Abstract
Hsp90 is an important cellular chaperone and attractive target for therapeutics against both cancer and infectious organisms. The Hsp90 protein from the parasite Plasmodium falciparum, the causative agent of malaria, is critical for this organism’s survival; the anti-Hsp90 drug geldanamycin is toxic to P. falciparum growth. We have solved the structure of the N-terminal ATP-binding domain of P. falciparum Hsp90, which contains a principal drug-binding pocket, in both apo and ADP-bound states at 2.3 Å resolution. The structure shows that P. falciparum Hsp90 is highly similar to human Hsp90, and likely binds agents such as geldanamycin in an identical manner. Our results should aid in the structural understanding of Hsp90-drug interactions in P. falciparum, and provide a scaffold for future drug-discovery efforts.
Keywords: GHKL fold, ATPase, drug design, malaria
Introduction
The Hsp90 protein chaperone is a highly conserved protein critical for the proper functioning of various cellular signaling pathways, and serves to mediate appropriate folding and protein-protein interactions for many important client proteins1. Hsp90 exists as a homodimer, and contains three domains: An N-terminal GKHL ATP-binding domain (found also in type II topoisomerases, CheA-family histidine kinases, and the MutL DNA repair factor); a middle domain that supports ATP turnover (and which is also shared with type II topoisomerases and MutL); and a C-terminal dimerization domain1–3. Hsp90 undergoes large conformational changes during its ATPase cycle. Before ATP binding, the N-terminal and middle domains splay open like a pair of jaws, exposing various hydrophobic patches for client protein binding. Upon ATP binding, the N-terminal domains associate to form an extended dimer; following hydrolysis, the protein undergoes a compaction that rearranges this dimer, likely mediating the release of bound client proteins. Upon nucleotide release, the N-terminal domains dissociate to restart the cycle1–3.
Because of its importance to cell signaling and proliferation, Hsp90 is under investigation as a target for therapies against both cancer and infectious organisms4,5. Hsp90 is the target of several natural products, including geldanamycin and radiciol6,7. A derivative of geldanamycin, 17-(allylamino)-17-demethoxygeldanamycin (17-AAG) is currently in clinical trials as a treatment for various cancers, with promising initial results8. High-resolution structures of the eukaryotic Hsp90 N-terminal domain bound to both geldanamycin and radiciol have been solved, showing that both of these drugs bind the ATP-binding pocket in the protein’s N-terminal domain and compete with the nucleotide substrate for binding6,9.
Hsp90 is highly conserved across eukaryotes, and its proper function has been shown to be important for the growth of infectious agents such as Plasmodium falciparum, the causative agent of malaria10,11. P. falciparum has a complex life cycle with two host organisms (the Anopheles mosquito and humans), and must properly respond to developmental cues, as well as sudden changes in its environment, to survive4. Treatment with geldanamycin effectively inhibits the growth of P. falciparum in culture, presumably through its interaction with Hsp9010,11. In addition, a recent study has shown that PfHsp90 may be more sensitive to geldanamycin treatment than its human homolog12. Although P. falciparum Hsp90 (PfHsp90) is highly conserved with its human homolog and represents a promising drug target, the high-resolution structure of this protein has not been determined, hampering efforts to further understand or optimize protein-drug interactions. Here, we present the structure of the PfHsp90 N-terminal ATP-binding domain, in both apo and ADP-bound states. These results allow detailed comparisons between human and P. falciparum Hsp90, and provide a physical scaffold for future studies of drug interactions with this protein.
