributed to a sulfite complex of the Mo center (51). Experimental evidence for a Mo(V) species with a sulfur-containing ligand was obtained only recently from pulsed EPR investigations of samples reduced with sulfite labeled with 33S (I=3/2) at low pH, which allowed, for the first time, direct detection of coupling to 33S. This coupling was initially observed in PSO from A. thaliana and was assigned to a Mo(V)–SO4 complex (52). It was suggested that a conformational change of some of the nearby amino acid residues blocked hydrolysis of the sulfate ligand, and a structure similar to the Pi form was proposed (42), with a monodentate sulfate ligand (52). Subsequently, numerous examples of blocked forms have been observed for mutants of HSO and of SDH (22, 53). More recently, it was shown that the blocked form is also produced in chloride-depleted samples of wt HSO. Addition of excess chloride to the blocked forms of SOEs converts them, at least partially, to the familiar lpH form (54).
Scheme 1.
Possible Reaction Pathways for the Catalytic Oxidation of Sulfite by SDH or SOa
aFor the sake of simplicity, the enzyme and substrate are depicted as MoVIO and SO32-, respectively. The pathways differ in the sequence of steps which transform the enzyme-substrate complex, MoIV(OSO32-), into product (sulfate) and the reoxidized Mo(VI) state of the enzyme. The pathway colored red is the one commonly proposed, in which product release precedes reoxidation of the enzyme. For the pathway colored black, the enzyme-substate complex is oxidized by two electrons prior to product release (34). See the text for additional discussion.
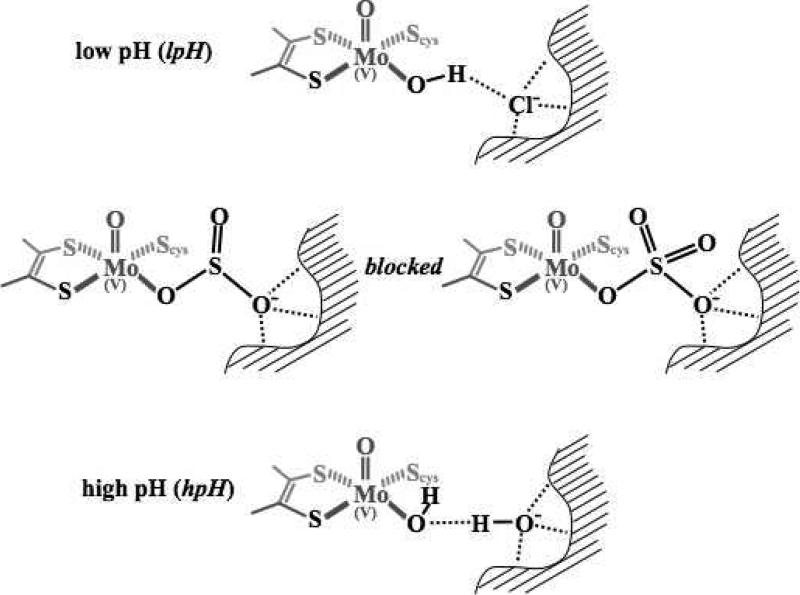
The hydrolysis of the product (sulfate) with H217O in the reductive half reaction (Figure 4) provides a convenient method for introducing 17O (I = 5/2) into the Mo(V) center of SOEs (46). It has long been known that the lpH and hpH forms show distinctly different 17O hfi (55), but it was not possible to investigate the details of their hfi and nqi parameters prior to the development of high resolution variable frequency pulsed EPR spectrometers (56) and sophisticated DFT programs to relate experimental results to electronic and molecular structure (57, 58). At high pH, SOEs exhibit two types of 17O atoms in pulsed EPR experiments. One has a large hfi of ~30-40 MHz and can be associated with the equatorially coordinated O-containing ligand. The second has a much smaller hfi (~ 5-6 MHz). For hpH SO, this latter 17O signal was assigned to the axial oxo-Mo(V) group from comparison to the results for model compounds and from extensive DFT calculations (39, 59). At low pH (~6) the blocked forms of SOEs also exhibit two types of 17O signals (22). The larger hfi (~18 MHz) is again associated with the equatorial O-containing ligand, but the origin of the smaller 17O hfi (5-6 MHz) is less clear because its nqi value (~ 4-5 MHz) is substantially larger than those for known axial oxo ligands in model compounds (1.5 MHz) (59). For studies of blocked forms of SOEs in H217O, it is also important to note that the oxygen atoms of sulfite exchange very rapidly (60). Consequently, all of the O atoms of a coordinated sulfite (reactant) or sulfate (product) anion will be labeled with 17O in the reductive half reaction of Figure 4. Recent extensive DFT calculations suggest that for the blocked form the smaller 17O hfi arises from remote O atoms on the sulfite (or sulfate) group bound to Mo(V) (61). The DFT calculations for the sulfite-coordinated structure originally proposed by Bray et al. (51) give somewhat better agreement with the experimental nqi parameters for this weakly coupled 17O and for the central 33S atom compared to the sulfate-coordinated structure proposed previously by us (52). Figure 5 summarizes the current view of the structures of the lpH, hpH and blocked forms of SOEs.
Figure 5.
Proposed structures of the molybdenum active sites of SOEs from pulsed EPR experiments and DFT calculations for the lpH (50), blocked (61), and hpH forms (58). The blocked form shown on the left has bound reactant (sulfite); the form shown on the right has bound product (sulfate). See text and (61) for details.
The availability of more than 4 million genome sequences from databases and more than 40,000 protein structures from the protein data bank (PDB) has made it possible to analyze protein folds of SOE's in all domains of life using bioinformatics (62, 63). After multiple sequence alignments, Dahl and Kappler determined a phylogenetic relationship with three distinguishable groups of SOEs (63). Group 1 includes the pathogen enzymes; group 2 contains the classic SOEs and nitrate reductases; and group 3 contains enzymes from Archaea, phototrophic and soil bacteria. Chicken, human, and plant SO, along with SDH are all included in group 2. This group contains a dimerization domain and a molybdenum domain (26, 29, 32).
Similar bioinformatic analyses performed by Weiner and co-workers identified an E. coli protein, YedY as a part of the SOEs family (62, 63). The crystal structure of YedY has been solved and has revealed that this protein has only the molydopterin binding SO fold (64). Yed Y is categorized in group 3. Its heme counterpart is a separate monoheme cytchrome b protein known as YedZ (64). To date, very little is known about the function of YedY. Sequence comparison of plant, chicken, nitrate reductase, S. novella SDH and YedY reveal that there are fifteen residues that are conserved throughout these five structures (64). One of these conserved residues is the molybdenum coordinating cysteine residue (64).
Intramolecular Electron Transfer (IET) Kinetic Studies in Sulfite Oxidizing Enzymes
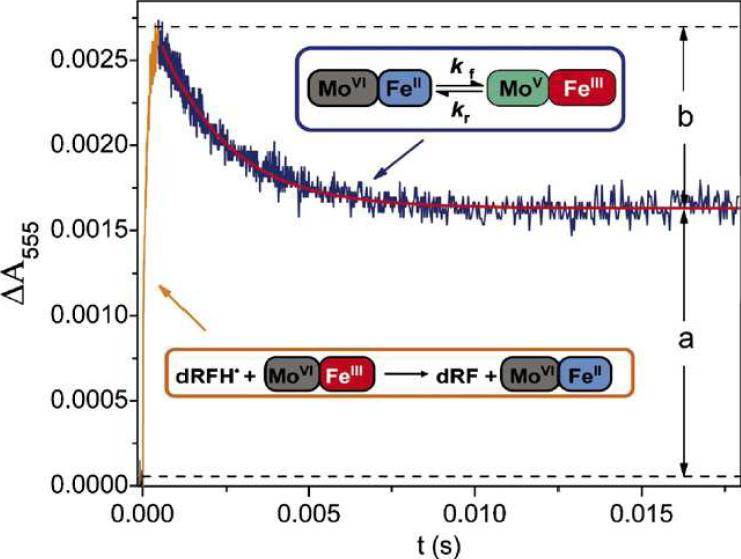
Intramolecular electron transfer (IET) is essential for the catalyzed reaction of SO. In animal SO and SDH, IET occurs between the heme and the molybdenum centers of the enzyme (Figure 4). The IET process can be studied using laser flash photolysis techniques. Laser flash photolysis experiments can be performed anaerobically in solutions containing 5-deazariboflavin (dRF) and freshly prepared semicarbazide or EDTA (AH2) as a sacrificial reductant. It has been previously shown that dRF radicals are generated with a laser pulse and will then rapidly reduce the Fe(III) heme center of animal SO and S. novella SDH by one electron (Figures 4 and 6). This process corresponds to the reverse of the second physiologically essential IET step that is boxed in Figure 4. The initial rapid increase in absorbance at 555 nm is due to the reduction of the heme from the Fe(III) to the Fe(II) form (Eq. (4)) by the dRF radical. This process is followed by a slower absorption decay that is due to the net IET from the Fe(II) to Mo(VI), i.e., the intramolecular re-oxidation of Fe(II) (forming the Fe(III)/Mo(V) species (Eq. (5)). The laser flash photolysis technique follows the IET process in the reverse direction of the enzymatic turnover.
Figure 6.
Kinetic transient obtained at 555 nm upon photoexcitation of a solution containing wild type human SO, dRF, and 0.5 mM fresh semicarbazide hydrochloride (pH 7.4). The portion of the figure outlined by the orange box points to the initial heme reduction by dRFH·; this process is pseudo first order, and its rate depends on protein concentration. The dark blue box points to heme reoxidation due to the subsequent IET between the Mo and Fe centers; this process is independent of protein concentration, consistent with its intraprotein nature. The red solid line indicates a single-exponential fit to the IET phase. Keq=b/a. Copyright Elsevier publishing; reproduced from Feng, C., Tollin, G., and Enemark, J. H. (2007) Sulfite Oxidizing Enzymes, Biochim. Biophys. Acta 1774, 527-539, with permission.
The methodologies used to obtain rate and equilibrium constants for IET in SO have been described previously (65) and are summarized below. The laser flash photolysis apparatus system has been extensively described (66) as has been the basic photochemical process by which 5-deazariboflavin semiquinone (dRFH•) is generated by reaction between triplet state dRF and the sacrificial reductant and used to reduce redox-active proteins (eqs. 2-5) (67-69).
| (2) |
| (3) |
| (4) |
| (5) |
The IET rate constant can be calculated by fitting the heme reoxidation curve with an exponential function (eq. 6) where the IET rate constant is the sum of the forward and the reverse electron transfer rate constants (kf and kr, respectively, in eq. 7).
| (6) |
| (7) |
The equilibrium constant can then be calculated using the parameters a and b, which are determined from the kinetic traces. A0 is the absorbance extrapolated to t = 0, assuming that the photochemically-induced reduction of SO is instantaneous.
| (8) |
| (9) |
| (10) |
The forward and reverse rate constants (kf and kr, respectively) of IET can then be calculated from the equilibrium constant (eq. 10), thereby providing quantitative information about interdomain electron transfer in the enzyme. Note that kf in these flash photolysis experiments is actually the reverse of the net physiological IET direction.
Tether Linking the Heme and Molybdenum Domains of SO: Past and Recent Studies
Until recently, little has been known about the role of the tether within animal SO (Figure 2), and this linkage is not present in the plant and bacterial SOE's. The initial work on this tether was performed by Feng and co-workers and has been discussed previously (24, 70). They proposed that interdomain motion that decreases the Mo to Fe distance is essential for rapid IET and that the flexible tether linking the two domains of SO facilitates this motion. Consistent with this, laser photolysis revealed that the IET rate constant in CSO decreased with increasing solvent viscosity. Steady-state kinetics and EPR measurements on SO were also consistent with this hypothesis (70).
Kawatsu and Beratan have used a computational model to explore the mechanisms of IET in proteins such as SO, in which domains containing two cofactors are linked by a flexible tether (71). They found that the constrained conformational flexibility of the tether introduces an entropic component to the effective donor–acceptor interaction potential that produces a kinetic regime intermediate between “unimolecular” and “bimolecular”. Their calculations predicted that for SO the tether length may control the transition between the electron tunneling and diffusion-limited regimes. Recently, Pushie and George used molecular dynamics calculations to investigate the large-scale domain motions of CSO (72). They observed motion of the N-terminal heme domain into an orientation similar to that postulated for rapid electron transfer. The simulations also probed the dynamics of specific active site residues to further understand the structural and thermodynamic details of SO (72). Very recently, Utesch and Mroginski investigated the interdomain motion of CSO by combining steered molecular dynamics with all-atom molecular dynamics (73). They obtained a stable 3D “docked” structure with a relatively short Mo···Fe distance (~19 Å), similar to that proposed previously from the rapid IET rates measured experimentally (24, 65). The best theoretically calculated IET pathways for the “docked” model involved 1-3 interfacial water molecules and gave IET rates that were within an order of magnitude of the experimental values.
Prior to 2010 there were no experimental data from site-specific mutants to investigate the role of conformational changes of the tether that had been proposed from the effects of viscosity on IET (70) and from theoretical calculations (71). Recently, the proposed role(s) of the tether in facilitating conformational change and reactivity have been explored by altering both the length and flexibility of the tether in HSO by site-specific mutagenesis, and the reactivities of the resulting variants were studied using laser flash photolysis and steady-state kinetics assays (79).
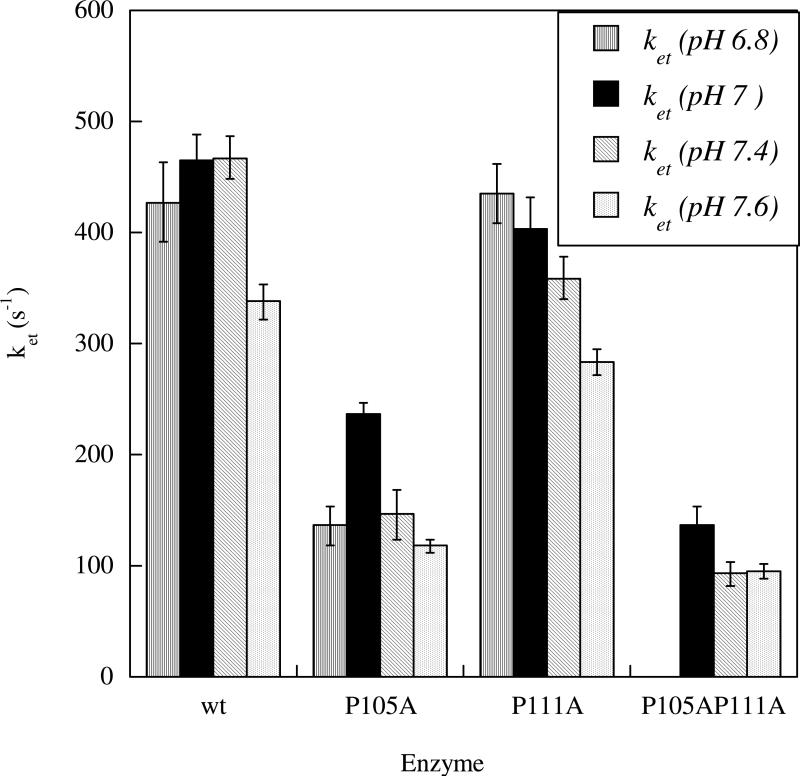
In order to change the flexibility of the tether, two conserved proline residues were mutated to alanines, P105 (adjacent the heme domain) and P111 (at the center of the tether) (Figure 2). The double mutant, P105A/P111A has also been made. While P105 is conserved within CSO and HSO, P111 is not. However, P111 is conserved within other mammalian forms. Figure 7 illustrates the dependence of ket on pH and mutation. The data show that the P105A mutation decreases ket by approximately 3-fold at pH 7.4. In contrast, the P111A mutation has only a minimal effect on ket. The IET rate constants for the P105A/P111A double mutant are similar to those for P105A, suggesting that the P105A change is the primary cause of the decrease in IET that is observed in this mutant.
Figure 7.
IET Rate Constants for the Proline to Alanine Tether Mutants (79).
The equilibrium constants (Keq) for the P111A mutant show a pH dependence with a trend that is similar to wt, i.e. Keq increases with increasing pH. However, this trend is not seen with P105A. The kinetic data indicate that the P111A mutation in the middle of the tether has little effect upon the IET kinetics. However, the much larger effects on the IET kinetics for the P105A mutation suggest that this conserved proline, which is adjacent to the heme domain, promotes a tether conformation that results in optimal IET.