Materials and Methods
Cloning and Expression
A construct consisting of amino acids 1–215 of P. falciparum Hsp90 (PF07_0029) was cloned from a cDNA library into a derivative of pET28b with an N-terminal, TEV protease-cleavable 6xHis tag. Site-directed mutagenesis was subsequently performed to remove residues 212–215 and add the sequence “GGGVEHEWEELN” (a three-glycine linker followed by amino acids 302–310 of the native protein) to the C-terminus (Hsp90N). This modification improved protein expression and solubility, and has previously been successful in expressing the N-terminal domain of Grp94, an Hsp90 paralog13. Hsp90N was expressed at 20°C for 18 hours in Rosetta(DE3) pLysS cells (Novagen) using auto-inducing media14. Cells were harvested by centrifugation and resuspended in Buffer A (20 mM HEPES pH 7.5, 10% glycerol, 2 mM β-mercaptoethanol) plus 20 mM imidazole and 300 mM NaCl prior to flash freezing in liquid nitrogen. For lysis, cells were thawed and sonicated, after which the cell debris was removed by centrifugation and the clarified lysate purified over a Ni2+ affinity column (GE Biosciences). His-tagged TEV protease15 was added at a ratio of 1:50 TEV protease: Hsp90N (wt:wt), and the mixture incubated overnight at 4°C. The cleavage reaction was then repassaged over a Ni2+ affinity column to remove the TEV protease, uncleaved Hsp90N, and cleaved 6xHis tags, and the flow-through was further purified by gel filtration (Superdex 200, GE Biosciences). Fractions were concentrated to ~20 mg/mL by ultrafiltration (Amicon Ultra, Millipore) and stored at 4°C.
Crystallization and Structure Solution
For crystallization trials, purified Hsp90N at ~20 mg/mL was dialyzed overnight against 20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM DTT, 5 mM MgCl2, with or without 1 mM ADP, or other ATP analogs (AMP-PNP, ADP·BeF3, ADP·AlFx) or drugs (radicicol, 17-AAG). Only samples dialyzed with ADP produced crystals; for these conditions, dialyzed Hsp90N was mixed 1:1 with 1.8–2.0 M Li2SO4, 0.1 M HEPES pH 7.5, and 1–2% xylitol at 20°C and allowed to equilibrate by hanging-drop vapor diffusion. Following crystallization, the drop was flooded with cryoprotectant containing well solution plus 15% glycerol, and crystals (small hexagonal rods ~ 0.1 × 0.1 × 0.2 mm) were looped and flash-frozen in liquid nitrogen. Data were collected on NE-CAT beamline 24ID-E at the Advanced Photon Source at Argonne National Laboratory. Diffraction data were indexed and reduced with HKL200016. Molecular replacement was performed with PHASER17, using the previously-reported ADP-bound structure of the S. cerevisiae Hsp90 N-domain (PDB ID 1AMW)18 as a search model, with the nucleotide and external loops removed. Manual rebuilding of the ‘ATP lid’ region of both monomers in the asymmetric unit was required. Electron density was clearly observed for residues 304–310 in both monomers, and for ADP bound to monomer A (Figure 1A). Positional, B-factor, and TLS refinement were performed with phenix.refine19, and ordered water molecules were placed automatically into difference maps using COOT20. The final model was refined to an R-factor of 16.3% and a free-R factor of 19.0%, with excellent geometry (Table 1). Figures were produced with PyMOL21.
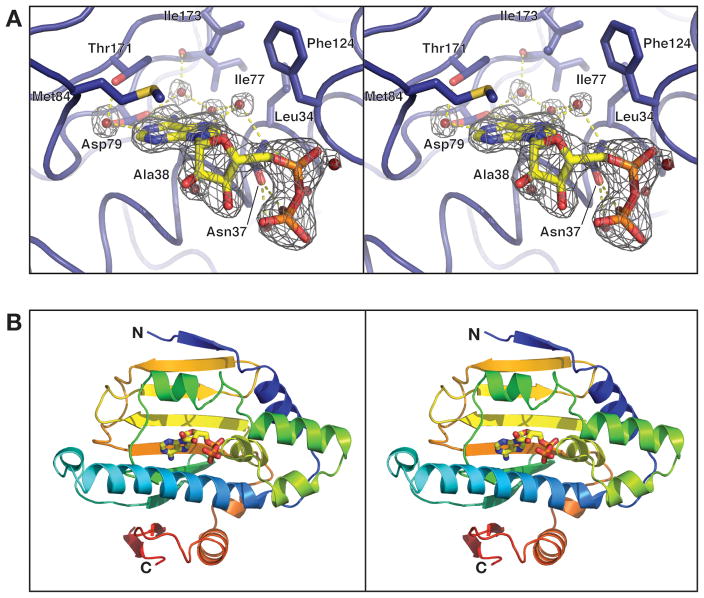
Figure 1. Overall structure and nucleotide binding by Hsp90N.