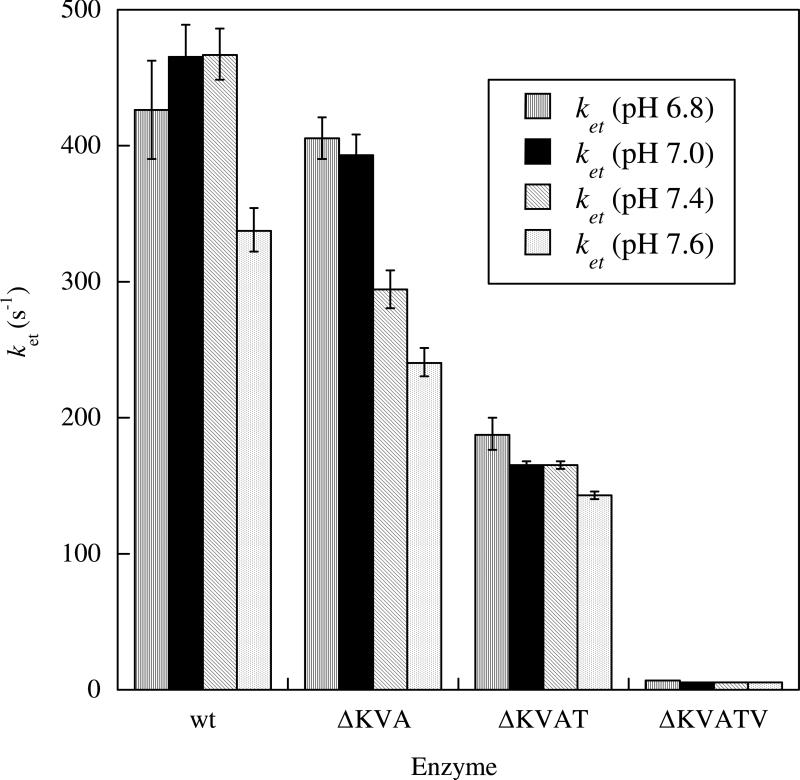
In addition to these proline to alanine mutations, several non-conserved residues (K108, V109, A110, T112 and V113, Figure 2) have been deleted from the tether in order to determine the effects of tether length on the IET kinetics of HSO. Deletions of three (ΔK108V109A110 (ΔKVA)), four (ΔK108V109A110T112 (ΔKVAT)), and five (ΔK108V109A110T112V113 (ΔKVATV)) non-conserved amino acids from the tether result in steadily decreasing IET rate constants, from 467 ± 19 s-1 in wt HSO down to 5.59 ± 0.03 s-1 in ΔKVATV HSO at pH 7.4 (Figure 8). Interestingly, the largest decrease occurs on deletion of the fifth residue (ΔKVATV). Additionally, all three deletion mutants showed decreasing IET rates with increasing pH from 6.8 to 7.6. Keq was significantly larger for ΔKVATV HSO (0.91 ± 0.03) in comparison to the wt HSO value (0.46 ± 0.02) and the other tether deletion mutants, but all the enzymes have a maximal Keq around pH 7.4. To further understand the cause of this large increase in Keq for ΔKVATV, spectroelectrochemistry studies were performed on wt and ΔKVATV HSO (Table 1), which will be discussed in the spectroelectrochemistry section of this review.
Figure 8.
IET Rate Constants for Tether Deletion Mutants (79).
Table 1.
Electrochemical Midpoint Potentials of SOEs cofactorsa
| HSO | CSO | SDH | P105A HSO | ΔKVATV HSO | |
|---|---|---|---|---|---|
| pH | 7.5 | 7.5 | 7.1 | 7.5 | 7.5 |
| Heme Cofactor (Fe (III/II)) | + 62 ± 2c (79) | ~+ 69d (78) | + 177d (82) | + 56 ± 3c (79) | + 42 ± 2c (79) |
| Molybdenum Cofactor (VI/V) | + 41 ± 1b (79) | n.d. | + 264d (82) | + 36 ± 1b (79) | + 41 ± 1b (79) |
All potentials are in mV vs SHE.
Calculated from ΔEoReaction (Heme-Mo) and the experimental heme potential.
Determined by spectroelectrochemistry, see Figure 9.
Determined by Protein Film Voltammetry
In parentheses are the cited references.
The steady-state kinetics revealed that the largest effect observed was on the Michaelis-Menten constants (Km) for P111A and P105A/P111A, which were approximately five-fold higher than for wt (data not shown). The increase in Km caused a 30% decrease in the catalytic efficiency (kcat/Km) of the enzyme. From these results it was concluded that the P111A mutation may affect the binding of sulfite to the enzyme. In addition, the steady-state data revealed that the kcat values for each of these mutations are much smaller than ket. Therefore, the IET process is not rate-limiting in catalysis, which is also true of the wt enzyme.
The steady-state kinetic parameters for ΔKVA and ΔKVAT HSO, are slightly higher in comparison to the wt HSO. However, the turnover number for ΔKVATV HSO (10.6 ± 0.3 s-1) is approximately one-third that of wt (26.9 ± 0.5) at pH 8.0. Unlike the proline to alanine mutations, IET for the ΔKVATV deletion mutant became rate limiting in catalysis. Thus, decreasing the length of the tether apparently influences the docking of the heme domain onto the molybdenum domain, thereby changing the distance and relative orientation of the two redox centers. This effect is the probable cause for the decrease in ket.
Very recently the tether sequence of CSO (Figure 2) was introduced into HSO. The expectation was that this chimeric HSO protein would show IET kinetics that were faster than wt HSO, and more similar to those of CSO. Surprisingly, however, IET for the chimeric HSO was actually slower than wt HSO (74). This result suggests that the composition of the non-conserved tether sequence of animal SOs may have become optimized for individual species. The kinetic results for all of the tether mutants point to a critical role for the tether in facilitating the IET reaction between the heme and molybdenum centers.
Mechanistic considerations based upon conserved arginine residues in the active site of SOE's
A new analysis of possible mechanistic pathways of sulfite oxidizing enzymes has emerged from laser flash photolysis and EPR studies on wt and R55 variants of SDH (34). Three variants of SDH (R55M, R55Q and R55K) were made to explore the role of R55 in the pathway for IET in SDH. The SDHR55Q variant had the same Arg to Gln substitution that is observed in R160Q in humans, which is a fatal mutation, causing sulfite oxidase deficiency. HSOR160Q shows a substantial decrease in IET, which is almost 400 fold lower than HSOwt (122 ± 5 s-1) (21). For SDHR55Q the laser flash photolysis data revealed that ket was 3-fold larger than for SDHwt. Both Keq and ket were mildly pH dependent within SDHR55Q. At pH 5.1, there was a 2-fold increase in Keq for this variant (34).
The IET rate constant for SDHR55K was unchanged with varying pH, which is a similar trend to SDHwt. However, the IET rate could not be determined at pH 6.0. IET in SDHR55K is much slower (15 ± 0.7 s-1) when compared to SDHwt. Between pH 5.1 (0.59 ± 0.02) and 5.5 (0.34 ± 0.03) the SDHR55K variant showed a decrease in Keq. These data are much different than those for HSOR160K, which showed a decrease in IET.
In the SDHR55M variant the hydrogen bonding interactions within the active site are disrupted. The IET rate constant for SDHR55M (217 ± 25 s-1) was 2-fold larger than SDHwt (122 ± 5 s-1) at pH 6, and increased 4-fold (535 ± 44) by pH 5.1. While the IET rate constant for SDHR55M was pH-dependent; Keq was nearly pH independent. These behaviors are the opposite of those observed for SDHwt. Therefore, both the forward and the reverse IET rate constants for SDHR55M increase with decreasing pH, raising the possibility that SDHR55M has a more efficient tunneling pathway for IET (21, 34).
The structure of SDHR55M shows that there is a sulfate anion bound within the active site, which is different from SDHwt, SDHY236F (30) and SDHH57A (31). The pulsed EPR studies of SDHR55M show only a hpH–type signal, similar to SDHwt enzyme, with no pH dependence (53). SDHR55Q exhibits the blocked form, with bound sulfate at low pH, which was verified by 33S couplings observed upon reduction with 33S-labeled sulfite (53). This blocked feature has also been observed in the fatal mutant, HSOR160Q (22).
SDHR55M and SDHR55Q are the first SOE active site variants to display an increase in the IET rate constant. This effect is the opposite of that observed in HSOR160Q (21). These results indicate that the conserved arginine within the active site of SOEs is not a required part of the electron transfer pathway between the heme and molybdenum domains. However, the result is consistent with the positively charged arginine influencing the docking between the two domains in animal SO. Additional kinetic studies of SDH catalysis have shown that the R55 residue is important for effective substrate binding, and that KMsulfite,app for SDHR55M is 2-3 orders of magnitude larger than that for SDHwt (31). This result is also consistent with that data for SDHR55K, which showed a KMsulfite,app factor of ~10 times larger than that for SDHwt (31).
Based upon the kinetic data and the pulsed EPR studies of SDHwt and variants, and similar studies on HSOR160Q, alternative mechanistic possibilities for the catalytic cycle of SOEs must be considered. Scheme 1, proposed by Emesh et al., summarizes alternative possibilities for the catalytic cycle of SOEs (34). The sequence colored in red is the prevailing view (1, 24, 75), in which product is released (k2) from the enzyme-substrate complex. This is followed by two sequential one-electron oxidations of the Mo center (k4 and k7). This mechanism was discussed previously above (see Figure 4).
A second alternative (colored in blue) would be for the enzyme-substrate complex to be oxidized by one electron (k3 in Scheme 1). This is followed by product dissociation (k5) and then another oxidation by one electron (k7). Emesh et al. suggested that this pathway is operative in SDHwt and the R55 variants because IET rates are significantly faster than catalytic turnover (34). Direct evidence for the one-electron oxidized enzyme-substrate complex, i.e., the MoV(OSO32-) species of Scheme 1, has been obtained by pulsed EPR studies of HSOR160Q that has been reduced by sulfite labeled with 33S (I=3/2) (22) and other blocked forms of SOEs (31, 52, 53).
A third possibility shown in black in Scheme 1 is for the enzyme-substrate complex to be oxidized by two electrons back to the resting state Mo(VI) (k3 and k6) before release of sulfate (k8). The IET and steady-state kinetics of the SDHY236F variant are consistent with this pathway. This variant is catalytically competent even though it does not exhibit IET by laser flash photolysis (steps k7 and k-7) (30).
Kinetics of Plant SO
Hille and co-workers have recently reported in-depth rapid kinetics and steady-state studies of PSO, using stopped-flow methods. The data demonstrated that superoxide is produced during the course of the reaction between the reduced enzyme and molecular oxygen and then decays to hydrogen peroxide (76). The rate constant was linearly dependent on the concentration of molecular oxygen, and the second-order rate constant was kox = 8.7 ± 0.5 × 104 M-1s-1. They also found that the reaction, in the presence of cyt c was biphasic, with a fast and a slow phase. This result indicated that the first equivalent of superoxide was generated from the rapid one-electron oxidation of the fully reduced Mo(IV) state to Mo(V). This reaction was followed by the slower oxidation of the Mo(V) to the Mo(VI) state.
Electrochemistry
Studies of Animal SO
As discussed above, SO is a multi-domain enzyme, containing heme and molybdenum redox centers. An extensive micro-coulometric study of native CSO by Spence et al. investigated the reduction potentials of the Fe(III/II), Mo(VI/V) and Mo(V/IV) couples as a function of pH and anions in the medium (77). The Fe(III/II) couple was nearly independent of pH, whereas the Mo(VI/V) couple showed a pH dependence of ~60 mV per pH unit, characteristic of an accompanying proton transfer. More recently, Elliott et al. investigated native CSO by protein film voltammetry (PFV) (78). A catalytic current was observed in the “potential domain” of CSO upon addition of sulfite. From the limiting current, they obtained a turnover number, kcat of 2-4 s-1, which is significantly slower than the reported turnover numbers of SO in solution (100 s-1) (38). The PFV data suggested that a large fraction of the SO molecules immobilized on the electrode surface do not engage in catalysis, but undergo non-catalytic electron transfer. The observed behavior is independent of the electrode and does not seem to be an artifact of the surface-protein interaction. The midpoints of the catalytic waves (Emid) in the PFV experiments show very small pH dependence, which is only consistent with the wave arising from action at the Fe(III/II) center (77) and identifies it as the central distributive site for catalytic ET. The redox potential at pH 7.5 is approximately +65 mV vs SHE, which is similar to the heme potential of HSOwt determined from spectroelectrochemistry (see discussion below) (79).
The reduction potential of the b5 heme of wt and mutant HSO has recently been measured using spectroelectrochemistry, following procedures described previously for heme proteins (77, 79-81). Since the ratio of oxidized to reduced forms of each redox species is directly related to the absorbances of the optical spectra via Beer's law (assuming the extinction coefficients of the reduced and oxidized species are different at the chosen wavelength), the change in absorbance with respect to applied potential can be fit to the Nernst equation (eq. 11) using the nonlinear-least-squares fitting algorithm in the software Origin©.
| (11) |
where Eapp is the applied potential, E0 is the midpoint potential determined from these data, and [Ox] and [Red] are, respectively, the concentrations of the Fe(III) and Fe(II) states of the b5 heme of HSO.
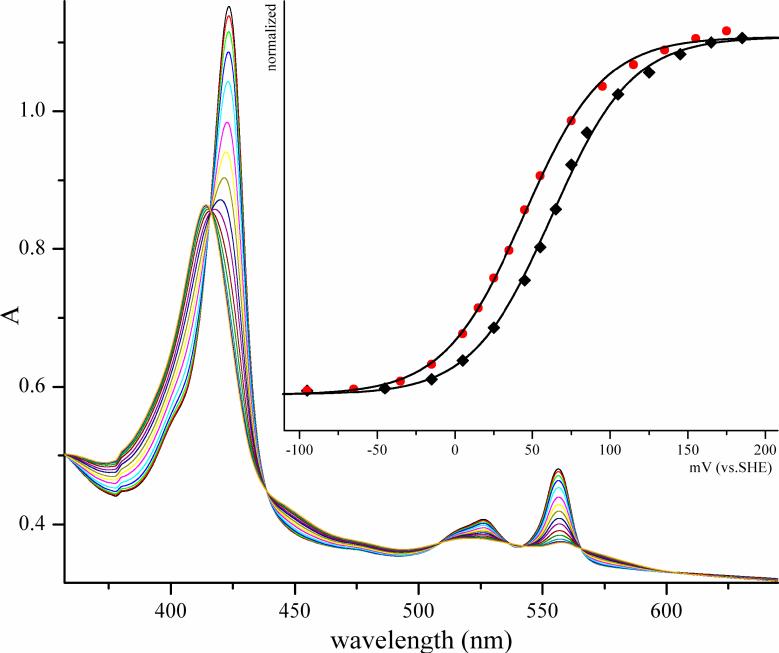
Based upon kinetic results by Johnson-Winters et al. (79) the laser flash photolysis data for ΔKVATV resulted in Keq values that were approximately 0.91, compared to 0.46 for the wt. To confirm whether this was a result of the change in the midpoint potential between the heme and molybdenum, spectroelectrochemistry studies were performed to determine the Fe(III/II) potentials for wt and ΔKVATV HSO. The corresponding midpoint potentials vs. SHE are +62 ± 2 mV for wt HSO and +44 ± 2 mV for ΔKVATV HSO (Figure 9, Table 1). Thus, the Fe (III/II) potential for ΔKVATV HSO is 18 ± 3 mV more negative than wt, supporting the laser flash photolysis kinetic data and favoring the Fe(III)/Mo(V) species in eq. 5. Using the Keq value from the laser flash photolysis data and the heme potential, the potential of the molybdenum center can be calculated, as well. The calculated potentials of the Mo(VI)/Mo(V) couple for ΔKVATV and wt HSO are +41 ± 2 and +42 ± 2 mV, respectively (Table 1). Therefore, it was concluded that the change in Keq for ΔKVATV was primarily due to a change in the potential of the heme. Perhaps the deletion of five amino acids from the tether changes the exposure of the heme to solvent, thereby causing a shift in the potential and in Keq.
Figure 9.
Spectroelectrochemical titration of the b5 heme of wild-type HSO at pH 7.5 and 27°C (79). The inset shows the fit of the data to eq. 11 at 413 nm (78); black = wt; red = ΔKVATV mutation.
Spectroelectrochemistry has also been used to measure the Fe(III/II) potential of the P105A mutant which shows much slower rates of IET compared to wt, but similar Keq values (Figure 7). Table 1 shows that the Fe(III/II) potential for P105A is essentially identical to wt, as is the calculated Mo(VI/V) potential. Thus, the P105A mutation does not change the thermodynamic driving force for IET (as seen from Keq) or the individual potentials of the two redox centers. Therefore, the 2-4 fold decrease in ket for P105A mutant compared to wt (Figure 7) presumably reflects differences in their IET pathways.