(A) Close-up of the Hsp90N nucleotide-binding site bound to ADP. Amino acid residues surrounding the active site are shown in stick view, and water molecules involved in hydrogen bonding are shown as spheres. Fo-Fc simulated-annealing omit electron density is shown for ADP and surrounding water molecules, contoured at 6.0 σ. (B) Stereo view of monomer A of Hsp90N, bound to ADP. The protein is colored in a rainbow scheme, with the N-terminus blue and the C-terminus red. Bound ADP is shown in stick view.
Table 1.
Data collection, refinement and stereochemistry
| Data collection | |
|---|---|
| Resolution (Å) | 50 – 2.3 |
| Wavelength (Å) | 0.97918 |
| Space Group | P3221 |
| Unit Cell Dimensions (a, b, c) Å | 118.16, 118.16, 105.90 |
| I/σ(last shell) | 17.9 (2.1) |
| 1Rsym (last shell) % | 0.107 (0.824) |
| Completeness (last shell) % | 99.9 (100.0) |
| Redundancy (last shell) | 5.5 (5.3) |
| Refinement | |
| Resolution (Å) | 50 – 2.3 |
| No. of reflections | 35497 |
| working | 33646 |
| free (% total) | 1851 (5.2%) |
| 2Rwork (last shell) (%) | 16.3 (19.8) |
| 2Rfree (last shell) (%) | 19.0 (21.2) |
| Structure and Stereochemistry | |
| No. of atoms | 3771 |
| protein | 3452 |
| solvent | 262 |
| nucleotide | 27 |
| SO4 | 30 |
| Ramachandran – most favored | 98.4% |
| Ramachandran – allowed | 100% |
| r.m.s.d. bond lengths (Å) | 0.004 |
| r.m.s.d. bond angles (°) | 0.901 |
Rsym = ΣΣj|Ij – 〈I〉|/ΣIj, where Ij is the intensity measurement for reflection j and 〈I〉 is the mean intensity for multiply recorded reflections.
Rwork, free = Σ||Fobs| – |Fcalc||/|Fobs|, where the working and free R-factors are calculated using the working and free reflection sets, respectively. The free reflections were held aside throughout refinement.
Results and Discussion
Since geldanamycin and other anti-Hsp90 compounds are known to bind the protein’s N-terminal ATP-binding domain, we initially cloned only this domain of PfHsp90 (residues 1–215, PlasmoDB ID PF07_0029). To overcome poor expression and solubility for this fragment, we subsequently engineered a variant construct that comprised residues 1–211, a tri-glycine linker, and residues 302–310 of PfHsp90. This strategy removed an extended, disordered segment from the principal GHKL domain (residues 212–301), but also added back a single β-strand predicted to pair with the N-terminal region. This construct (hereafter referred to as Hsp90N) expressed at high levels, and was well behaved in solution after purification.
Crystal screens of Hsp90N resulted in one crystallization condition that required the presence of adenosine 5’-diphosphate (ADP). Crystals were not reproducible without nucleotide, with other nucleotides/analogs, or with Hsp90-targeting drugs (radicicol or 17-AAG). Diffraction data collected from these crystals to 2.3 Å resolution permitted solution of the structure by molecular replacement, followed by refinement of the final model to R/Rfree values of 16.3%/19.0%, with excellent geometry (Table 1). There are two molecules of Hsp90N per asymmetric unit: one in an apo state, and the other with clear density for an ADP molecule bound in the active site (Figure 1A). This asymmetric configuration is reinforced by crystal packing, which precludes ADP binding in one protomer by stabilizing the packing of a flexible loop (the ‘ATP lid’) into the ATP-binding pocket. The two molecules in the asymmetric unit do not appear to form a physiological association with each other.