Studies of SDH
From PFV studies of SDHwt on a pyrolytic graphite working electrode, McEwan and coworkers identified the heme and molybdenum potentials as: Em,8 (Fe(III/II)) +177 mV; Em,8 (Mo(VI/V)) +211 mV; and Em,8 (Mo(V/IV)) -118 mV vs SHE (82). The Michaelis-Menten constant (Km) for SDHwt obtained upon saturation of the catalytic current with sulfite was similar to previous steady-state assay values (26 μM) (13). Also, using square wave voltammetry, they were able to show that the SDHwt potentials of the heme and molybdenum exhibit a pH dependence.
Attempts to extend the PFV measurements to the SDHR55 variants, however, were not successful (34). Also, it was not feasible to directly determine the Mo(VI/V) potentials of the SDHR55 variants by EPR titrations, because of the large amount of protein required for such experiments. However, the Mo(VI/V) potentials of SDHwt can be calculated by using Keq (1.57 at pH 5.2) from the laser flash photolysis experiments and the Fe(III/II) potential determined from spectroelectrochemistry, as described above for HSO. For SDHwt this procedure gave a ΔE° of 12 mV and a calculated Mo(VI/V) potential of 196 mV (34). This calculated Mo(VI/V) potential is very different from the +381 mV reported for the direct experimental measurement for SDHwt at pH 5.2 by PFV (82). It is unknown why these potential values vary by so much. However, it could be due to the fact that the technique of PFV requires adsorption of the protein on an electrode surface and immobilization within a surfactant film. This is very different than spectroelectrochemical determination of the potential in solution (34).
Conclusions
As noted above, a review of SOEs in 2007 by Feng (24) and coworkers concluded with a list of topics to be addressed in future research in this area. Substantial progress has already been made on several of these topics by the combined approaches of molecular biology, rapid kinetics, advanced spectroscopy, protein crystallography, and computational modeling. Scheme 2 summarizes the current status of research on SOEs and some of the remaining challenges. Specifically, recent advances include:
Flash photolysis studies of variants of HSO and of bacterial SDH have provided new insights concerning the amino acids involved in direct IET through the protein medium. In humans, the HSOR160Q mutation is fatal, and the rate of IET in the purified protein is dramatically reduced relative to HSOwt. However, the analogous SDHR55Q mutation of the conserved active site arginine in bacterial SDH does not decrease the rate of IET. In SDH the heme and Mo domains are locked in place by strong interactions between the two subunits, whereas in HSO the heme and Mo domains are linked by a flexible tether and domain reorientation is required in order to accomplish IET. These combined flash photolysis results suggest that in HSO the primary role of the positively charged R160 residue is to aid in docking the heme domain with the Mo domain for efficient IET and catalysis (34). The relative unimportance of R55 in SDH implies that this conserved active site arginine is not directly on the pathway for IET. Calculations of the IET pathway(s) for SDH should provide further insight concerning this hypothesis.
Substantial progress has been made in elucidating the effect of the tether linking the heme and Mo domains of HSO on the rates of IET and steady-state kinetics. We have shown that both the length and composition of the tether strongly affect the rate of IET (79). Theoretical calculations of the effects of interdomain motion on IET are beginning to appear (71), and the first molecular mechanics calculations on the time evolution of the structure of CSO have recently been described (72, 73). The increasing availability of GPU-based computer clusters should make molecular mechanics calculations of longer time trajectories of interdomain motions in SO more computationally tractable in the future (83). This is critical since IET occurs on the millisecond time scale, which is presently computationally inaccessible.
Pulsed EPR spectroscopy of “difficult” nuclei (17O, 33S, 35,37Cl) with I > ½ has been combined with DFT calculations to elucidate the structures of the Mo(V) states of SOEs and to gain insight concerning their catalytic mechanisms. The long-standing question of the structure of the lpH form of SO has been resolved from 35,37Cl pulsed EPR and DFT calculations; the chloride ion is in the second coordination sphere and hydrogen-bonded to the equatorial Mo-OH group and surrounding water molecules and proton donors from amino acid side chains (50). Observation of 33S couplings in SOEs reduced by 33S-labeled sulfite at low pH in chloride-depleted solutions has shown that the blocked form is a common Mo(V) species in SOEs. Addition of excess chloride converts the blocked form to the lpH form in most cases (54). DFT calculations on models for 33S- and 17O-labeled blocked SO suggest that a structure with O-bound sulfite is most likely. The weakly coupled 17O atom detected in the blocked form is assigned to remote oxygen atoms of the sulfite (sulfate) ligand (61). Detection of the nqi parameters for strongly coupled equatorial 17O ligands remains difficult, but recent advances in W-band ESEEM methods hold promise for future advances (84).
X-ray crystal structures of several variants of SDH (30, 31) have appeared, and structures of the Mo domain of recombinant CSO and CSOR138Q have been described (28). However, to date the only structure of an intact form of vertebrate SO is the original report of native CSO at 1.9 Å resolution (26). The crystallization and structure determination of recombinant HSO, both wt and mutants, remain as challenges for future research on SO.
Scheme 2.
Overview of the current state of research on sulfite oxidizing enzymes (SOEs). From bioinformatic analyses, the diverse sulfite oxidase family of proteins that contains the same molybdenum cofactor center (1) can be classified into at least three groups (63, 85). This review has focused on the SOEs. Animal sulfite oxidases possess two redox active domains and present fundamental biophysical problems relating to intramolecular electron transfer (IET), the relationship of IET rates to steady-state kinetics, the overall conformation of the protein, and the molecular dynamics of the motion of the two domains relative to one another. The effects of extensive mutations of the tether connecting the heme and molybdenum domains of human SO have been discussed here. However, as yet there are no X-ray structural results of the intact protein for any of these variants. The detailed structures of the molybdenum centers of these variants have been determined from analysis of high resolution pulsed EPR spectra of their Mo(V) states as a function of pH, anions in the media, and mutations of nearby amino acid residues. A long term hope is that these studies will contribute to the continuing development of molecular medicine to treat sulfite oxidase deficiency (13, 16).
Acknowledgements
We thank Drs. Andrei Astashkin, Robert E. Berry, Changjian Feng, James T. Hazzard, Russ Hille, Ulrike Kappler, Eric L. Klein, Arnold Raitsimring, Asha Rajapakshe, Mr. Robert Byrne and Ms. Safia Emesh for helpful discussions.