The overall structure of Hsp90N consists of a nine-stranded, antiparallel β-sheet backed on one side with seven α-helices (Figure 1B). Residues 304–310 are clearly visible in both monomers and form the ninth β-strand. As predicted on the basis of amino acid sequence, the domain belongs to the GHKL ATPase fold family22. The nucleotide-binding site nestles against one face of the β-sheet, between several α-helices, and is mostly solvent-exposed in the current structure. In other GHKL ATPases, including homologous nucleotide-bound Hsp90 N-domain structures, a Mg2+ ion has been observed to coordinate β- and γ-phosphate oxygens (if present), along with the side-chain oxygen of Asn37; Mg2+ is universally required for ATP hydrolysis in 2+ these proteins. Despite the presence of 5 mM MgCl2 in the crystallization conditions, no Mg ion was observed in our structure, possibly due to the high concentration of sulfate ions in the crystallization solution. In the absence of Mg2+, the δ-nitrogen of Asn37 directly hydrogen-bonds to an α-phosphate oxygen of ADP and the δ-oxygen hydrogen-bonds to a β-phosphate oxygen. This direct bonding results in a ~0.5 Å shift of both the side-chain of Asn37 and the α- and βphosphates of ADP toward one another in our structure, when compared to the human Hsp90 complex with Mg2+·ADP23. Other than the lack of a metal cofactor, the nucleotide-binding mode in Hsp90N is largely identical to that previously observed in other Hsp90 proteins (Figure 2A).
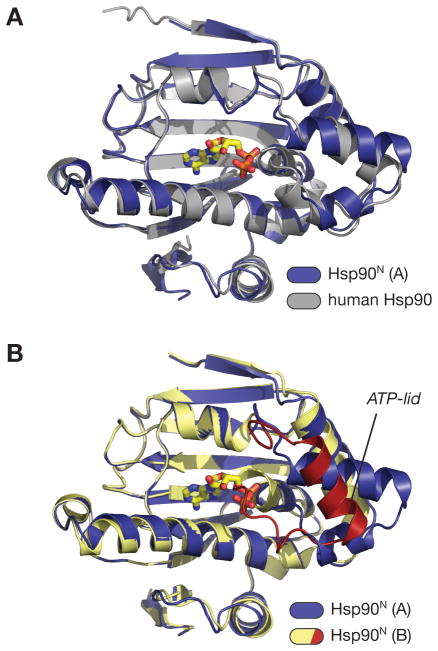
Figure 2. Hsp90N comparisons.
(A) Superposition of the A (ADP-bound) monomer of Hsp90N (blue; bound ADP in yellow) with the structure of the human Hsp90 N-terminal domain bound to geldanamycin (PDB ID 1YET) (gray; bound geldanamycin not shown). The overall r.m.s.d. of the two domains is 0.79 Å. (B) Superposition of the A (ADP-bound) and B (apo) monomers of Hsp90N. Monomer A is colored blue, and monomer B is colored yellow with the ATP-lid red. The conformation of the ATP-lid in monomer B precludes nucleotide binding, as residues 118–121 would directly overlap with the α- and β-phosphates of ADP.
GHKL-family ATP-binding domains are characterized by a glycine-rich loop, known as the ATP lid, which wraps around the γ-phosphate of a bound nucleotide, positioning this moiety for hydrolysis. The ATP lid also is intimately involved in the packing arrangement of the ATP-binding domain dimer that forms in response to nucleotide binding. In previous structures of isolated ATP-binding domains of various Hsp90 proteins, the ATP lid’s position is variable, and has not been observed in a hydrolytically competent conformation (This conformation, with the ATP lid wrapped tightly around the γ-phosphate of ATP, has only been observed in a structure of full-length S. cerevisiae Hsp90 bound to a non-hydrolyzable ATP analog)2,6,7. In monomer A of our structure, the ATP lid is rotated away from the active site to accommodate ADP binding. In monomer B, this segment’s position is constrained by crystal packing contacts, such that amino acids 118–121 overlap the ADP binding site and preclude nucleotide binding (Figure 2B). This ‘collapsed’ conformation of the ATP lid is similar to one previously observed in the human Hsp90 N-terminal domain, also in the apo state9.