This research was supported by NIH Grant GM-037773 (to JHE); Ruth L. Kirchstein-NIH Fellowship 1F32GM082136-01 (to KJW)
Abbreviations
- SO
sulfite oxidase
- SOE
sulfite oxidizing enzymes
- dRF
5-deazariboflavin
- DFT
density functional theory
- EPR
electron paramagnetic resonance
- SDH
sulfite dehydrogenase
- NR
nitrate reductase
- CSO
chicken sulfite oxidase
- HSO
human sulfite oxidase
- PSO
plant sulfite oxidase
- cyt c
cytochrome c
- IET
intramolecular electron transfer
- ket
electron transfer rate constant
- Keq
equilibrium constant for intramolecular electron transfer
- kf and kr
microscopic rate constants for the forward and reverse directions, respectively, of intramolecular electron transfer
- Moco
molybdopterin cofactor
- wt
wild type
- cPMP
cyclic pyranopterin monophosphate
- nqi
nuclear quadrupole interaction
- hfi
hyperfine interaction
- lpH
low pH
- hpH
high pH
- Pi
phosphate inhibited
References
- 1.Hille R. The Mononuclear Molybdenum Enzymes. Chem. Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopalan KV, Johnson JL. Sulfite Oxidase. In: Creighton TE, editor. Wiley Encyclopedia of Molecular Medicine. Wiley; New York: 2002. pp. 3048–3051. [Google Scholar]
- 3.Kisker C. Sulfite Oxidase. In: Messerschmidt A, Huber R, Poulos T, Wieghardt K, editors. Handbook of Metalloproteins. John Wiley and Sons, Ltd; New York: 2001. pp. 1121–1135. [Google Scholar]
- 4.Schindelin H, Kisker C, Rajagopalan KV. Molybdopterin from Molybdenum and Tungsten Enzymes. Adv. Protein Chem. 2001;58:47–94. doi: 10.1016/s0065-3233(01)58002-x. [DOI] [PubMed] [Google Scholar]
- 5.Cohen HJ, Betcher-Lange S, Kessler DL, Rajagopalan KV. Hepatic Sulfite Oxidase. Congruency in Mitochondria of Prosthetic Groups and Activity. J. Biol. Chem. 1972;247:7759–7766. [PubMed] [Google Scholar]
- 6.Kessler DL, Johnson JL, Cohen HJ, Rajagopalan KV. Visualization of Hepatic Sulfite Oxidase in Crude Tissue Preparations by Electron Paramagnetic Resonance Spectroscopy. Biochim. Biophys. Acta. 1974;334:86–96. [Google Scholar]
- 7.Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, Koch B, Hille R, Hansch R, Mendel RR. Identification and Biochemical Characterization of Arabidopsis thaliana Sulfite Oxidase. A New Player in Plant Sulfur Metabolism. J. Biol. Chem. 2001;276:46989–46994. doi: 10.1074/jbc.M108078200. [DOI] [PubMed] [Google Scholar]
- 8.Hansch R, Lang C, Riebeseel E, Lindigkeit R, Gessler A, Rennenberg H, Mendel RR. Plant Sulfite Oxidase as Novel Producer of H2O2—Combination of Enzyme Catalysis With a Subsequent Nonenzymatic Reaction Step. J. Biol. Chem. 2006;281:6884–6888. doi: 10.1074/jbc.M513054200. [DOI] [PubMed] [Google Scholar]
- 9.Kappler U, Bennett B, Rethmeier J, Schwarz G, Rainer-Deutzmann R, McEwan AG, Dahl C. Sulfite: Cytochrome c oxidoreductase from Thiobacillus novellus. Purification, Characterization, and Molecular Biology of a Heterodimeric Member of the Sulfite Oxidase Family. J. Biol. Chem. 2000;275:13202–13212. doi: 10.1074/jbc.275.18.13202. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JL. Prenatal Diagnosis of Molybdenum Cofactor Deficiency and Isolated Sulfite Oxidase Deficiency. Prenat. Diagn. 2003;23:6–8. doi: 10.1002/pd.505. [DOI] [PubMed] [Google Scholar]
- 11.Dublin AB, Hald JK, Wootton-Gorges SL. Isolated Sulfite Oxidase Deficiency: MR Imaging Features. Am. J. Neuroradiol. 2002;23:484–485. [PMC free article] [PubMed] [Google Scholar]
- 12.Sass JO, Gunduz A, Araujo C, Funayama R, Korkmaz B, Giselle K, Pinto D, Tuysuz B, Lam C-W, Reiss J, Walter M, Yalcinkaya C, Camelo Junior JS. Functional Deficiencies of Sulfite Oxidase: Differential Diagnoses in Neonates Presenting with Intractable Seizures and Cystic Encephalomalacia. Brain & Development. 2009 doi: 10.1016/j.braindev.2009.09.005. in press. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz G, Santamaria-Araujo JA, Wolf S, Lee JH, Adham IM, Grone HJ, Schwegler H, Sass JO, Otte T, Hanzelmann P, Mendel RR, Engel W, Reiss J. Rescue of Lethal Molybdenum Cofactor Deficiency by a Biosynthetic Precursor from Escherichia coli. Hum. Mol. Genet. 2004;13:1249–1255. doi: 10.1093/hmg/ddh136. [DOI] [PubMed] [Google Scholar]
- 14.Matthies A, Rajagopalan KV, Mendel RR, Leimkuhler S. Evidence for the Physiological Role of a Rhodanese-Like Protein for the Biosynthesis of the Molybdenum Cofactor in Humans. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5946–5951. doi: 10.1073/pnas.0308191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JL, Duran M. Molybdenum Cofactor Deficiency and Isolated Sulfite Oxidase Deficiency. McGraw-Hill; New York: 2001. pp. 3163–3177. [Google Scholar]
- 16.Veldman A, Santamaria-Araujo JA, Sollazzo S, Pitt J, Gianello R, Yaplito-Lee J, Wong F, Ramsden CA, Reiss J, Cook I, Fairweather J, Schwarz G. Successful Treatment of Molybdenum Cofactor Deficiency Type A with cPMP. Pediatrics. 2010;125:e1249–54. doi: 10.1542/peds.2009-2192. [DOI] [PubMed] [Google Scholar]
- 17.Johnson JL, Waud WR, Rajagopalan KV, Duran M, Beemer FA, Wadman SK. Inborn Errors of Molybdenum Metabolism: Combined Deficiencies of Sulfite Oxidase and Xanthine Dehydrogenase in a Patient Lacking the Molybdenum Cofactor. Proc. Natl. Acad. Sci. U. S. A. 1980;77:3715–3719. doi: 10.1073/pnas.77.6.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendel RR, Bittner F. Cell Biology of Molybdenum. Biochim. Biophys. Acta. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Temple CA, Graf TN, Rajagopalan KV. Optimization of Expression of Human Sulfite Oxidase and its Molybdenum Domain. Arch. Biochem. Biophys. 2000;383:281–287. doi: 10.1006/abbi.2000.2089. [DOI] [PubMed] [Google Scholar]
- 20.Feng C, Wilson HL, Tollin G, Astashkin AV, Hazzard JT, Rajagopalan KV, Enemark JH. The Pathogenic Human Sulfite Oxidase Mutants G473D and A208D are Defective in Intramolecular Electron Transfer. Biochemistry. 2005;44:13734–13743. doi: 10.1021/bi050907f. [DOI] [PubMed] [Google Scholar]
- 21.Feng C, Wilson HL, Hurley JK, Hazzard JT, Tollin G, Rajagopalan KV, Enemark JH. Essential Role of Conserved Arginine 160 in Intramolecular Electron Transfer in Human Sulfite Oxidase. Biochemistry. 2003;42:12235–12242. doi: 10.1021/bi0350194. [DOI] [PubMed] [Google Scholar]
- 22.Astashkin AV, Johnson-Winters K, Klein EL, Feng C, Wilson HL, Rajagopalan KV, Raitsimring AM, Enemark JH. Structural Studies of the Molybdenum Center of the Pathogenic R160Q Mutant of Human Sulfite Oxidase by Pulsed EPR Spectroscopy and 17O and 33S Labeling. J. Am. Chem. Soc. 2008;130:8471–8480. doi: 10.1021/ja801406f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson HL, Wilkinson SR, Rajagopalan KV. The G473D Mutation Impairs Dimerization and Catalysis in Human Sulfite Oxidase. Biochemistry. 2006;45:2149–2160. doi: 10.1021/bi051609l. [DOI] [PubMed] [Google Scholar]
- 24.Feng C, Tollin G, Enemark JH. Sulfite Oxidizing Enzymes. Biochim. Biophys. Acta. 2007;1774:527–539. doi: 10.1016/j.bbapap.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hille R. Plants have SOX: the Structure of Sulfite Oxidase from Arabidopsis thaliana. Structure. 2003;11:1189–1190. doi: 10.1016/j.str.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Kisker C, Schindelin H, Pacheco A, Wehbi WA, Garrett RM, Rajagopalan KV, Enemark JH, Rees DC. Molecular Basis of Sulfite Oxidase Deficiency from the Structure of Sulfite Oxidase. Cell. 1997;91:973–983. doi: 10.1016/s0092-8674(00)80488-2. [DOI] [PubMed] [Google Scholar]
- 27.Kappler U, Bailey S. Crystallization and Preliminary X-ray Analysis of Sulfite Dehydrogenase from Starkeya novella. Acta. Crystallogr. D. Biol. Crystallogr. 2004;60:2070–2072. doi: 10.1107/S0907444904022395. [DOI] [PubMed] [Google Scholar]
- 28.Karakas E, Wilson HL, Graf TN, Xiang S, Jaramillo-Busquets S, Rajagopalan KV, Kisker C. Structural Insights Into Sulfite Oxidase Deficiency. J. Biol. Chem. 2005;280:33506–33515. doi: 10.1074/jbc.M505035200. [DOI] [PubMed] [Google Scholar]
- 29.Kappler U, Bailey S. Molecular Basis of Intramolecular Electron Transfer in Sulfite-Oxidizing Enzymes is Revealed by High Resolution Structure of a Heterodimeric Complex of the Catalytic Molybdopterin Subunit and a c-Type Cytochrome Subunit. J. Biol. Chem. 2005;280:24999–25007. doi: 10.1074/jbc.M503237200. [DOI] [PubMed] [Google Scholar]
- 30.Kappler U, Bailey S, Feng C, Honeychurch MJ, Hanson GR, Bernhardt PV, Tollin G, Enemark JH. Kinetic and Structural Evidence for the Importance of Tyr236 for the Integrity of the Mo Active Site in a Bacterial Sulfite Dehydrogenase. Biochemistry. 2006;45:9696–9705. doi: 10.1021/bi060058b. [DOI] [PubMed] [Google Scholar]
- 31.Bailey S, Rapson T, Johnson-Winters K, Astashkin AV, Enemark JH, Kappler U. Molecular Basis for Enzymatic Sulfite Oxidation: How Three Conserved Active Site Residues Shape Enzyme Activity. J. Biol. Chem. 2009;284:2053–2063. doi: 10.1074/jbc.M807718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrader N, Fischer K, Theis K, Mendel RR, Schwarz G, Kisker C. The Crystal Structure of Plant Sulfite Oxidase Provides Insights into Sulfite Oxidation in Plants and Animals. Structure. 2003;11:1251–1263. doi: 10.1016/j.str.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Hänsch R, Lang C, Rennenberg H, Mendel RR. Significance of Plant Sulfite Oxidase. Plant Biol. 2007;9:589–595. doi: 10.1055/s-2007-965433. [DOI] [PubMed] [Google Scholar]
- 34.Emesh S, Rapson TD, Rajapakshe A, Kappler U, Bernhardt PV, Tollin G, Enemark JH. Intramolecular Electron Transfer in Sulfite-Oxidizing Enzymes: Elucidating the Role of a Conserved Active Site Arginine. Biochemistry. 2009;48:2156–2163. doi: 10.1021/bi801553q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hille R. Molybdenum Enzymes. Essays Biochem. 1999;34:125–137. doi: 10.1042/bse0340125. [DOI] [PubMed] [Google Scholar]
- 36.Hille R. The Reaction Mechanism of Oxomolybdenum Enzymes. Biochim. Biophys. Acta. (BBA)-Bioenerg. 1994;1184:143–169. doi: 10.1016/0005-2728(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 37.Brody MS, Hille R. The Reaction of Chicken Liver Sulfite Oxidase with Dimethylsulfite. Biochim. Biophys. Acta. 1995;1253:133–135. doi: 10.1016/0167-4838(95)00194-4. [DOI] [PubMed] [Google Scholar]
- 38.Brody MS, Hille R. The Kinetic Behavior of Chicken Liver Sulfite Oxidase. Biochemistry. 1999;38:6668–6677. doi: 10.1021/bi9902539. [DOI] [PubMed] [Google Scholar]
- 39.Astashkin AV, Feng C, Raitsimring AM, Enemark JH. 17O ESEEM Evidence for Exchange of the Axial Oxo Ligand in the Molybdenum Center of the High pH Form of Sulfite Oxidase. J. Am. Chem. Soc. 2005;127:502–503. doi: 10.1021/ja0461466. [DOI] [PubMed] [Google Scholar]
- 40.Astashkin AV, Raitsimring AM, Feng C, Johnson JL, Rajagopalan KV, Enemark JH. Pulsed EPR Studies of Nonexchangeable Protons Near the Mo(V) Center of Sulfite Oxidase: Direct Detection of the Alpha-Proton of the Coordinated Cysteinyl Residue and Structural Implications for the Active Site. J. Am. Chem. Soc. 2002;124:6109–6118. doi: 10.1021/ja0115417. [DOI] [PubMed] [Google Scholar]
- 41.Astashkin AV, Mader ML, Pacheco A, Enemark JH, Raitsimring AM. Direct Detection of the Proton-Containing Group Coordinated to Mo(V) in the High pH Form of Chicken Liver Sulfite Oxidase by Refocused Primary ESEEM Spectroscopy: Structural and Mechanistic Implications. J. Am. Chem. Soc. 2000;122:5294–5302. [Google Scholar]
- 42.Pacheco A, Basu P, Borbat P, Raitsimring AM, Enemark JH. Multifrequency ESEEM Spectroscopy of Sulfite Oxidase in Phosphate Buffer: Direct Evidence for Coordinated Phosphate. Inorg. Chem. 1996;35:7001–7008. doi: 10.1021/ic9607806. [DOI] [PubMed] [Google Scholar]
- 43.Lamy MT, Gutteridge S, Bray RC. Electron-Paramagnetic-Resonance Parameters of Molybdenum(V) in Sulphite Oxidase from Chicken Liver. Biochem. J. 1980;185:397–403. doi: 10.1042/bj1850397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutteridge S, Lamy MT, Bray RC. The Nature of the Phosphate Inhibitor Complex of Sulphite Oxidase from Electron-Paramagnetic-Resonance Studies Using Oxygen-17. Biochem. J. 1980;191:285–288. doi: 10.1042/bj1910285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bray RC, Gutteridge S, Lamy MT, Wilkinson T. Equilibria Amongst Different Molybdenum (V)-Containing Species from Sulphite Oxidase. Evidence for a Halide Ligand of Molybdenum in the Low-pH Species. Biochem. J. 1983;211:227–236. doi: 10.1042/bj2110227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enemark JH, Astashkin AV, Raitsimring AM. Investigation of the Coordination Structures of the Molybdenum(V) Sites of Sulfite Oxidizing Enzymes by Pulsed EPR Spectroscopy. Dalton Trans. 2006:3501–3514. doi: 10.1039/b602919a. [DOI] [PubMed] [Google Scholar]
- 47.Enemark JH, Astashkin AV, Raitsimring AM. High Resolution EPR Spectroscopy of Mo-Enzymes. Sulfite Oxidases: Structural and Functional Implications” in Biological Magnetic Resonance. In: Hanson GR, Berliner LJ, editors. Metals in Biology: Applications of High Resolution EPR to Metalloenzymes. Vol. 29. Springer; New York: 2010. pp. 121–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray RC. The Inorganic Biochemistry of Molybdoenzymes. Q. Rev. Biophys. 1988;21:299–329. doi: 10.1017/s0033583500004479. [DOI] [PubMed] [Google Scholar]
- 49.Doonan CJ, Wilson HL, Bennett B, Prince RC, Rajagopalan KV, George GN. Mo(V) Electron Paramagnetic Resonance of Sulfite Oxidase Revisited: the Low-pH Chloride Signal. Inorg. Chem. 2008;47:2033–2038. doi: 10.1021/ic7017083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein EL, Astashkin AV, Ganyushin D, Riplinger C, Johnson-Winters K, Neese F, Enemark JH. Direct Detection and Characterization of Chloride in the Active Site of the Low-pH Form of Sulfite Oxidase Using Electron Spin Echo Envelope Modulation Spectroscopy, Isotopic Labeling, and Density Functional Theory Calculations. Inorg. Chem. 2009;48:4743–4752. doi: 10.1021/ic801787s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bray RC, Lamy MT, Gutteridge S, Wilkinson T. Evidence from Electron-Paramagnetic-Resonance Spectroscopy for a Complex of Sulphite Ions With the Molybdenum Centre of Sulphite Oxidase. Biochem. J. 1982;201:241–243. doi: 10.1042/bj2010241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Astashkin AV, Johnson-Winters K, Klein EL, Byrne RS, Hille R, Raitsimring AM, Enemark JH. Direct Demonstration of the Presence of Coordinated Sulfate in the Reaction Pathway of Arabidopsis thaliana Sulfite Oxidase Using 33S labeling and ESEEM Spectroscopy. J. Am. Chem. Soc. 2007;129:14800–14810. doi: 10.1021/ja0704885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rapson TD, Astashkin AV, Johnson-Winters K, Bernhardt PV, Kappler U, Raitsimring AM, Enemark JH. Pulsed EPR Investigations of the Mo(V) Centers of the R55Q and R55M Variants of Sulfite Dehydrogenase from Starkeya novella. J. Biol. Inorg. Chem. 2010;15:505–514. doi: 10.1007/s00775-009-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajapakshe A, Johnson-Winters K, Nordstrom AR, Meyers KT, Emesh S, Astashkin AV, Enemark JH. Biochemistry. 2010;49:5154–5159. doi: 10.1021/bi902172n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cramer SP, Johnson JL, Rajagopalan KV, Sorrell TN. Observation of 17O Effects on Mo(V) EPR Spectra in Sulfite Oxidase, Xanthine Dehydrogenase, and MoO(SC6H5)4−. Biochem. Biophys. Res. Commun. 1979;91:434–439. doi: 10.1016/0006-291x(79)91540-7. [DOI] [PubMed] [Google Scholar]
- 56.Astashkin AV, Enemark JH, Raitsimring AM. Concepts Magn. Reson. Part B, (Magn. Reson. Engineering) 2006;29B:125–136. [Google Scholar]
- 57. http://www.thch.uni-bonn.de/tc/orca.