When considering the entire N-terminal domain except for the ATP lid, the structure of PfHsp90 is very similar to the equivalent domain of other eukaryotic Hsp90 proteins (Figure 2A), displaying overall Cα r.m.s.d. values of 0.63 and 0.79 Å with S. cerevisiae and human Hsp90, respectively18,23. The similarities are even greater within the nucleotide-binding site, with the Hsp90N active site almost perfectly matching that of the human Hsp90 in both amino acid identity and side-chain rotamers (0.23 Å Cα r.m.s.d., Figure 3A). Because of this close similarity, we were able to confidently model the likely conformation of a complex between Hsp90N and geldanamycin, using the previously-solved complex between the drug and human Hsp90 (Figure 3B)9. This docking analysis, derived from structural superposition of protein backbone atoms, reveals an extremely good fit for the inhibitor in the PfHsp90 active site, with only one stereochemical clash (to the side chain of Asn92) that could be easily resolved by a small rotamer shift (Figure 3C). Asn92 of S. cerevisiae Hsp90 is known to undergo such a rotamer shift upon geldanamycin binding6.
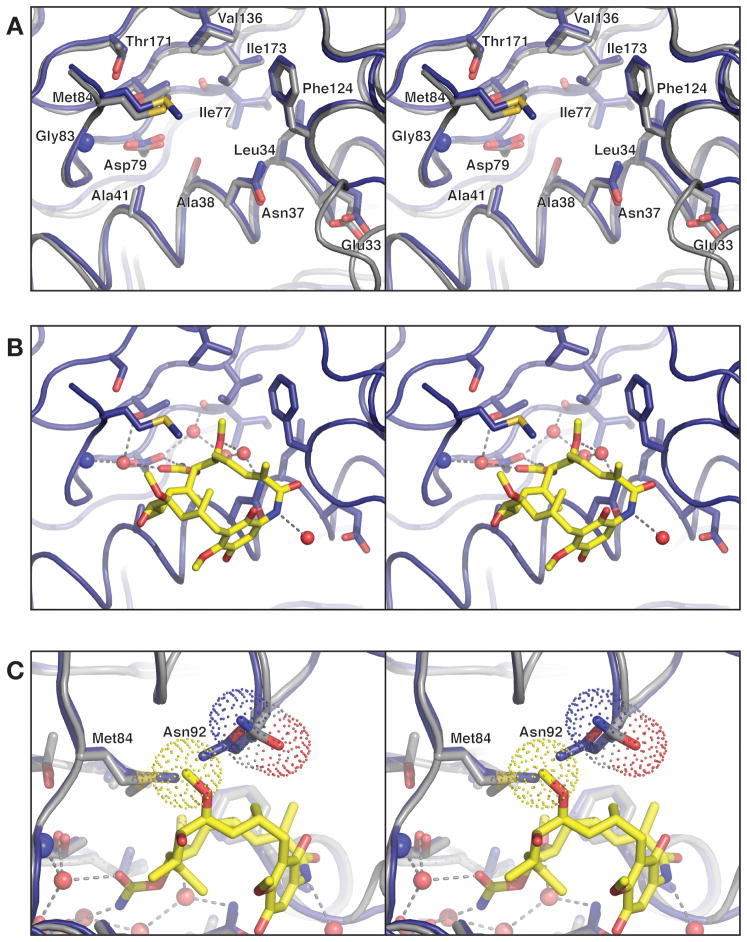
Figure 3. Prospective drug binding site of PfHsp90.
(A) Close-up of the Hsp90N nucleotide- binding site (blue) overlaid with the human Hsp90 N-terminal domain (gray), with residues surrounding the binding pocket show as sticks. The Cα r.m.s.d. for residues displayed is 0.23 Å. (B) Model for geldanamycin binding by PfHsp90, based on structural superposition of Hsp90N with the human Hsp90-geldanamycin complex (above). Geldanamycin and water molecules shown are present in the human Hsp90-geldanamycin complex structure. All chemical groups involved in hydrogen-bonding and Van der Waals interactions with geldanamycin are conserved between human and PfHsp90. (C) 90° rotated view from panel B, showing how Hsp90N residue Asn92 (blue model) clashes with C27 of modeled geldanamycin (yellow sticks, van der Waals surface for this atom is shown as yellow dots). The equivalent residue in human Hsp90, Asn106, is rotated slightly away to accommodate the ligand (gray model, van der Waals surface for the side chain is shown as dots).