- 58.Astashkin AV, Klein EL, Ganyushin D, Johnson-Winters K, Neese F, Kappler U, Enemark JH. Exchangeable Oxygens in the Vicinity of the Molybdenum Center of the High-pH Form of Sulfite Oxidase and Sulfite Dehydrogenase. Phys. Chem. Chem. Phys. 2009;11:6733–6742. doi: 10.1039/b907029j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Astashkin AV, Neese F, Raitsimring AM, Cooney JJ, Bultman E, Enemark JH. Pulsed EPR Investigations of Systems Modeling Molybdenum Enzymes: Hyperfine and Quadrupole Parameters of oxo-17O in [Mo17O(SPh)4]−. J. Am. Chem. Soc. 2005;127:16713–16722. doi: 10.1021/ja055472y. [DOI] [PubMed] [Google Scholar]
- 60.Betts RH, Voss RH. The Kinetics of Oxygen Exchange Between the Sulfite Ion and Water. Can. J. Chem. 1970;48:2035–2041. [Google Scholar]
- 61.Enemark JH, Raitsimring AM, Astashkin AV, Klein EL. Implications for the Mechanism of Sulfite Oxidizing Enzymes from Pulsed EPR Spectroscopy and DFT Calculations for “Difficult” Nuclei. Faraday Discussions. 2010;148 doi: 10.1039/c004404k. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Workun GJ, Moquin K, Rothery RA, Weiner JH. Evolutionary Persistence of the Molybdopyranopterin-Containing Sulfite Oxidase Protein Fold. Microbiol. Mol. Biol. R. 2008:228–248. doi: 10.1128/MMBR.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kappler U. Bacterial Sulfite-Oxidizing Enzymes – Enzymes for Chemolithotrophs Only? In: Dahl C, Friedrich CG, editors. Microbial Sulfur Metabolism. Springer; 2008. pp. 152–169. [Google Scholar]
- 64.Brokx SJ, Rothery RA, Zhang G, Ng DP, Weiner JH. Characterization of an Escherichia coli Sulfite Oxidase Homologue Reveals the Role of a Conserved Active Site Cysteine in Assembly and Function. Biochemistry. 2005;44:10339–10348. doi: 10.1021/bi050621a. [DOI] [PubMed] [Google Scholar]
- 65.Pacheco A, Hazzard JT, Tollin G, Enemark JH. The pH Dependence of Intramolecular Electron Transfer Rates in Sulfite Oxidase at High and Low Anion Concentrations. J. Biol. Inorg. Chem. 1999;4:390–401. doi: 10.1007/s007750050325. [DOI] [PubMed] [Google Scholar]
- 66.Hurley JH, Dean AM, Sohl JL, Koshland DE, Stroud RM. Regulation of an Enzyme by Phosphorylation at the Active Site. Science. 1990;249:1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- 67.Tollin G, Hurley JK, Hazzard JT, Meyer TE. Use of Laser Flash Photolysis Time-Resolved Spectrophotometry to Investigate Interprotein and Intraprotein Electron Transfer Mechanisms. Biophys. Chem. 1993;48:259–279. doi: 10.1016/0301-4622(93)85014-9. [DOI] [PubMed] [Google Scholar]
- 68.Tollin G. Use of Flavin Photochemistry to Probe Intraprotein and Interprotein Electron Transfer Mechanisms. J. Bioenerg. Biomembr. 1995;27:303–309. doi: 10.1007/BF02110100. [DOI] [PubMed] [Google Scholar]
- 69.Tollin G. In: In Electron Transfer in Chemistry. Balzani V, editor. IV. Wiley-VCH; Weinheim, Germany: 2001. pp. 202–231. [Google Scholar]
- 70.Feng C, Kedia RV, Hazzard JT, Hurley JK, Tollin G, Enemark JH. Effect of Solution Viscosity on Intramolecular Elelctron Transfer in Sulfite Oxidase. Biochemistry. 2002;41:5816–5821. doi: 10.1021/bi016059f. [DOI] [PubMed] [Google Scholar]
- 71.Kawatsu T, Beratan DN. Electron Transfer Between Cofactors in Protein Domains Linked by a Flexible Tether. Chem. Phys. 2006;326:259–269. [Google Scholar]
- 72.Pushie MJ, George GN. Active-Site Dynamics and Large-Scale Domain Motions of Sulfite Oxidase: A Molecular Dynamics Study. J. Phys. Chem. B. 2010;114:3266–3275. doi: 10.1021/jp908731f. [DOI] [PubMed] [Google Scholar]
- 73.Utesch T, Mroginski MA. Three-Dimensional Structural Model of Chicken Liver Sulfite Oxidase in its Activated Form. J. Phys. Chem. Lett. 2010;1:2159–2164. [Google Scholar]
- 74.Johnson-Winters K, Nordstrom AR, Davis AC, Tollin G, Enemark JH. Effects of Large Scale Amino Acid Substitution in the Polypeptide Tether Connecting the Heme and Molybdenum Domains on Catalysis in Human Sulfite Oxidase. Metallomics. 2010 doi: 10.1039/c0mt00021c. Accepted. [DOI] [PubMed] [Google Scholar]
- 75.Rajagopalan KV. Sulfite Oxidase (Sulfite:Ferricytochrome c Oxidoreductase). In: Coughlan MP, editor. Molybdenum and Molybdenum-Containing Enzymes. 1 ed. Pergamon Press; Oxford ; New York: 1980. [Google Scholar]
- 76.Byrne RS, Hansche R, Mendel RR, Hille R. Oxidative Half-reaction of Arabidopsis thaliana Sulfite Oxidase: Generation of Superoxide by a Peroxisomal Enzyme. J. Biol. Chem. 2009;284:35479–35484. doi: 10.1074/jbc.M109.067355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spence JT, Kipke CA, Enemark JH, Sunde RA. Stoichiometry of Electron Uptake and the Effect of Anions and pH on the Molybdenum and Heme Reduction Potentials of Sulfite Oxidase. Inorg. Chem. 1991;30:3011. [Google Scholar]
- 78.Elliott SJ, McElhaney AE, Feng C, Enemark JH, Armstrong FA. A Voltammetric Study of Interdomain Electron Transfer within Sulfite Oxidase. J. Am. Chem. Soc. 2002;124:11612–11613. doi: 10.1021/ja027776f. [DOI] [PubMed] [Google Scholar]
- 79.Johnson-Winters K, Nordstrom AR, Emesh S, Astashkin AV, Rajapakshe A, Berry RE, Tollin G, Enemark JH. Effects of Interdomain Tether Length and Flexibility on the Kinetics of Intramolecular Electron Transfer in Human Sulfite Oxidase. Biochemistry. 2010;49:1290–1296. doi: 10.1021/bi9020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berry RE, Shokhireva TK, Filippov I, Shokhirev MN, Zhang H, Walker FA. Effect of the N-Terminus on Heme Cavity Structure, Ligand Equilibrium, Rate Constants, and Reduction Potentials of Nitrophorin 2 from Rhodnius prolixus. Biochemistry. 2007;46:6830–6843. doi: 10.1021/bi7002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ding XD, Weichsel A, Andersen JF, Shokhireva TK, Balfour C, Pierik AJ, Averill BA, Montfort WR, Walker FA. Nitric Oxide Binding to the Ferri- and Ferroheme States of Nitrophorin 1, a Reversible NO-Binding Heme Protein from the Saliva of the Blood-Sucking Insect, Rhodnius prolixus. J. Am. Chem. Soc. 1999;121:128–138. [Google Scholar]
- 82.Aguey-Zinsou K-F, Bernhardt PV, Kappler U, McEwan AG. Direct Electrochemistry of a Bacterial Sulfite Dehydrogenase. J. Am. Chem. Soc. 2003;125:530–535. doi: 10.1021/ja028293e. [DOI] [PubMed] [Google Scholar]
- 83.Friedrichs MS, Eastman P, Vaidyanathan V, Houston M, Legrand S, Beberg AL, Ensign DL, Bruns CM, Pande VS. Accelerating Molecular Dynamic Simulation on Graphics Processing Units. J. Comput. Chem. 2009;30:864–872. doi: 10.1002/jcc.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goldfarb D, Lipkin Y, Potapov A, Gorodetsky Y, Epel B, Raitsimring AM, Radoul M, Kaminker I. HYSCORE and DEER with an Upgraded 95 GHz Pulse EPR Spectrometer. J. Magn. Reson. 2008;194:8–15. doi: 10.1016/j.jmr.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 85.Wilson JJ, Kappler U. Sulfite Oxidation in Sinorhizobium meliloti. Biochim. Biophys. Acta. 2009;1787:1516–1525. doi: 10.1016/j.bbabio.2009.07.005. [DOI] [PubMed] [Google Scholar]