A detailed comparison of the PfHsp90 and human Hsp90 N-terminal domain structures reveals several slight differences between their nucleotide and drug-binding sites, some of which may be exploitable in the design of drugs specific for PfHsp90 (Sequence alignment in Supplemental Figure S1). For example, PfHsp90 Met84 adopts a different side-chain rotamer than human Met98, altering the shape of the ‘ceiling’ of the binding pocket. Val186 of human Hsp90 is replaced by an isoleucine (Ile173) in PfHsp90, resulting in a slight constriction in the back of the pocket (Figure 2B). The most significant difference is the substitution of Ser52 (human Hsp90) with an alanine (Ala38) in the Plasmodium ortholog: this change enlarges the posterior end of the pocket in PfHsp90, and makes this region slightly more hydrophobic. Sequence alignments do not reveal a clear preference for serine or alanine at this position in higher eukaryotes. For instance, the α-isoform of human Hsp90 possesses a serine, whereas the β-isoform possesses an alanine, implying that this particular distinction may not be useful for drug design efforts. Nonetheless, the combination of modest differences between the human and P.falciparum Hsp90 ATP-binding domains suggests that it may be feasible to engineer small molecule agents with greater selectivity for PfHsp90.
Recently, a structure of the N-terminal domain of a second P. falciparum Hsp90 homolog (PF14_0417) has been solved in complex with an ADP analog by the Structural Genomics Consortium at the University of Toronto (PDB ID 3IED; no reference available). This structure overlays closely with our Hsp90N model, with an overall Cα r.m.s.d. of 0.62 Å, and binds nucleotide in an identical manner. Of the four Hsp90-family proteins encoded by the P. falciparum genome, the protein imaged in the present study (PF07_0029) has been assumed to be the major cytosolic Hsp90, based on its high similarity to other Hsp90’s (65% identical to human Hsp90 versus 35% for PF14_0417), as well as sequence hallmarks such as the C-terminal EEVD motif4,11. Of the other three proteins, the PFL1070c gene has been identified as Grp94, the endoplasmic reticulum Hsp90, while the remaining two, PF14_0417 and PF11_0188, have recently been tentatively assigned as homologs of Trap1, a mitochondrial Hsp9024. The relative importance of these four Hsp90 paralogs to this organism’s life cycle has not been fully explored; thus, future drug discovery efforts aimed at Hsp90 in P. falciparum will likely need to account for the activities of all four of these paralogs.
Conclusion
We have solved the structure of the N-terminal nucleotide- and drug-binding domain of Hsp90 from Plasmodium falciparum in apo and ADP-bound states. These structures provide a physical view of PfHsp90, a potential drug target in the fight against malaria. The structure shows that PfHsp90 is highly similar to the homologous human protein, but that it may be possible to find or design small molecules to specifically target the PfHsp90 nucleotide-binding site by exploiting several subtle differences. In addition, drugs such as geldanamycin and radicicol already represent potential anti-malarial drugs, as they effectively abrogate the growth of P. falciparum at levels far below those that result in toxicity in humans10,11. Further study, including examination of the affinities of existing drugs for this protein, will be needed to accurately assess the usefulness of PfHsp90 as a drug target for anti-malarial treatment. In this regard, the Hsp90N construct developed here may prove useful for future in vitro studies as a highly-expressed and soluble variant of P. falciparum Hsp90.
Supplementary Material
Sequence alignment (using MAFFT25) of the N-terminal domain of PfHsp90 with both α- and β-isoforms of human Hsp90, and S. cerevisiae Hsp90. Residues 2–123 of human α-Hsp90 have been removed for clarity (denoted by a double slash after Met1). Residues surrounding the ATP-binding site, shown in sticks in Figure 1, are highlighted in purple and their residue numbers in PfHsp90 are shown. Pairwise identity of full-length PfHsp90 with human α-Hsp90,β-Hsp90 and S. cervisiae Hsp90 are 65.2%, 64.8%, and 59.8%, respectively. For the region shown, pairwise identity of PfHsp90 with human α-Hsp90, β-Hsp90 and S. cervisiae Hsp90 are 71.5%, 72.7%, and 68.3%, respectively.
Acknowledgments
The authors thank the staff of APS NE-CAT beamline 24ID-E for assistance with data collection, D. Pillai and J. DeRisi for the original PfHsp90 clone and valuable advice, and members of the Berger lab for critical reading of the manuscript. K.D.C. acknowledges a Helen Hay Whitney postdoctoral fellowship, and J.M.B. acknowledges support from the NCI (CA077373). The structure of PfHsp90N has been submitted to the Protein Data Bank with PDB ID 3K60.
References
- 1.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Ann Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 2.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440(7087):1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 4.Acharya P, Kumar R, Tatu U. Chaperoning a cellular upheaval in malaria: heat shock proteins in Plasmodium falciparum. Mol & Biochem Parasitology. 2007;153:85–94. doi: 10.1016/j.molbiopara.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Mahalingam D, Swords R, Carew JS, Nawrocki ST, Bhalla K, Giles FJ. Targeting HSP90 for cancer therapy. Br J Cancer. 2009;100(10):1523–1529. doi: 10.1038/sj.bjc.6605066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roe SM, Prodromou C, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42(2):260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 7.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91(18):8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp S, Workman P. Inhibitors of the Hsp90 molecular chaperone: current status. Adv Cancer Res. 2006:323–348. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- 9.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89(2):239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 10.Banumathy G, Singh V, Pavithra SR, Tatu U. Heat shock protein 90 function is essential for Plasmodium falciparum growth in human erythrocytes. J Biol Chem. 2003;278(20):18336–18345. doi: 10.1074/jbc.M211309200. [DOI] [PubMed] [Google Scholar]
- 11.Kumar R, Musiyenko A, Barik S. The heat shock protein 90 of Plasmodium falciparum and antimalarial activity of its inhibitor, geldanamycin. Malaria J. 2003;2(1):30. doi: 10.1186/1475-2875-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wider D, Peli-Gulli MP, Briand PA, Tatu U, Picard D. The complementation of yeast with human or Plasmodium falciparum Hsp90 confers differential inhibitor sensitivities. Mol Biochem Parasitol. 2009;164(2):147–152. doi: 10.1016/j.molbiopara.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Immormino RM, Dollins DE, Shaffer PL, Soldano KL, Walker MA, Gewirth DT. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J Biol Chem. 2004;279(44):46162–46171. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- 14.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8(8):1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;D60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 18.Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90(1):65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 19.Afonine PV, Grosse-Kunstieve RW, Adams PD. CCP4 Newsl. 2005;42:8. [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.DeLano WL. PyMOL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 22.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25(1):24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 23.Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143(4):901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavithra SR, Kumar R, Tatu U. Systems analysis of chaperone networks in the malarial parasite Plasmodium falciparum. PLoS Comput Biol. 2007;3(9):1701–1715. doi: 10.1371/journal.pcbi.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment (using MAFFT25) of the N-terminal domain of PfHsp90 with both α- and β-isoforms of human Hsp90, and S. cerevisiae Hsp90. Residues 2–123 of human α-Hsp90 have been removed for clarity (denoted by a double slash after Met1). Residues surrounding the ATP-binding site, shown in sticks in Figure 1, are highlighted in purple and their residue numbers in PfHsp90 are shown. Pairwise identity of full-length PfHsp90 with human α-Hsp90,β-Hsp90 and S. cervisiae Hsp90 are 65.2%, 64.8%, and 59.8%, respectively. For the region shown, pairwise identity of PfHsp90 with human α-Hsp90, β-Hsp90 and S. cervisiae Hsp90 are 71.5%, 72.7%, and 68.3%, respectively.